Abstract
The unique property of stem cells to self-renew suggests specific mechanisms that regulate their cell-cycle progression. In the present study, we identify a novel protein, nucleostemin, found in the nucleoli of CNS stem cells, embryonic stem cells, and several cancer cell lines and preferentially expressed by other stem cell-enriched populations. It contains an N-terminal basic domain and two GTP-binding motifs. When stem cells differentiate, nucleostemin expression decreases rapidly prior to cell-cycle exit both in vitro and in vivo. Depletion or overexpression of nucleostemin reduces cell proliferation in CNS stem cells and transformed cells. Mutation analysis indicates that excessive nucleostemin, particularly mutants that lack the GTP-regulatory domain, prevents cells from entering mitosis and causes apoptosis in a p53-dependent manner. The N-terminal basic domain specifies nucleolar localization, the p53 interaction, and is required for the cell death caused by overexpression. This work describes a novel nucleolar mechanism that controls the cell-cycle progression in CNS stem cells and cancer cells.
Keywords: Stem cell, nucleostemin, nucleolus, GTP, cell proliferation, p53
Stem cells have been identified in several somatic tissues, including the nervous system, bone marrow, epidermis, skeletal muscle, and liver. This “set-aside” population of cells is believed to be responsible for maintaining homeostasis within individual tissues in adult animals. The number of stem cells and their decision to differentiate must be tightly controlled during embryonic development and in the adult animal to avoid premature aging or tumor formation. Different somatic stem cells share the properties of self-renewal and multidevelopmental potential, suggesting the presence of common cellular machinery. Understanding the molecular determinants that impart these properties will have major impact on the field. As a consequence, there is growing interest in the analysis of patterns of gene expression in stem cells by using microarray technology (Geschwind et al. 2001; Terskikh et al. 2001; Ivanova et al. 2002; Ramalho-Santos et al. 2002). However, few studies have taken the next step of showing how an individual gene might function in the complex network of signals in developing tissues.
To elucidate the mechanism that governs the proliferative state in stem cells, we looked for genes that are preferentially expressed by stem cells and involved in cell-proliferation regulation. In the present study, we report a new nucleolar protein named nucleostemin that is expressed in both embryonic and adult CNS stem cells in culture. Nucleostemin is also present in embryonic stem (ES) cells, primitive cells in the bone marrow, and cancer cells, but not in the differentiated cells of most adult tissues. The expression of nucleostemin is abruptly down-regulated during differentiation prior to terminal cell division. We also present data showing that nucleostemin participates in the control of stem and cancer cell proliferation.
Results
Cloning of nucleostemin
Clonal analysis shows that CNS stem cells can be isolated from the developing rat brain and preferentially enriched in basic fibroblast growth factor (bFGF)-expanded culture (Johe et al. 1996). Before differentiation, all stem cells divide and express nestin, an intermediate filament characteristic of neuroepithelial precursors (Lendahl et al. 1990). After withdrawal of bFGF and addition of 10% fetal bovine serum (FBS) for 8 d, these cells differentiate efficiently into neurons, astrocytes, and oligodendrocytes (Fig. 1A). Taking advantage of the rapid differentiation of CNS stem cells in vitro, a subtractive library was constructed by using the undifferentiated pool as tester and the differentiated pool as driver. From this library, two novel clones (SR204 and SR75) and a known gene, 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMG-CoA R), were identified as highly enriched in cortical stem cells. HMG-CoA R, a rate-limiting enzyme in cholesterol biosynthesis, has been shown to have a cholesterol-independent role in regulating primordial germ cell migration in Drosophila (Van Doren et al. 1998). The present study focuses on one of the two novel clones SR204 found to be expressed abundantly in the stem cell population and rapidly down-regulated during differentiation (Fig. 1B).
Figure 1.
Cloning of nucleostemin. (A) Rat embryonic cortical stem cells (Stem) were used as the tester and 8-d serum-differentiated cultures (S8) as the driver in a cDNA subtractive screen. (Left) nestin, red; BrdU, green. (Middle) Tuj1 for neurons, red; GFAP for astrocytes, green. (Right) O4 for oligodendrocytes, green. (B) Northern analysis of subtractive outputs and controls. Sc indicates stem cells; F2 and F8, 2-d and 8-d differentiation in 10% serum and bFGF (20 ng/mL); S2 and S8, 2-d and 8-d differentiation in 10% serum without bFGF; NS, nucleostemin; HMGCoR, 3-hydroxyl, 3-methyl glutaryl CoA reductase; Tβ-3, neuron-specific type III β-tubulin; GFAP, glial fibrillary acidic protein; MBP, myelin basic protein. (C) Schematic diagram of nucleostemin protein. CC, coiled-coil domain; G4 and G1, GTP-binding motifs; B and A, basic and acidic domains; filled boxes, NLS. (D) Sequence alignment of rodent, murine, and human nucleostemin. Shaded areas correspond to the structural domains indicated in C. Underline, NLS; red, conserved residues in all species; blue, conserved residues in two of the three species. Bars: A (left), 25 μm; A (middle and right), 50 μm.
SR204, named nucleostemin because of its nucleolar localization and preferential expression in cortical stem cells, hybridized to a 1.8-kb transcript. The complete cDNA clones comprised 1810 nucleotides (nt) for rats (raNS) and 1813 nt for mice (muNS). Sequence analysis revealed an open reading frame (ORF) of 1617 nt, an in-frame stop codon in the 5′ untranslated region (UTR), an optimal Kozak sequence, and polyadenylation signals in the 3′ UTR, representing the full-length mRNAs. The ORF encoded a 538-amino-acid protein of 61 kD (pI, 9.4). The raNS and muNS shared 90% identity and 94% similarity. PROSITE analysis and Conserved Domain search identified two consensus motifs that define GTP-binding proteins (the G4 motif KXDL, codons 177–180, and the G1 or P-loop GXXXXGK[S/T], codons 256–263), a highly basic region in the N terminus (codons 2–46; pI, 12.4) and an acidic domain in the carboxyl end (codons 460–503; pI, 3.7). PSORT analysis revealed multiple nuclear localization sequences and a coiled-coil domain (Fig. 1C,D). A BLAST search in the GenBank database identified a human homolog (accession no. AAH01024) that shares 81% identity with the rat sequence. The human gene was isolated based on its estrogen-induced increased expression in breast cancer cell lines.
Nucleostemin is a nucleolar protein
To define the intracellular distribution of nucleostemin protein, a chicken polyclonal antibody (2438) was raised against a peptide corresponding to murine nucleostemin codon 522–538 (DRASQEDETYDFTTDYI). Western analysis detected a 60- to 70-kD band of strong intensity in cortical stem cell lysates but only weakly in the 2-d and 7-d serum-differentiated cultures (Fig. 2A). Immunofluorescence revealed strong signals in one or several nodular structures within the nucleus of embryonic cortical stem cells (Fig. 2B,C). The specificity of the staining pattern was supported by a second antibody (1164) raised against a different epitope in rat nucleostemin (codon 32–49, LRKEAKKRGHKKPRKDPG; Fig. 2D,E). Nodular staining with nucleostemin antibody was also seen in cells derived from adult subventricular zone expanded as neurospheres (Fig. 2F; Reynolds and Weiss 1992) and pluripotent ES cells (Fig. 2G,H), and in Chinese hamster ovary (CHO) cells transfected with a construct encoding raNS-eGFP (green fluorescent protein) fusion protein (Fig. 2I,J). These nodular structures are devoid of DAPI staining and contain the known nucleolar protein nucleolin as seen in HEK293 cells (Fig. 2K,L). Additionally, nucleostemin was found in the nucleoli of the three human cancer cell lines we examined, including H1299 (a p53-null non-small cell lung cancer; Fig. 2M), U2OS (an osteosarcoma with wild-type p53; Fig. 2N), and Saos-2 (a p53-null osteosarcoma; Fig. 2O). During interphase, nucleostemin was easily seen in the large nuclei of CHO and HEK293 cells, in which it was predominantly localized in the nucleolus and at a lower level in the nucleoplasm. When cells entered mitosis, the protein dissociated from the nucleolar region. At prophase, a portion of nucleostemin was still associated with the nucleolus. During metaphase, anaphase, and telophase, nucleostemin was diffusely distributed (Fig. 2P).
Figure 2.
Protein expression and localization of nucleostemin. (A). Western blot with α-nucleostemin (2438) antiserum identified a major band in undifferentiated stem cells (Stem) but not in 2-d and 7-d serum-differentiated cultures (SeD2, SeD7). α-Tub indicates anti-α-tubulin antibody used as internal loading control. Immunodetection of endogenous nucleostemin in rat embryonic cortical stem cells (B,D), neurosphere cells derived from adult SVZ (F), ES cells (G), and corresponding DAPI counterstain (C,E,H and inset in F). CHO cells transfected with a nucleostemin-GFP fusion construct revealed strong nucleolar fluorescence (I,J). Nucleostemin is colocalized with nucleolin immunoreactivity in HEK293 cells (K,L). Immunostaining of nucleostemin in three human cancer cell lines: H1299 (M), U2OS (N), and Saos-2 (O). (P) Distribution of nucleostemin during cell cycle. CHO cells at prophase (Pro), metaphase (Meta), anaphase (Ana), and telophase (Tel) were identified by DAPI staining. Bars: B–E,I–P, 10μm; F, 25 μm; G,H, 50μm.
The expression of nucleostemin during CNS development
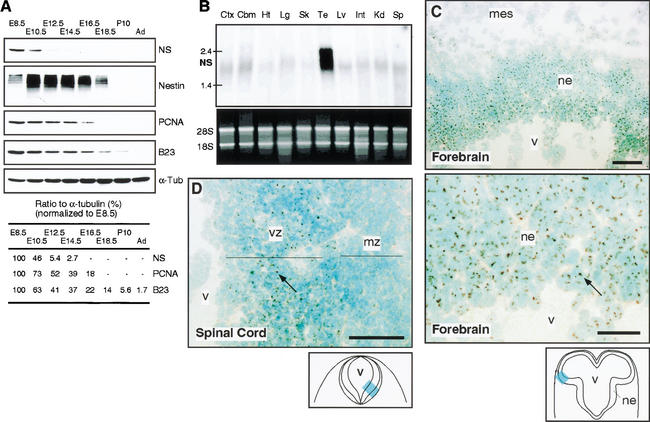
Western blot analysis shows that the expression of nestin peaked around embryonic day 10.5 (E10.5) to E14.5. The nucleostemin protein was elevated as early as E8.5 and declined considerably after E10.5 in the mouse cerebral cortex (Fig. 3A). In contrast, a proliferative marker PCNA and a nucleolar protein B23 decreased gradually and proportionally from E8.5 until birth, reflecting the increase in cells exiting cell cycle. These results are consistent with the notion that nucleostemin is preferentially expressed in vivo by neuroepithelial precursors at a stage before the peak of nestin expression and the precisely timed terminal division of neurons that correlates with the phase of PCNA and B23 expression. Northern analysis of adult rat tissues shows that the mRNA of nucleostemin is present at high levels only in the adult testis (Fig. 3B).
Figure 3.
In vivo distribution of nucleostemin. (A) Developmental Western blot of nucleostemin protein expressed in mouse forebrains. The same blot was reprobed with different antibodies. NS, nucleostemin; E8.5, embryonic day 8.5; P10, postnatal day 10; Ad, adult; PCNA, proliferating cell nuclear antigen; α-Tub, α-tubulin. (B) Multitissue Northern blot on adult rat. Ctx cortex; Cbm, cerebellum; Ht, heart; Lg, lung; Sk, skeletal muscle; Te, testis; Lv, liver; Int, intestine; Kd, kidney; Sp, spleen. Regional distribution of nucleostemin in transverse sections of the forebrain (C) and spinal cord (D) of E10.5 mouse embryos by immunohistochemistry (1643). Nucleostemin signals appear as brown dots (indicated by arrows) and are seen in the neuroepithelium (ne) and mesenchyme (mes). The signal is stronger in the forebrain than in the spinal cord. Within the spinal cord, the ventricular zone (vz) expressed higher levels of nucleostemin than the mantle zone (mz). v, ventricle. Bottom panel in C represents a high-power view of the top panel. Bars: C (top) and D, 50 μm; C (bottom), 25μm.
In E10.5 mouse embryos, nucleostemin staining was seen in the neuroepithelial cells in the forebrain (Fig. 3C), midbrain, hindbrain, and spinal cord (Fig. 3D). Although the E10.5 forebrain contains mainly neural precursors and few differentiated cells, many neurons have already differentiated in the spinal cord at this stage. Consistent with the different onset of differentiation along the CNS axis, nucleostemin signals were strongest in the forebrain compared with the spinal cord at E10.5. Within the spinal cord, the proliferating cells in the ventricular zone expressed nucleostemin at higher levels than those in the mantle zone, where differentiating neurons reside (Fig. 3D). These results show that nucleostemin is highly expressed by early stage CNS precursors in vivo.
Nucleostemin expression is down-regulated when stem cells differentiate into dividing progenitors prior to cell-cycle exit
The Western blot data indicate that the expression of nucleostemin is switched off while PCNA and B23 expression is maintained (Fig. 3A), suggesting that cells continue to proliferate for a period of time after nucleostemin expression is lost. In culture, nucleostemin was found in almost all rat embryonic cortical stem cells (Fig. 4A), but became undetectable after treatment with ciliary neurotrophic factor (CNTF) for 8 d, when 98% of the cells turn into astrocytes (Fig. 4B,C; Johe et al. 1996). To examine if nucleostemin expression is turned off at the stem cell-to-dividing progenitor or the dividing progenitor-to-postmitotic progeny transition, the expression of nucleostemin was analyzed in 2- and 8-d CNTF-differentiated cultures labeled with a 15-min pulse of bromodeoxyuridine (BrdU). If nucleostemin reflects the actual state of proliferation and is turned off when progenitors become postmitotic, the level of nucleostemin should be higher in the S-phase cells labeled with BrdU. After 2- and 8-d differentiation in CNTF, the expression level of nucleostemin was significantly reduced in both the dividing and nondividing cells (CNTF/D2, CNTF/D8; Fig. 4D), indicating that this down-regulation occurs as an early step in the differentiation of proliferating glial precursors. Such an abrupt change of expression is not restricted to the glial lineage. When cortical stem cells differentiated into neurons, astrocytes, and oligodendrocytes in 10% FBS, the expression of nucleostemin was also rapidly attenuated in all cells (SeD2, SeD8; Fig. 4D). These results show that both the dividing cells and the terminally differentiated cells in all lineages present in the 8-d differentiated cultures were nucleostemin-negative. Thus nucleostemin expression is not simply a reflection of the proliferative state but is characteristic of an early multipotential state.
Figure 4.
Nucleostemin (NS) protein is down-regulated in both dividing and nondividing progeny during differentiation. Nucleostemin is expressed in all the rat cortical stem cells (A, Stem) but not in the 8-d CNTF-differentiated culture (B, CNTF/D8). (C) Bar graph showing the percentage of nucleostemin+ cells in A and B. (D) Nucleostemin-BrdU double-labeled immunofluorescence in undifferentiated stem cells (Stem), 2-d and 8-d differentiated cultures with CNTF (10 ng/mL, CNTF/D2 and CNTF/D8), or 10% FBS (SeD2 and SeD8). All cultures are pulse-labeled with 15 min of BrdU (40 μM) before fixation. Arrow and asterisk indicate BrdU-labeled and nonlabeled cells. Both species have low level of nucleostemin expression in the differentiated cultures. (E) Western analysis of lineage-depleted, c-kit+ bone marrow cells (Kit), B-lymphocytes (B-Lo), and granulocytes (Gran) from adult mice. Bar: D, 20 μm.
We also compared nucleostemin expression level in adult bone marrow cells by Western analysis (Fig. 4E). Consistent with our findings in the nervous system, nucleostemin protein could be detected in the c-kit+/lin− population that is enriched for adult hematopoietic stem cells, but not in the lineage-committed B-lymphocytes (B220-positive) or granulocytes (Gr-1-positive). The sustained expression of PCNA suggests that the decrease in nucleostemin expression occurs in cells that are still dividing. These results strengthen the view that nucleostemin is expressed in uncommitted cells, and the level of expression does not simply reflect the number of dividing cells.
Roles of nucleostemin in maintaining the proliferation of cortical stem cells and U2OS cancer cell line
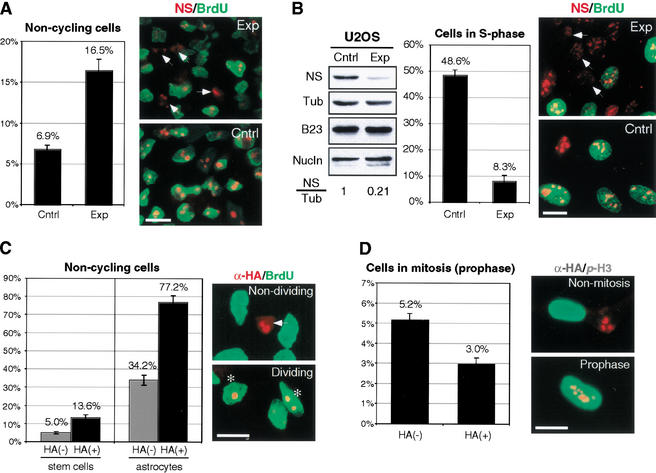
To establish the functional importance of nucleostemin, cortical stem cells were transfected with small inhibitory RNAs (siRNA) for 18 h and assayed for BrdU labeling 72 h later (Elbashir et al. 2001). As shown in Figure 5A (left), cultures transfected with nucleostemin-specific siRNA consistently showed a higher percentage of noncycling cells, represented by nonlabeled cells after an 18-h BrdU pulse, than did the control group (16.5% ± 1.4% versus 6.9% ± 0.5%, mean ± S.E.M.; p < 0.001). Immunostaining revealed a consistent decrease in the nucleostemin-staining intensity in cells treated with nucleostemin-specific siRNA compared with the control siRNA-treated cells (right). To obtain a better transfection efficiency, siRNA knockdown experiments were conducted in U2OS cells (Fig. 5B). Western analysis showed an 80% reduction in nucleostemin protein in cultures treated with nucleostemin-specific siRNA (Fig. 5B, left) compared with cultures transfected with control siRNA 2 d after transfection. In addition, only in nucleostemin-specific siRNA-treated culture were interphase cells seen with diminished nucleostemin staining (Fig. 5B, right, indicated by arrows). The percentage of cells in S phase of these nucleoste-min-negative cells was considerably less than that of control siRNA-transfected cultures (8.3% ± 2.1% versus 48.6% ± 2.1%, mean ± S.E.M.; p < 0.001; middle). These experiments demonstrate that knockdown of nucleostemin protein results in a loss-of-function phenotype as evidenced by a decrease in proliferation and support a role for nucleostemin in maintaining the cell division of CNS stem cells and U2OS cells.
Figure 5.
Perturbed expression of nucleostemin drives cells out of the cell cycle. (A) siRNA knockdown of nucleostemin (NS) in cortical stem cells. (Left) The percentage of noncycling cells increases in cultures transfected with nucleostemin-specific siRNA (Exp) compared with cultures treated with control siRNA (Cntrl). (Right) Nucleostemin-BrdU double-labeled immunofluorescence of culture treated with nucleostemin-specific (Exp) or control (Cntrl) siRNA. Arrows indicate nondividing cells. (B) siRNA knockdown of nucleostemin (NS) in U2OS cells. (Left) Western analysis of the expression level of nucleostemin, α-tubulin (α-Tub), B23, and nucleolin (Nucln). The amounts of nucleostemin protein (2 d after transfection) were standardized to the amounts of α-tubulin on the same blot and expressed as ratios to the control. (Middle) The percentage of cells in S phase is lower in cultures transfected with nucleostemin-specific siRNA (Exp) than the control (Cntrl). (Right) Nucleostemin-BrdU double-labeled immunofluorescence of control and experimental cultures. Arrows indicate cells without nucleostemin staining. (C) Overexpression of HA-tagged nucleostemin in bFGF-maintained, undifferentiated culture (stem cells) or CNTF-induced, 2-d differentiated culture (astrocytes). The percentage of nondividing cells among raNS-expressing cells [HA(+)] is higher than the control-transfected cells [HA(−)] in both the bFGF and 2-d CNTF cultures. (Right) Arrow indicates nondividing raNS-expressing cells and asterisk indicates dividing raNS-expressing cells. (D) Overexpression of HA-tagged nucleostemin [left panel, HA(+); right panel, red] in undifferentiated stem cell culture decreases the percentage of cells in prophase, revealed by α-p-H3 staining (right panel, green). In all the studies in this figure, statistical analyses were carried out by scoring randomly chosen HPFs (high power fields) from two independent triplicate experiments (n = 6); error bars represent S.E.M. Bars: A,C,D, 10 μm; B, 20 μm.
Overexpression of nucleostemin elicits loss-of-function phenotypes in embryonic cortical stem cells
To gain more insight into the function of nucleostemin, we increased the gene dosage in cortical stem cells and analyzed the consequences (Fig. 5C,D). Exogenous nucleostemin was introduced by transfecting stem cells with the plasmid encoding hemagglutinin (HA)-tagged raNS under the control of a constitutively active EF-1α promoter (Mizushima and Nagata 1990). The percentage of noncycling cells (Fig. 5C, right, indicated by arrows) was measured in two different paradigms: an undifferentiated culture, in which stem cells were maintained in bFGF (Fig. 5C, bar graph, left), and a differentiating culture, in which stem cells were differentiated into astrocytes by 2-d CNTF treatment (Fig. 5C, bar graph, right). In both conditions, more of the raNS-overexpressing cells became nonmitotic than did control transfected cells (13.6% ± 1.4% versus 5.0% ± 0.7% in bFGF culture; 77.2% ± 3.4% versus 34.2% ± 2.7% in 2-d CNTF culture, mean ± S.E.M.; p < 0.001). To estimate the number of cells in mitosis, anti-phospho-histone H3 (α-p-H3) staining was used (Fig. 5D, right panel). We only included cells in prophase in this analysis because the nucleolar structures disperse in later stages of mitosis, which made it impossible to determine if those cells were expressing low level of exogenous protein. In accord with the increase in nondividing cells, the population overexpressing nucleostemin contained less cells in prophase than did the control transfected population (3.0% ± 0.3% versus 5.2% ± 0.3%, mean ± S.E.M.; p < 0.001; Fig. 5D, left). These results show that the misexpression of nucleostemin at a high level causes cells to exit cell cycle, a loss-of-function phenotype similar to the knockdown experiment. This is consistent with previous studies showing that elevated levels of gene dosage in a complex pathway may perturb the normal development and/or function of cells overexpressing the gene (Hay et al. 1997; Yang et al. 1999).
Functional importance of the basic and GTP-binding domains in nucleostemin
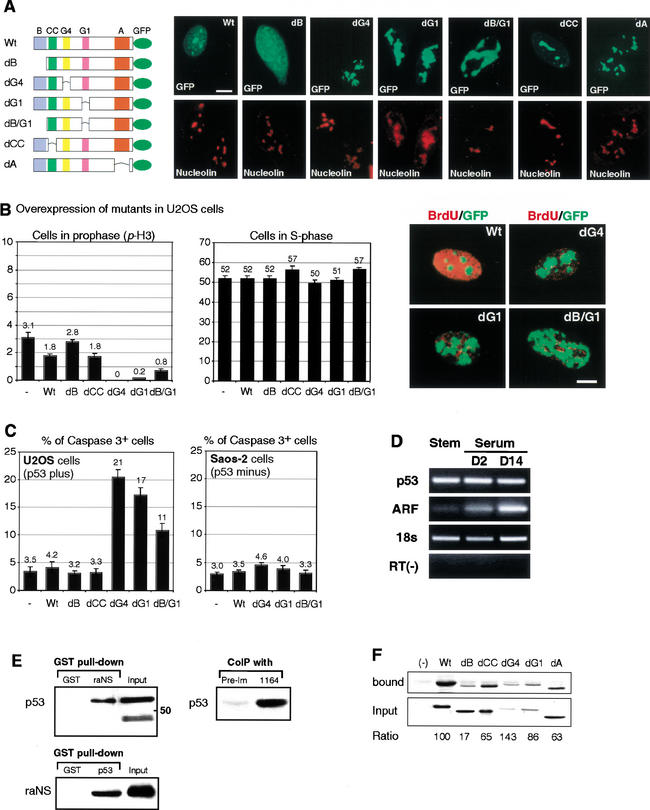
To illuminate how excessive nucleostemin blocks cell-cycle progression and to further dissect the molecular mechanism of nucleostemin function, we conducted a series of deletion analyses. Constructs with engineered C-terminal GFP-fusion proteins were generated with deletion of the basic domain (dB; codons 2–46), the coiled-coil domain (dCC; codons 61–87), the G4 GTP-binding motif (dG4; codons 177–180), the G1 GTP-binding motif (dG1; codons 256–263), the acidic domain (codons 460–503), or both the basic and G1 domains (dB/G1; Figs. 6A, 1C,D). When introduced into U2OS cells, all mutants were localized in the nucleus but displayed different patterns of distribution within the nucleus (Fig. 6A, right). The nucleolar distribution of nucleostemin depends mostly on the N-terminal basic domain. Without this region, the mutant protein (dB) becomes more diffuse in the nucleoplasm. On the other hand, mutants lacking either G4 or G1-GTP-binding motif (dG4, dG1) form irregular aggregates associated with the nucleolar region labeled by α-nucleolin staining, and distort the nucleolar structure. To test whether the ability of these mutants to form aggregates depends on their nucleolar localization, a double mutant (dB/G1) was made that lacked both the basic and the G1 domain. Interestingly, although this mutant protein still forms aggregates, it is now dissociated from the nucleolus. Deletion of the coil-coiled domain or the acidic domain does not produces a consistent change in the intracellular distribution of nucleostemin or the morphology of nucleoli (Fig. 6A). These results demonstrate that the basic domain is required for the nucleolar localization and that the GTP-binding motifs are required to form appropriate nucleolar structures.
Figure 6.
Deletion analysis and regulatory control of nucleostemin. (A) Schematic diagram (left) and intracellular distribution (right) of mutant nucleostemins lacking individual structural domains. (B) Overexpressing mutant nucleostemin decreases the number of cells in prophase (left, revealed by p-H3 staining), particularly with the dG4 and dG1 mutants, but not the number of cells in S phase (middle) in U2OS cells. Statistical analyses were carried out by scoring randomly chosen HPFs from two independent triplicate experiments (n = 6). (Right) BrdU-GFP immunofluorescence shows the patterns of BrdU labeling in wild-type or mutants-transfected cells during S phase. (C) Overexpressing dG4, dG1, and, to a lesser extent, dB/G1 mutant nucleostemin increases the number of apoptotic cells in U2OS cells (left) but not in Saos-2 cells (right). Statistical analyses were carried out by scoring randomly chosen HPFs from two independent triplicate experiments (n = 6). (D). The expression levels of p53 and ARF in CNS stem cells and cultures treated with 10% serum for 2 d and 14 d were measured by semiquantitative RT–PCR. 18S rRNA with 2:8 primer to Competimer ratio was used as a positive control; RT(−) reaction, carried out without RT in the first-strand synthesis reaction, was used as a negative control. Amplication cycles are p53 (29×), ARF (35×), 18s (23×), and RT(−) (33×). (E). Nucleostemin and p53 exist in a protein complex. In GST pull-down assays, p53 is specifically retained in the GST-raNS fraction and later detected by α-nucleostemin (2438) Western blot analysis (left, top) and vice versa (left, bottom). Such interaction can also be shown by coimmunoprecipitation of endogenous p53 and nucleostemin (right) in HEK293 cell lysate by α-nucleostemin (1164) antiserum but not by preimmune serum (Pre-Im). (F). Mapping of p53-interacting region in nucleostemin by GST pull-down assay. Same amount of GST-p53 and comparable amount of HA-tagged nucleostemin proteins (with the exception of dG4 and dG1) was used in each reaction. The retained proteins (bound) were detected by α-HA Western blot and expressed as ratios to the input. Error bars, S.E.M. Bars: A,B, 10 μm.
Overexpression of nucleostemin, especially mutants lacking GTP-binding motif, disrupts cell-cycle progression and induce cell death
In Figure 5, we show that perturbation of nucleostemin expression disrupts cell-cycle progression. To better define this effect, we analyzed cell-cycle kinetics in cells expressing wild-type and mutated nucleostemin. The proportion of cells in prophase was estimated by an antibody that recognizes the phosphorylated form of histone H3 (α-p-H3), and the percentage of S-phase cells was given by the number of BrdU-labeled cells after a 15-min pulse. Consistent with the previous results in cortical stem cells (Fig. 5D), overexpression of wild-type nucleostemin, as well as coil-coiled domain mutant, caused a significant decrease in the percentage of cells in prophase (1.8% ± 0.1% and 1.8% ± 0.2% respectively, versus 3.1% ± 0.4% in control, mean ± S.E.M.; p < 0.01). In contrast to the wild-type protein, deletion of the N-terminal basic domain had no effect on the number of prophase cells (2.8% ± 0.1%), suggesting that nucleolar localization was required for the overexpression phenotype seen with the wild-type protein. Expression of mutants lacking the G4 or G1 domains caused a dramatic decrease in the number of cells in prophase (0.0% ± 0.0% and 0.2% ± 0.0% respectively). Again, this effect depends partially on the basic domain, as overexpression of dB/G1 resulted in a higher number of cells in prophase than the G1 deletion alone (0.2% ± 0.0% versus 0.8% ± 0.1%; p < 0.05; Fig. 6B, left).
The 15-min pulse with BrdU allowed us to measure the number of cells in S phase. Surprisingly, there were no changes in the number of S-phase cells among different mutants, wild-type nucleostemin, and control (Fig. 6B, middle). However, the BrdU labeling pattern showed a clear difference between cells transfected with the GTP-deleted mutants and the wild-type protein. In the mutants lacking the GTP-binding motifs, BrdU labeling was restricted to small regions in the nucleus, indicating that chromatin replication was blocked (dG1, dG4, dB/G1; Fig. 6B, right). None of the other deletions had this effect. These findings suggest that dysregulation of GTP binding in nucleostemin blocks cell-cycle progression in the late S phase.
To determine the outcome of cell-cycle arrest in the overexpression experiments, we measured cell death in cells expressing the mutant proteins. Cell death was assayed by measuring the expression of the proteolytically activated form of Caspase3 as an indicator of apoptosis (CM1; Srinivasan et al. 1998). Shown in Figure 6C (left), the percentage of Caspase3+ cells increased significantly in all mutants lacking the GTP-binding domains when expressed in U2OS cells. The increase in cell death is partially dependent on the N-terminal basic domain, as the apoptotic cells in the dB/G1 mutant were diminished compared with the dG1 mutant (10.9% ± 1.3% versus 17.3% ± 1.3%; p < 0.005). Interestingly, the increase in cell death by these mutants also depended on the presence of wild-type p53 as the same mutants expressed in Saos-2 cells, a p53-null osteosarcoma cell line, exhibited no significant changes in the percentage of apoptotic cells from the wild-type protein or control (Fig. 6C, right).
Nucleostemin and p53 exist in a protein complex
p53 is a key cell-cycle checkpoint protein that is involved in stress response, tumor suppression, and premature aging. To elucidate the role of p53 in mediating the overexpression phenotypes of nucleostemin, we first examined changes in the expression level of p53 and ARF in CNS stem cells and differentiated cultures (Fig. 6D). ARF is a tumor suppressor that stabilizes p53 by acting through MDM2 (Kamijo et al. 1998; Zhang et al. 1998; Tao and Levine 1999) and, like nucleostemin, is localized in the nucleolus. Using semiquantitative RT–PCR, we showed that p53 was present in the stem cell population, and its level of expression remained unchanged throughout differentiation. On the other hand, the expression of ARF was low in the CNS stem cells and up-regulated as they differentiated, a pattern opposite to that seen with nucleostemin.
Despite its nucleolar distribution, ARF has been shown to physically interact with p53 (Kamijo et al. 1998). Next, we examined if nucleostemin, like ARF, could also bind p53. In GST-raNS pull-down experiments, raNS retained specifically the full-length p53 (Fig. 6E, upper left) but not a proteolytic product of p53 (lower band in the input lane). This binding was not seen with the negative control that used the backbone GST protein to pull down p53. The interaction between p53 and nucleostemin could be further demonstrated by (1) GST pull-down assay using p53 as the GST fusion protein (lower left) and (2) coimmunoprecipitation of the endogenous p53 and nucleostemin protein complex by 1164 antibody in nontransfected HEK293 cells (right). The specificity of this interaction was emphasized by its requirement for both nucleostemin protein and α-nucleostemin antibody. The presence of p53 in the precipitated endogenous protein complex suggests that the association between nucleostemin and p53 occurs under physiological conditions.
The basic region of nucleostemin mediates its interaction with p53
To map the interacting region between nucleostemin and p53, HA-tagged mutant proteins were expressed in H1299 cells and incubated with agarose-bound GST-p53 fusion protein in GST pull-down experiments. The amount of mutant proteins retained by GST-p53 fusion was measured by Western blot and presented as the ratio to the input proteins. As shown in Figure 6F, deletion of the basic domain significantly reduced the binding of nucleostemin to p53 (17%), whereas the amount of p53 bound by other mutants either had little change (dG1) or a mild 1.5-fold increase (dG4) or reduction (coiled-coil domain, acidic domain). This data showed that the interaction of nucleostemin with p53 is mediated through its N-terminal basic domain.
Discussion
Nucleostemin expression in embryonic and somatic stem cells
In the present study, we have taken advantage of the precise kinetics of differentiation in cell culture to identify a novel nucleolar protein expressed by developing CNS stem cells. In addition, nucleostemin is present in adult subventricular zone CNS cells expanded as neurospheres, ES cells, other tissues from developing embryos, and several human cancer cell lines, but is absent in most adult cells. In the hematopoietic system, nucleostemin is found in a database of genes enriched in Sca-1+, AA4.1+, c-kit+ fetal liver hematopoietic stem cells (SC|LL2GRID-78|14254; Phillips et al. 2000), and is preferentially expressed in the c-kit+/lin− adult bone marrow population that is enriched for hematopoietic stem cells.
The expression of nucleostemin is reduced considerably when the expression of PCNA (a proliferative marker) and B23 (a major nucleolar protein) still remains high during CNS stem cell differentiation both in vitro and in vivo. When cortical stem cells differentiate into neurons, astrocytes, and oligodendrocytes, nucleostemin levels are reduced in both dividing and postmitotic progeny. In the hematopoietic system, B-lymphocytes and granulocytes are nucleostemin-negative, but they express the same level of PCNA as that of the stem cell-enriched population. These results demonstrate that nucleostemin is expressed predominantly in primitive but not differentiated cells, and the down-regulation of nucleostemin occurs prior to terminal differentiation.
Regulation of cell-cycle progression by nucleostemin
The protein knockdown experiments suggest that nucleostemin functions to maintain the proliferative capacity of stem cells and cancer cells. After siRNA knockdown, 8% of U2OS cells remain in the cycle even when immunostaining fails to detect nucleostemin signals. The retained proliferative activity may be driven by residual nucleostemin, or perhaps cells that lack nucleostemin can continue to proliferate for a short period and resemble the dividing progeny seen during the differentiation of CNS stem cells.
Overexpression of nucleostemin also reduces the proliferative rate of stem cells and U2OS cells. Deletion analysis provides an explanation for the overexpression phenotype, as well as new insights into the regulatory control of nucleostemin. The deletion study shows that mutants missing the N-terminal basic region distribute diffusely in the nucleoplasm, and mutants lacking the GTP-binding sites accumulate as nucleolar aggregates and disrupt the nucleolar structure. When both the basic region and the G1 GTP-binding motif are deleted, nucleostemin is aggregated outside the nucleolus. These results indicate that the N-terminal basic domain determines the nucleolar localization and the GTP-binding motifs regulate the formation of protein aggregates.
Overexpression of nucleostemin causes a decrease in prophase but not S-phase cells, and an increase in cell death that is dependent on the N-terminal basic domain. The effect of overexpression on the numbers of cells in mitosis and cell survival is particularly augmented in the case of mutants lacking the GTP-binding sites. Although the number of cells in S phase remains the same, the BrdU-labeling patterns are quite different in cells expressing these GTP-deleted mutants compared with the wild-type protein. The punctate pattern of BrdU labeling occurs between aggregates of the mutant proteins, indicating that DNA replication is initiated but does not progress through the chromatin regions associated with high levels of nucleostemin. In agreement with this finding, the percentage of cells in prophase is underrepresented in cells expressing the same mutants, and they also show a dramatic effect on cell survival. These data suggest that excess nucleostemin may directly disrupt the nucleolar complex or indirectly cause a block in cell-cycle progression in late S phase, leading to cell death (Fig. 7).
Figure 7.
Schematic diagram illustrating a model of the GTP-regulated control of nucleostemin on stem cell proliferation. During normal cell proliferation in stem cells or cancer cells, nucleostemin is distributed predominantly in the nucleolus (represented by white polygons) and less in the nucleoplasm (represented by dark gray polygons). The association of nucleostemin with the nucleolar complex (represented by gray circles) is mediated through the N-terminal basic domain of nucleostemin. The down-regulation of nucleostemin, during stem cell differentiation or in knockdown experiments, leads to cell-cycle exit. Overexpression of mutants lacking GTP-binding motifs causes nucleolar aggregates, which leads to cell-cycle arrest in the late S phase and cell death, suggesting the GTP-binding domain plays a role in the dynamic shutting of nucleostemin between in the nucleolar and nucleoplasmic compartments and is important for its normal function. The interaction of nucleostemin with p53 is mediated through its N-terminal basic domain and presumably takes place in the nucleoplasm.
Small GTP binding proteins use the conserved GTP-binding structure as a molecular switch to regulate fundamental cellular functions. Drawing analogy from other small GTPases, one can infer that the dynamic shuttling of nucleostemin between the nucleolar and the nucleoplasmic compartment may be regulated by the GTP-binding state of nucleostemin. We hypothesize that nucleostemin forms a complex with other nucleolar proteins when it is in a non-GTP bound form and becomes dissociated on binding with GTP. Although it is presently unknown whether the GTP-bound or non-GTP-bound nucleostemin represents the active form, our data demonstrate that disruption of this regulatory mechanism can result in cell-cycle arrest and cell death. The increased cell death in cells expressing the GTP-deleted mutants is abolished when the same constructs are expressed in p53-null Saos-2 cells, indicating that the cell-cycle arrest and cell death effects are mediated by a p53-dependent pathway. In accordance with this idea, these overexpression phenotypes can be partially rescued by deletion of the N-terminal basic domain, which is required for p53 binding. These experiments suggest that nucleostemin may permit a GTP-regulated and stem cell-specific control of p53 regulation.
The role of nucleoli in the control of stem and cancer cell proliferation
Nucleolar proteins have been shown to regulate cell proliferation and growth by controling ribosome biosynthesis and p53 functions (Drayton and Peters 2002; Du and Stillman 2002; Michael and Oren 2002). The activation of p53 has been linked to cell-cycle arrest and apoptosis in response to stress. Although nucleolar localization may not be essential (LaBonne and Bronner-Fraser 1998), p53 function can be modulated by several nucleolar proteins, including ARF (Kamijo et al. 1998; Zhang et al. 1998; Tao and Levine 1999) and Werner syndrome protein (Blander et al. 1999; Spillare et al. 1999). ARF knockout mice show increased resistance to senescence (Kamijo et al. 1997), and patients carrying WRN gene mutations exhibit increased genomic instability and premature aging. A recent report demonstrates that in mice with a gain-of-function p53 mutation, there is a remarkable acceleration of aging and reduced longevity (Tyner et al. 2002). Our work presents a novel control mechanism of cell-cycle progression in stem cells and cancer cells, involving a new nucleolar protein that may function through known tumor suppressor genes in late S and G2. The complementary expression patterns of nucleostemin and ARF suggest a switch in the nucleolar control of cell proliferation during stem cell differentiation. It will be interesting to determine whether nucleostemin is a stem cell-specific component of the G2/M checkpoint system that monitors genotoxic stress (Taylor and Stark 2001), and what other molecules are involved in this pathway.
Materials and methods
Cortical stem cell culture and transfection protocol
Primary cortical stem cell culture was prepared from rat cerebral cortices of E14.5 embryos (Johe et al. 1996; Tsai and McKay 2000). Cells were expanded in the presence of bFGF (20 ng/mL) for 4 d and were replated at 2 × 105 per 10-cm plate for 4 d before induction of differentiation. Neurospheres were derived from the subventricular zone of the lateral ventricles of adult rats and cultured in suspension. Transfection was carried out on cells grown as monolayers on glass coverslips using LipofectAmine Plus reagent (Invitrogen).
Molecular cloning and RT–PCR
For PCR-select cDNA subtraction (Clontech), polyA+ RNA (2 μg) was used in the cDNA synthesis process. Because of the nature of our screening methodology, genes that are part of a family with close homology or of low abundance may be missed. The complete cDNAs for murine (muNS) were isolated from a rapid-screen cDNA library panel derived from E12.5 Swiss Webster mice (OriGene Technologies); the rat nucleostemin (raNS; Sprague-Dawley), from RACE PCR on first-stranded cDNA derived from cortical stem cells. Semiquantitative PCR reactions were carried out in 25 μL with 1 μL of the first-strand cDNA solution and 0.4 μM of each primer. The linear range was predetermined by pilot experiments.
Antibody synthesis and immunocytohistochemistry
The 2438 antibody was raised in chicken (Aves Labs); 1164 antibody, in rabbit (Biosynthesis Inc.). The 2438 antibody recognized primarily rat and murine nucleostemin, and the 1164 antibody recognized human, mouse, and rat proteins. Primary antibodies and their dilution used in this study were chicken (2438; 1:500) and rabbit (1164; 1:1000) α-nucleostemin, rat α-BrdU (Harlan SeraLab; 1:200), α-HA (mouse or rabbit, covance; 1:1000), α-nucleolin (RDI; 1:1000), α-phospho-histone H3 (Upstate; 1:400), and α-activated caspase3 (CM1; 1:5000). Secondary antibodies were rhodamine red or FITC-conjugated (Jackson). Cells were fixed with ice-cold methanol (for α-nucleostemin and nucleolin) or 4% paraformaldehyde. For immunohistochemistry, embryos were harvested from time-pregnant CD1 mice and embedded in OCT embedding material (Tissue-Tek) without fixation. Frozen sections (16 μm) were fixed in 4% paraformaldehyde for 2 min and incubated with 1164 (1:1500). Immunohistochemistry was performed with ABC kit (Vector) and 3,3′-diaminobenzidine (DAB) color reaction (SigmaFast). For BrdU double-labeled immunocytochemistry, a 15-min pulse of BrdU (40 μM) was administered to mark S-phase cells, or an 18-h pulse of BrdU (10 μM) was used to label all cells in cell cycle. After carrying out immunostaining of the first antibody, cells were fixed, treated with 4 N HCl for 10 min to denature the DNA, and continued with α-BrdU staining.
siRNA knockdown experiment
Cells were incubated with siRNA (100 nM, Dharmacon Research) complexed with oligofectamine (Invitrogen) for 18 h. BrdU labeling (40 μM for 15-min pulse or 10 μm for 18-h pulse) was measured 72 h after transfection. The sequences for siRNA are as follows: nucleostemin-specific siRNA, AAGAACUAA AACAGCAGCAGAdTdT; control siRNA, AACAUUCAGA CUGGGAAAUGGdTdT (for rat cells) and AAUCAGACGUG GACCAGAAGAdTdT (for human cells).
GST binding, coimmunoprecipitation, and Western blot analysis
Two hundred ninety-three cells (100-mm dish) were transfected with 5–10 μg of DNA plasmid and harvested 36 h later. Cells were lysed in extraction buffer (PBS/1% Triton X-100/1mM PMSF/protease inhibitors) and assayed for protein concentration (Bradford colorimetric assay; Bio-Rad). Lysates (500 μg) were incubated with (1) immobilized purified GST or GST fusion protein, and glutathione agarose in 1 mL volume for GST pull-down experiment, or (2) antibody (1 μg for purified antibody or 10 μL for crude serum) and protein A-Sepharose (50 μL) for coimmunoprecipitation for 2 h at 4°C. Beads were washed with PBS once, PBS/500 mM NaCl twice, and PBS twice. The antibody used in Western blot detection are nucleostemin (2438 and 1164; 1:1000) and p53 (FL393, 1:500, Santa Cruz; Ab7, 1:2000, Calbiochem). The antibody used in developmental Western blot detection are nucleostemin (2438 and 1164; 1:1000), B23 (C-19; 1:200; Santa Cruz), and PCNA (1:500, Novocastra).
Flow cytometry
To enrich hematopoietic stem cells, bone marrow cells were harvested from adult C57BL/6J mice, depleted of cells with lineage-specific markers (CD4, CD8α, B220, Gr-1, and Mac-1) by magnetic sorting, incubated with FITC-conjugated α-c-kit antibody, and sorted on a FACS Vantage II flow cytometer (Beckman Coulter). For B-lymphocyte and granulocytes lineages, bone marrow cells were depleted of the other hematopoietic lineages and then positively sorted with B220 or Gr-1 antibody respectively by flow cytometry.
Acknowledgments
We are particularly grateful to Florian Then Bergh and NINDS DNA sequencing facility for their help with FACS and DNA sequencing. We also thank Dave Owens, Heather Cameron, David Panchision, Howard Nash, and members in McKay's lab for insightful discussion and help with the manuscript. Caspase3 antibody (CM1) was supplied by agreement with Idun Pharmaceuticals.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
EMAIL mckay@codon.nih.gov; FAX (301) 402-4738.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.55671.
References
- Blander G, Kipnis J, Leal JF, Yu CE, Schellenberg GD, Oren M. Physical and functional interaction between p53 and the Werner's syndrome protein. J Biol Chem. 1999;274:29463–29469. doi: 10.1074/jbc.274.41.29463. [DOI] [PubMed] [Google Scholar]
- Drayton S, Peters G. Immortalisation and transformation revisited. Curr Opin Genet Dev. 2002;12:98–104. doi: 10.1016/s0959-437x(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Du YC, Stillman B. Yph1p, an ORC-interacting protein: Potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Ou J, Easterday MC, Dougherty JD, Jackson RL, Chen Z, Antoine H, Terskikh A, Weissman IL, Nelson SF, et al. A genetic analysis of neural progenitor differentiation. Neuron. 2001;29:325–339. doi: 10.1016/s0896-6273(01)00209-4. [DOI] [PubMed] [Google Scholar]
- Hay BA, Maile R, Rubin GM. P element insertion-dependent gene activation in the Drosophilaeye. Proc Natl Acad Sci. 1997;94:5195–5200. doi: 10.1073/pnas.94.10.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes & Dev. 1996;10:3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Induction and patterning of the neural crest, a stem cell-like precursor population. J Neurobiol. 1998;36:175–189. doi: 10.1002/(sici)1097-4695(199808)36:2<175::aid-neu6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Lendahl U, Zimmerman LB, McKay RD. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- Michael D, Oren M. The p53 and Mdm2 families in cancer. Curr Opin Genet Dev. 2002;12:53–59. doi: 10.1016/s0959-437x(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Mizushima S, Nagata S. pEF-BOS: A powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RL, Ernst RE, Brunk B, Ivanova N, Mahan MA, Deanehan JK, Moore KA, Overton GC, Lemischka IR. The genetic program of hematopoietic stem cells. Science. 2000;288:1635–1640. doi: 10.1126/science.288.5471.1635. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Spillare EA, Robles AI, Wang XW, Shen JC, Yu CE, Schellenberg GD, Harris CC. p53-mediated apoptosis is attenuated in Werner syndrome cells. Genes & Dev. 1999;13:1355–1360. doi: 10.1101/gad.13.11.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, Roth KA, Sayers RO, Shindler KS, Wong AM, Fritz LC, Tomaselli KJ. In situ immunodetection of activated caspase-3 in apoptotic neurons in the developing nervous system. Cell Death Differ. 1998;5:1004–1016. doi: 10.1038/sj.cdd.4400449. [DOI] [PubMed] [Google Scholar]
- Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- Terskikh AV, Easterday MC, Li L, Hood L, Kornblum HI, Geschwind DH, Weissman IL. From hematopoiesis to neuropoiesis: Evidence of overlapping genetic programs. Proc Natl Acad Sci. 2001;98:7934–7949. doi: 10.1073/pnas.131200898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, McKay RD. Cell contact regulates fate choice by cortical stem cells. J Neurosci. 2000;20:3725–3735. doi: 10.1523/JNEUROSCI.20-10-03725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Broihier HT, Moore LA, Lehmann R. HMG-CoA reductase guides migrating primordial germ cells. Nature. 1998;396:466–469. doi: 10.1038/24871. [DOI] [PubMed] [Google Scholar]
- Yang XW, Wynder C, Doughty ML, Heintz N. BAC-mediated gene-dosage analysis reveals a role for Zipro1 (Ru49/Zfp38) in progenitor cell proliferation in cerebellum and skin. Nat Genet. 1999;22:327–335. doi: 10.1038/11896. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]