Abstract
Virulence in the human fungal pathogen Cryptococcus neoformans is associated with the α mating type. Studies to identify the properties of α cells that enhance pathogenesis have led to the identification of a mating-type locus of unusually large size and distinct architecture. Here, we demonstrate that the previously identified MATα components are insufficient to regulate sexual differentiation, and we identify a novel α-specific regulator, SXI1α. Our data show that SXI1α establishes α cell identity and controls progression through the sexual cycle, and we discover that ectopic expression of SXI1α in a cells is sufficient to drive a/α sexual development. SXI1α is the first example of a key regulator of cell identity and sexual differentiation in C. neoformans, and its identification and characterization lead to a new model of how cell fate and the sexual cycle are controlled in C. neoformans.
Keywords: Basidiomycete, fungal pathogenesis, gene regulation, sex determination, mating-type locus, virulence
A fundamental question for organisms with different cell types is how unique cell identities are established and maintained. Sexual organisms from algae to mammals have evolved mechanisms that direct cells of different types to adopt unique sexual identities. Two common mechanisms govern cell identity: those based on sensing ploidy or dosage and those based on expression of a sex-determining gene (Capel 1998). Dosage-dependent regulation establishes sexual identity in both flies and worms (Parkhurst and Meneely 1994). Although the molecular mechanisms by which dosage dictates cell fate differ, the principle is the same; X animals develop into males, whereas XX animals develop into females. By sensing the ratio of X chromosomes to autosomes, these organisms initiate different developmental programs that result in opposing sexual identities.
On the other hand, sexual differentiation in mice and humans is regulated by the presence of a sex-determining gene (Haqq and Donahoe 1998). In this case, the Sry gene on the Y chromosome designates maleness and determines the fate of the organism; XX individuals develop into females, whereas XY individuals develop into males. This mechanism appears to be an ancient one because even the single-celled, green algae Chlamydomonas reinhardtii uses a differentially expressed gene to control cell identity. In this case, the Gsp1 homeodomain protein controls sexual cycle progression by triggering a change from gamete to zygote cell type (Zhao et al. 2001). The conservation of these mechanisms illustrates the importance of sexual identity and cell fate determination in both uni- and multicellular organisms.
Unlike mammals, most single-celled eukaryotes do not have sex chromosomes dedicated to maintaining cell identity, but they do contain a specialized region of the genome that differs between cell types. This region, called the mating-type (MAT) locus, has been studied extensively in fungi. Genes located in the MAT locus control fungal mating, and different mating types contain dissimilar information at this locus. That is, MAT loci differ from most of the genome because they contain information that is not the same between homologous chromosomes (Coppin et al. 1997; Casselton and Olesnicky 1998; Turgeon 1998). In a typical fungus, one haploid mating type contains genes encoding a information, and the other contains genes encoding α information. In the diploid cell that forms after mating, homologous chromosomes contain a region of nonhomologous sequence (a situation that is reminiscent of the X and Y sex chromosomes of multicellular eukaryotes). This arrangement of the MAT locus allows cells of opposing mating types to respond differentially to each other and to their environments.
How the MAT locus determines cell fate has been elucidated in Saccharomyces cerevisiae (Herskowitz et al. 1992). This budding yeast has two mating types, a and α, which can mate to form a third cell type, the a/α diploid cell. Changes in gene expression in the different cell types control their sexual identities. In α cells, the α1 and α2 proteins are expressed from MATα and are responsible for activating α-specific genes and repressing a-specific genes, respectively. In a cells, the a1 protein is expressed from MATa and has no known function in a cells. When a and α cells mate to form a diploid cell, a1 and α2 interact to form a new regulatory activity in the cell that is responsible for repressing haploid-specific genes. This repression prevents diploid cells from mating but allows them to undergo meiosis and sporulation in response to specific environmental signals. Much like sexual identity determinants in multicellular organisms (such as SRY), these cell-type-specific proteins establish and maintain cell identity in this single-celled organism (Johnson 1995).
Cell identity is also important in other, less well-known fungi. Like S. cerevisiae, the fungal pathogen Cryptococcus neoformans is amenable to genetic study because it is a haploid organism with two mating types (a and α) that engage in a sexual cycle that can be reproduced and manipulated in the laboratory (Alspaugh et al. 2000). What distinguishes C. neoformans from S. cerevisiae and most other fungi, however, is its ability to cause disease in humans. C. neoformans infects primarily immunosuppressed individuals, including AIDS patients, transplant recipients, and patients undergoing chemotherapy (Casadevall and Perfect 1998). It appears that the haploid spores of C. neoformans lodge in the lung, where they can disseminate and lead to life-threatening cryptococcal meningitis.
Several virulence factors that contribute to the success of C. neoformans as a pathogen have been identified, and one of these is mating type (Casadevall and Perfect 1998). More than 95% of all C. neoformans isolates are MATα, and in murine experiments α cells have been shown to be more virulent than a cells (Kwon-Chung et al. 1992; Casadevall and Perfect 1998). It is clear that cell type plays an important role in C. neoformans virulence, but the exact mechanism by which it contributes remains to be determined. Thus, analyzing differences between the a and α cells of C. neoformans will prove useful in understanding the characteristics of the fungus required for virulence. It is likely that the important differences between these two cell types will be directly related to the genes in the MAT locus and their targets.
Mating information in the basidiomycete phylum of fungi to which C. neoformans belongs is generally found in two MAT loci, wherein one locus encodes pheromones and pheromone receptors and the other locus encodes homeodomain transcriptional regulators that control mating genes (Kronstad and Staben 1997; Casselton and Olesnicky 1998). A MAT locus has been defined in the C. neoformans α cell type (Moore and Edman 1993; Karos et al. 2000), and it is distinct from any previously identified basidiomycete or other fungal MAT locus. When we began our work, MATα was thought to encompass a single, ∼50-kb region that contains 12 genes, most of which have not been found previously in fungal MAT loci. Interestingly, this single locus contains pheromones and pheromone receptors but no obvious homeodomain proteins; thus, it has been unclear how mating genes are controlled in C. neoformans. Recent work, however, has shown the MAT locus to be much larger than ∼50 kb and to contain more genes than previously thought (Lengeler et al. 2002).
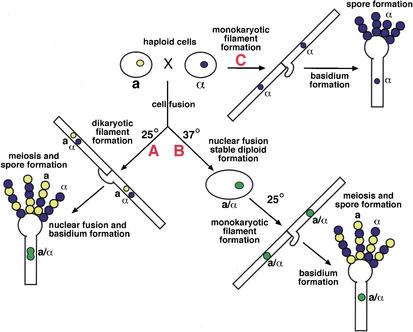
The MAT locus controls three developmental pathways in the C. neoformans sexual cycle (Fig. 1). During the process of mating, the locus is important for sensing a mating partner, forming projections called conjugation tubes, and making subsequent filamentation and sporulation structures (Davidson et al. 2000; Lengeler et al. 2000). The process of diploid filamentation, in which diploid cells form filaments and sporulation structures in response to a decrease in temperature, is similarly controlled by MAT (Sia et al. 2000). Components of the MAT locus are also responsible for regulating the process of filament and spore production in haploid cells known as haploid filamentation (Wickes et al. 1996). This process is specific to α cells, and several single gene deletions in MATα abolish this haploid differentiation (Yue et al. 1999; Chang et al. 2000; Clarke et al. 2001). Recent data suggest, however, that this previously defined locus is not sufficient for carrying out all of the functions of the sexual cycle and that additional regulatory components outside of MATα are required for specifying the α cell fate.
Figure 1.
The Cryptococcus neoformans life cycle. There are three developmental pathways for C. neoformans cells. The first pathway, known as mating (A), begins when two haploid cells of opposite mating types fuse at 25°C. The haploid cells fuse and grow as filaments with distinct nuclei (dikaryons) and special clamp cells that are fused to the filaments. In response to unknown signals, the dikaryon produces a specialized sporulation structure known as a basidium. It is in the basidium that nuclear fusion and meiosis take place. Many rounds of duplication and mitosis lead to the production of haploid a and α spore products that extend in long chains from the basidial head. The second developmental pathway, known as diploid filamentation (B), occurs when cells of opposite mating types fuse and are maintained at 37°C. In this case, the haploid cells and their nuclei fuse to create a yeast-form, diploid cell that grows as a yeast at 37°C. In response to a decrease in temperature (25°C), this a/α diploid differentiates to form monokaryotic filaments with unfused clamp cells, basidia, and haploid a and α spores that are indistinguishable from those produced during mating. The third possible fate occurs in cells of the α mating type. This pathway is known as haploid fruiting (C) and occurs when α cells are grown under nutrient-limited, desiccating conditions. In this pathway, haploid α cells form monokaryotic filaments with unfused clamp cells, basidia, and haploid α spores. In each case, the resulting spores are competent to germinate and grow as vegetative haploid cells or initiate one of the three developmental pathways (A–C).
Here, we present the discovery and analysis of a C. neoformans sexual cycle regulator that does not reside within the previously identified ∼50-kb MAT locus. Unexpectedly, we found that a deletion of the known MATα allele did not abolish a/α cell identity. This finding made it clear that additional components control the sexual cycle. To test the hypothesis that cell ploidy could control sexual development, we created homozygous a/a and α/α diploid strains of C. neoformans and tested their ability to differentiate. These experiments proved that ploidy alone is not sufficient for a/α diploid behavior and that both a and α information are required for proper sexual development. This result prompted us to use a bioinformatics approach to identify genes of interest and led to the discovery of a gene encoding an additional regulatory factor, SXI1α, a homeodomain protein found outside the previously identified MATα region. Our analysis of sxi1α deletion strains revealed a crucial role for this gene in establishing a/α cell identity. Strikingly, we also discovered that ectopic expression of SXI1α in haploid a cells caused them to adopt an a/α cell fate. This finding, along with expression data, reveals that this gene is a key transcriptional regulator responsible for controlling the C. neoformans sexual cycle. This discovery makes it clear that although C. neoformans is unique among fungi in its MAT locus architecture, it has maintained a conserved homeodomain regulator that both establishes cell identity and drives sexual differentiation.
Results
The previously identified MATα allele is insufficient to specify the α cell type
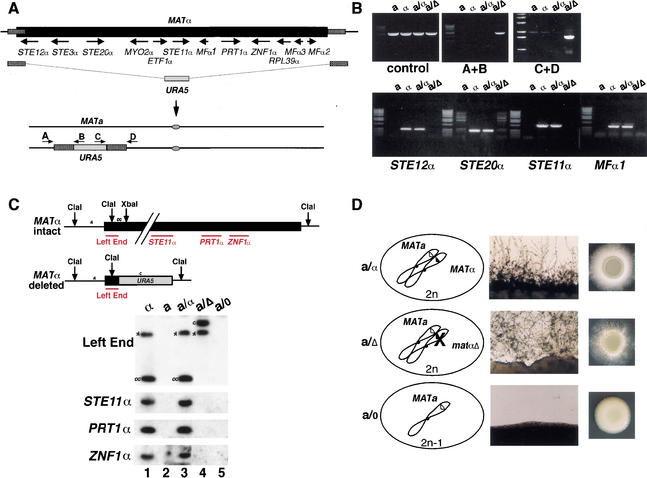
To evaluate the role of MATα in specifying the α cell type, we created a deletion of the entire previously identified MATα allele (Fig. 2A). Using a polymerase chain reaction (PCR)-based deletion strategy, we attempted to delete the entire 48.8-kb region of the known MATα region in an α haploid strain. This effort to disrupt the entire α locus failed (0/300 Ura+ transformants) presumably because of an essential gene(s) in the locus; however, in the first use of a stable C. neoformans a/α diploid strain for gene disruption, we deleted the entire MAT region on the α chromosome. MATa/matα deletion strains (a/Δ) were confirmed by PCR analysis (Fig. 2B) and by Southern blot analysis (Fig. 2C) and shown to contain precise deletions of the MATα region. As represented in Figure 1, pathway B, a/α diploid strains of C. neoformans undergo a temperature-dependent developmental process referred to as diploid filamentation (Fig. 2D, top panel; Sia et al. 2000). We predicted that the deletion of α information from a/α cells would lead to a loss of a/α identity and result in a cell behavior (i.e., the diploid strains would no longer be self-filamentous). Surprisingly, the a/Δ strain retained its a/α identity and remained competent to undergo diploid filamentation and produce viable haploid spores (Fig. 2D, middle panel). These findings suggested that the deleted α information was not required for diploid filamentation and that some other region of the genome was responsible for establishing a/α cell identity. This idea was further substantiated by the finding that a presumptive 2n − 1 diploid strain (a/0; in which the chromosome containing the URA5-marked matα deletion had been selected against by growth on 5-FOA medium) no longer retains its a/α cell identity and behaves instead like an a cell (exhibiting no filamentation behavior; Fig. 2D, bottom panel). The mating type of this strain was also confirmed in assays in which the a/0 strain mated well with an α tester strain but showed no mating with an equivalent a tester strain (data not shown). Thus, α identity was abolished by loss of the entire MATα-containing chromosome but not by loss of the known MATα region alone.
Figure 2.
Deletion of MATα does not abolish a/α cell identity. (A) A schematic diagram of the MATα allele (black box) shows the arrangements of genes in the locus, with arrows indicating the directions of transcription. Hatched boxes indicate the regions of DNA used as sites for recombination in a PCR-generated, URA5 deletion construct. A schematic of the a and α chromosomes shows the predicted matα deletion configuration, and arrows designated A–D represent primers used in PCR of B. (B) PCR results confirm the presence of the proper sequences at the junctions of the matα deletion and the absence of genes located in MATα. Each set of reactions shows the results of PCR with different primer pairs carried out on genomic DNA from wild-type a and α strains, a wild-type a/α diploid strain, and a diploid matα deletion strain (a/Δ). The first lane in each set of reactions contains size standards. (C) Southern blot analysis confirms the matα deletion. Genomic DNA from α and a wild-type strains (lanes 1,2), a wild-type a/α diploid strain (lane 3), a MATa/matα strain (a/Δ, lane 4), and a strain presumably missing the MATα chromosome (a/0, lane 5) was restriction-digested. A probe to the left end of the locus hybridized to two fragments in the expected strains. Symbols to the left of the Southern bands correspond to symbols in the accompanying schematic drawing of the intact and deleted MATα alleles. (D) The panels from left to right show the configuration of the MAT locus in each strain, a magnified view of the filaments formed during sexual differentiation, and a macroscopic view of the filaments on solid agar medium. The top row shows the pattern for wild-type a/α diploid cells, the middle row shows the MATa/matα strain (a/Δ), and the bottom row shows the MATα chromosome loss strain (a/0). Note: Analysis of spores from the a/Δ strain yielded only MATa ura5 meiotic segregants, proving the presence of an essential gene or genes in MATα.
The surprising behavior of the a/Δ strain lacking the entire known MATα allele led us to consider two possible hypotheses to explain how the process of filamentation could be regulated. In the first hypothesis, filamentation of diploid cells could be regulated in a ploidy- or dosage-dependent manner. a/α diploids would filament because the ploidy-sensing mechanism detects the diploid state and activates filamentation independent of MATα. The a/0 strain would fail to filament because it is not fully diploid, or the dose of a specific gene product from the MAT chromosome is diminished. In the second hypothesis, the process of filamentation in diploid cells could be regulated in a gene-dependent manner. Specific genes from a and α cells could function together to combinatorially promote filamentation, and the loss of either set of genes would result in a loss of filamentation. In the a/Δ strain the α gene(s) is still intact, so the strain filaments, but in the presumptive chromosome loss strain (a/0), the α gene(s) has been lost, and with it, the ability to filament.
Diploid filamentation is dependent on the presence of information from both a and α cells
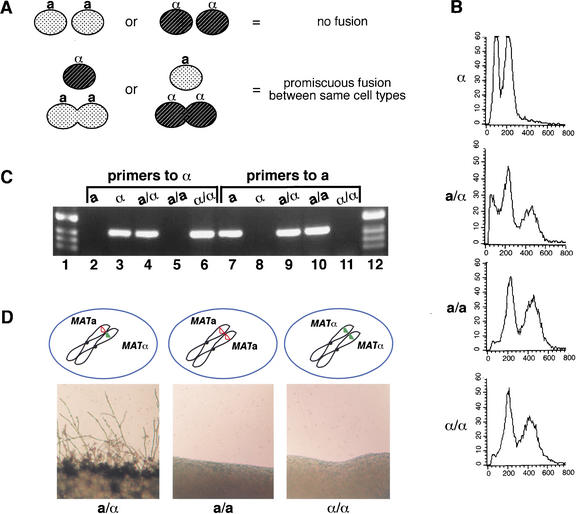
We distinguished between ploidy-dependent and gene-dependent regulation by constructing a/a and α/α diploids and testing their filamentation behavior. We predicted that if ploidy were an inducer of filamentation independent of a- and α-specific signals, then homozygous a/a and α/α diploid strains would be self-filamentous. Homozygous diploid strains of other fungi have been created using a variety of means from UV irradiation (Banuett and Herskowitz 1989) to forced gene conversion in a/α diploids (Hartwell 1980), but we were able to create these strains in C. neoformans by using a novel approach to take advantage of the propensity of C. neoformans for promiscuous fusion. Auxotrophic strains of the same mating type (e.g., a ura5 lys1 and a ade2 lys2) were mixed in a mating reaction in the presence of a multiply marked, auxotrophic opposite mating partner (e.g., α ura5 ade2 lys1). These mixes were then subjected to selective growth conditions in which only a/a or α/α diploid strains (but not a/α) strains could grow (e.g., medium lacking adenine, uracil, and lysine). At a low frequency, strains of the same mating type engaged in promiscuous fusion events in the presence of the opposite mating type (Fig. 3A). The resulting prototrophic strains were recovered and subjected to fluorescence-activated cell scanning (FACS) analysis that confirmed a DNA content consistent with diploidy (Fig. 3B). The prototrophic strains were also shown by PCR with mating-type-specific primers to contain only a or α information (Fig. 3C). These confirmed a/a and α/α homozygous diploid strains behaved like their haploid counterparts in mating (data not shown), haploid fruiting (data not shown), and diploid filamentation. Neither of the homozygous diploid strains exhibited filamentous growth on V8 medium after 48 h at 25°C, whereas an a/α heterozygous control strain filamented well (Fig. 3D). Because diploid cells that were a/a or α/α did not undergo diploid filamentation, we conclude that diploidy alone is not sufficient to activate filamentation.
Figure 3.
Ploidy alone does not regulate diploid filamentation. (A) Homozygous diploid strains of C. neoformans were created by incubating strains of the same mating type in the presence of a helper strain as a pheromone donor (see Materials and Methods for details). Shaded ovals represent a cells, and hatched ovals represent α cells. (B) FACS analysis of prototrophic products is consistent with diploidy. From top to bottom, panels show the FACS profile for an α haploid, an a/α diploid, an a/a diploid, and an α/α diploid. (C) PCR confirms the presence of only a or α information in homozygous diploid strains. Genomic DNA from five different strains (including those used in FACS analysis) was used in a PCR as designated. (Lanes 2–6) α-Specific primers to STE20α. (Lanes 7–11) a-Specific primers to STE20a. (D) Filamentation of diploid strains is not dependent on ploidy alone. Each panel shows a schematic representation of MAT and the results of a self-filamentation assay. (Left to right) a/α, a/a, α/α.
This finding rules out a ploidy-only model of diploid regulation, and indicates that both a and α information are necessary for diploid sporulation. This result is consistent with the hypothesis that a-specific and α-specific genes are responsible for regulating the filamentation process through the specification of cell type. Under this model, the ability of the a/Δ strain to filament makes it clear that the required information for filamentation from α cells is not located entirely within the previously defined MATα allele. In addition, the behavior of the a/0 strain indicates that at least one of the required components resides in a different location on the same chromosome as MATα.
Identification of an α-specific homeodomain protein
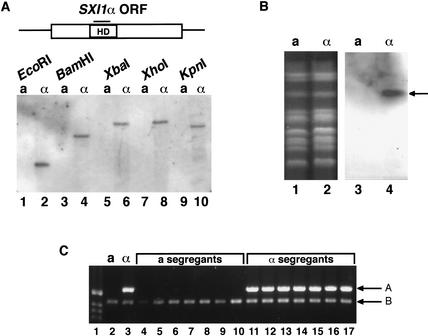
These results suggested strongly that additional regulatory factors not located in MATα are responsible for mediating diploid filamentation. Based on the structures of other fungal MAT loci (Casselton and Olesnicky 1998), and given that the previously defined locus in C. neoformans contained pheromones and a pheromone receptor but no clear DNA-binding proteins (Karos et al. 2000), it was logical to propose that another region of the MATα-containing chromosome encoded DNA-binding proteins. Using sequences from the Candida albicans α2 homeodomain protein (Hull and Johnson 1999), we searched the Stanford C. neoformans genome sequence for similar proteins. Using the BLAST algorithm in conjunction with homeodomain structural data (Li et al. 1995; Altschul et al. 1997), six possible homeodomain proteins were identified, and each sequence was evaluated in two ways. First, Southern blot analysis of DNA from both a and α cells was performed using a probe to the sequence of interest. A chromosome blot was then probed to determine the chromosome location of the sequence. One of the six genes (now known as SXI1α, for Sex Inducer 1α) was specific to α cells (Fig. 4A), and it was located on the MATα-containing chromosome (Fig. 4B). The other five homeodomain-containing sequences were not α-specific (data not shown). In addition, PCR analysis of multiple a and α segregants showed that the SXI1α DNA sequence was found only in the α mating type after a genetic cross (Fig. 4C), confirming linkage to the α mating type.
Figure 4.
SXI1α is an α-specific gene. (A) The open box represents the SXI1α open reading frame, and the box within represents the predicted homeodomain (HD). The bar above HD represents the probe used in Southern hybridization. A Southern blot was generated using DNA from wild-type a and α cells digested with several different restriction enzymes, as indicated. (B, lanes 1,2) An ethidium bromide-stained gel containing the C. neoformans chromosomes from both a and α cells. The MAT locus is located on the third visible chromosome from the top. (Lanes 3,4) A chromoblot filter to which the homeodomain probe of A has been hybridized; the arrow designates the α-specific band corresponding to the third visible chromosome. (C) Genomic DNA from segregants of a genetic cross was subjected to PCR analysis using primers to the homeodomain region of SXI1α. (Lane 1) marker; (lanes 2,3) DNA from control a cells and α cells, respectively; (lanes 4–10) DNA from segregants that mate as a cells; (lanes 11–17) DNA from segregants that mate as α cells. Arrow A indicates the SXI1α PCR product. Arrow B indicates a control product generated with non-cell-type-specific primers.
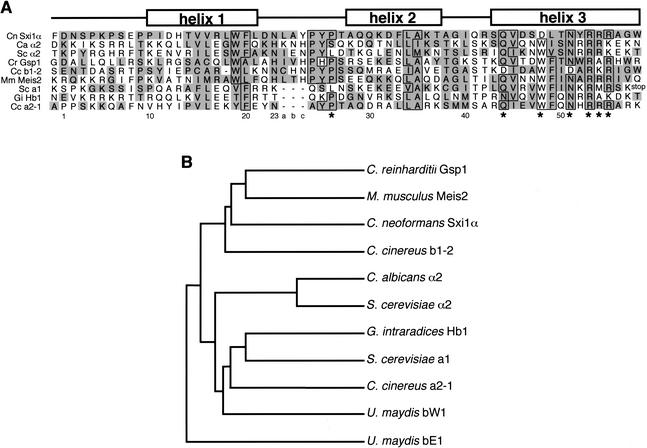
Alignment of the predicted SXI1α homeodomain region with other homeodomain proteins reveals that many residues known to be important for DNA interactions are conserved (Fig. 5A). Sxi1α contains an arginine-rich region in the predicted helix 3, consistent with other homeodomain proteins. Several key residues within this predicted helix are conserved. In particular, asparagine-51, which is invariant among almost all homeodomain proteins (Burglin 1997), and glutamine-44, arginine-53, arginine-54, and arginine-55, which are important for base contacts during DNA binding by a1–α2 (Li et al. 1995), are all conserved. Other important base contact residues are not strictly conserved in Sxi1α (i.e., tryptophan-48), but variation among homeodomains is not unusual, and the overall similarity of Sxi1α with other homeodomains is high enough to construct a phylogenetic tree. Of the fungal regulators, the Sxi1α homeodomain protein is most closely related to a mating protein from Coprinus cinereus (b1-2), a distant but related member of the basidiomycete family of fungi (Fig. 5B).
Figure 5.
Sxi1α is predicted to be a homeodomain protein. (A) The predicted homeodomain region of Sxi1α is aligned with known homeodomains of other proteins, Ca (Candida albicans) α2, Sc (Saccharomyces cerevisiae) α2, Cr (Chlamydomonas reinhardtii) Gsp1, Cc (Coprinus cinereus) b1-2, Mm (Mus musculus) Meis-2, Sc a1, Gi (Glomus intraradices) Hb1, and Cc a2-1. A schematic representation of the homeodomain region showing the helices of a classic three-helix structure found in many homeodomains is above the sequences. Identical residues are outlined, and similar residues are highlighted in gray. Stars highlight key DNA-binding contacts. (B) Phylogenetic tree constructed using the homeodomain regions of the represented proteins. Proteins are the same as in A with the exception of the Ustilago maydis proteins bW1 and bE1, which have been included in the tree.
SXI1α establishes cell identity in C. neoformans
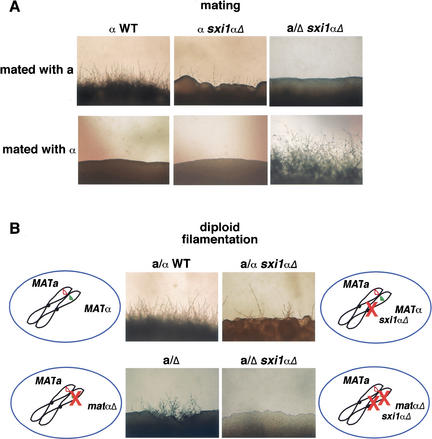
To assess what role this apparent homeodomain protein might play in the life cycle of C. neoformans, we created a sxi1α deletion strain. Most of the predicted SXI1α open reading frame, including the predicted homeodomain region, was deleted in both haploid and diploid cells using a PCR-based disruption strategy (Davidson et al. 2002). The resulting deletion strains were tested for their behavior in a series of phenotypic assays. We were most interested in determining what role SXI1α plays in regulating the C. neoformans sexual cycle, so haploid and diploid deletion strains were exposed to mating and filamentation conditions and evaluated for their ability to form spores. As shown in Figure 6A, mating in the sxi1α deletion strain is severely impaired, as evidenced by diminished filament formation in the presence of an a mating partner (Fig. 6A, top, middle) compared with a wild-type α-by-a cross (Fig. 6A, top, left). The few filaments that do form are grossly abnormal and fail to develop basidia or spores and are rapidly overgrown by the vegetative cell mass. This mating defect does not, however, stem from a defect in cell fusion between mating partners. Experiments in which prototrophs were selected in a cross with marked strains resulted in no decrease in prototroph formation when the sxi1α deletion strain was mated to a cells (data not shown). In summary, Sxi1α is dispensable for cell fusion but necessary for filament formation and development after cell fusion.
Figure 6.
Sxi1α is a key regulator of cell fate. (A) Panels show the filamentation results of a mating assay in which different strains were mixed and incubated under mating conditions. (Top panels, from left to right) The results of crosses between wild-type a cells and wild-type α cells, between wild-type a cells and the sxi1α deletion strain, and between wild-type a cells and the sxi1α matα double deletion strain. (Bottom panels, from left to right) The results of crosses between wild-type α cells, between wild-type α cells and the sxi1α deletion strain, and between wild-type α cells and the sxi1α matα double deletion strain. (B) Panels show the results of diploid filamentation after several days under filamentation conditions, and schematic drawings show the configuration of the MAT locus for each strain tested.
Experiments in a/α diploids also support a role for Sxi1α activity after cell fusion. Even though cell fusion has already occurred in a/α cells and is therefore not required for diploid self-filamentation, diploid a/α sxi1αΔ strains still fail to filament properly. As shown in Figure 6B, the a/α sxi1α deletion strain (Fig. 6B, top, right) is severely compromised for filament formation compared with a wild-type a/α strain (Fig. 6B, top left). In addition, the few filaments that do form are enlarged, arrested in development, and defective in basidia and spore production. Thus, Sxi1α is important for diploid cells to progress through the sexual cycle. This is very different from the a/Δ strain, in which filament formation is intact (although somewhat diminished), and the filaments that form produce normal basidia and spores (Fig. 6B, bottom, left).
To test the hypothesis that SXI1α is the “missing regulator” required for establishing α cell identity, SXI1α was deleted from the a/Δ strain to create a MATa/matαΔ sxi1αΔ strain in which filament and spore formation were tested (Fig. 6B, bottom, right). In this strain, all filament formation is eliminated, indicating that this combination of mutations abolishes the sexual cycle. More importantly, this strain now has a changed cell identity. By deleting both SXI1α and the ∼50-kb MATα region, the a/α diploid cell identity has been changed to that of an a cell. In filamentation assays, the diploid cells with either the matα deletion (a/Δ) or the sxi1α deletion (a/α sxi1αΔ) are partially self-filamentous, and neither responds to a or α mating partners. However, the strain containing both deletions (a/Δ sxi1αΔ) mates as an a cell. As shown in Figure 6A (right panels), the double deletion strain forms mating filaments in the presence of an α mating partner but does not respond to an a partner. Deletion of both the known MATα region and the SXI1α gene resulted in the phenotype previously anticipated for a MATa/matα deletion strain, suggesting that SXI1α is either part of a second, previously unidentified MAT locus in C. neoformans or that the known MATα allele is much larger than previously suspected and, in fact, contains SXI1α. Recent studies from our laboratory presented elsewhere prove the latter hypothesis to be correct, and SXI1α is located ∼55 kb away from the originally identified MATα region in a single, contiguous MATα allele with 20 other genes of diverse function (Lengeler et al. 2002). No other DNA-binding proteins resembling those of other fungal MAT loci are present in MATα, but it is clear that SXI1α and a gene (or genes) located in the ∼50-kb deletion are necessary to maintain α cell identity. Deletion of either one alone results in diminished α behavior but does not abolish α identity. Deletion of both SXI1α and a gene(s) in the MATα region results in a complete loss of α identity in a/α cells, consistent with having removed all of the genes responsible for designating the α cell fate.
Ectopic expression of SXI1α in a cells results in a/α cell development
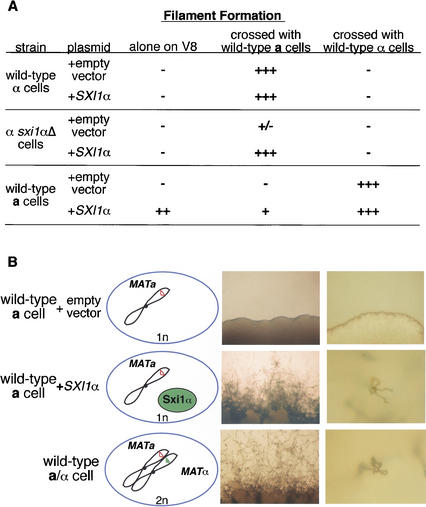
Having established that Sxi1α was a necessary contributor to α identity, we then tested whether Sxi1α is sufficient to confer α identity by expressing Sxi1α in haploid a cells. Under the control of its own promoter, SXI1α was not expressed in a cells (data not shown); therefore, SXI1α was placed under the control of a strong, constitutive promoter (glyceraldehyde-3-phosphate dehydrogenase), and the expression plasmid (GPD1–SXI1α) was transformed into wild-type α cells, sxi1α deletion cells, and wild-type a cells. Transformants were then tested for the ability to form filaments alone and in test crosses. As summarized in Figure 7A, sxi1α deletion cells transformed with GPD1–SXI1α behaved like wild-type α cells. That is, they were not self-filamentous under mating conditions, they mated and sporulated efficiently with a cells, and they were not induced to develop in the presence of other α cells. The expression of SXI1α in wild-type α cells did not have a noticeable effect on these cells. However, the expression of GPD–SXI1α in a cells resulted in a striking phenotype: a + SXI1α cells are self-filamentous (Fig. 7B, middle panel). These cells behave like a/α diploid cells; on mating medium, they form filaments, basidia, and viable spores that are indistinguishable from diploid filamentation. Thus, Sxi1α is sufficient to confer α identity and change an a cell into an a/α cell.
Figure 7.
Sxi1α induces sexual development in a cells. (A) The table summarizes the results of SXI1α overexpression in wild-type α cells, sxi1α deletion cells, and wild-type a cells alone and during mating with a and α cells. (B) Panels show the results of a filamentation assay in which strains were tested for the ability to produce filaments in the absence of a mating partner. Each row contains from left to right a schematic representation of the genotype of the test strain, the strain under low magnification, and individual cells under higher magnification. The top row shows wild-type a cells; the middle row shows wild-type a cells overexpressing SXI1α; and the bottom row shows wild-type a/α diploid filamentation.
SXI1α represses α pheromone transcription during mating and diploid filamentation
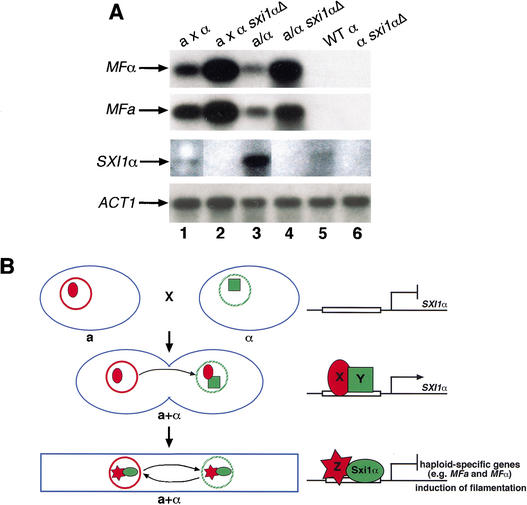
SXI1α is an important regulator of the C. neoformans sexual cycle, and we were interested in identifying targets of SXI1α. In many other fungi, the regulatory complexes that control cell identity repress the expression of haploid-specific genes such as pheromone, making this a logical target for repression by Sxi1α. To evaluate regulation of pheromone transcription by SXI1α, Northern blot analysis was carried out on wild-type and sxi1αΔ strains under filamentation conditions. Figure 8A shows the levels of MFα pheromone in the different test strains. Pheromone transcript levels are very low in haploid wild-type and sxi1αΔ strains on filamentation medium in the absence of a mating partner (Fig. 8A, first row, lanes 5,6). However, when mixed with a wild-type a mating partner, pheromone transcript levels in both wild-type and sxi1αΔ strains are greatly induced (Fig. 8A, first row, lanes 1,2), and the induction in the sxi1αΔ strain is much higher than in wild-type α cells. This increased expression of MFα is also observed during diploid filamentation. Wild-type a/α strains produce substantially more pheromone than haploid cells on filament-inducing medium, but the induction in an a/sxi1αΔ strain is even higher (Fig. 8A, first row, lanes 3,4). This expression pattern is mirrored in the pheromone transcripts from a cells as well (MFa; Fig. 8A, second row). Our data show increased pheromone transcript levels and are consistent with a model in which Sxi1α represses pheromone expression after cell fusion. Perhaps high levels of pheromone after cell fusion inhibit proper filament formation, and the role of Sxi1α is to repress pheromone levels after cell fusion to allow normal filament formation.
Figure 8.
Sxi1α represses pheromone gene expression. (A) A Northern blot probed with the MFα open reading frame (first row) shows the levels of MFα transcripts in wild-type and sxi1α deletion strains under different conditions. The second, third, and fourth rows show the same blot probed with portions of the MFa, SXI1α, and ACT1 open reading frames, respectively. (Lane 1) Wild-type a strain cocultured with wild-type α strain under mating conditions; (lane 2) wild-type a strain cocultured with α sxi1αΔ mutant under mating conditions; (lane 3) wild-type a/α diploid under filamentation conditions; (lane 4) a/α sxi1αΔ diploid under filamentation conditions; (lane 5) wild-type α cells alone under mating conditions; (lane 6) α sxi1αΔ cells alone under mating conditions. (B) Model for the role of Sxi1α in controlling sexual development. The blue objects represent cells. The solid, red circle represents the a nucleus, and the hatched, green circle represents the α nucleus. The red oval X and the red star Z represent unknown a-specific factors. The green square Y represents an unknown α-specific factor. The green oval represents Sxi1α. (Top) SXI1α is not expressed in haploid α cells. (Middle) Following cell fusion, SXI1α expression is dependent on factors from both a and α cells (X and Y). (Bottom) After induction, Sxi1α is depicted forming a heterodimeric complex with an unknown a-specific factor Z that establishes the dikaryotic state and induces sexual development through the repression of haploid-specific genes.
The expression pattern of SXI1α is consistent with this idea as well. As shown in Figure 8A (third row, lanes 5,6), the SXI1α transcript is not detectable in haploid cells in the absence of a mating partner or in the sxi1α deletion strain under the same conditions. However, in mating mixes of a and α cells, the transcript is induced in wild-type strains but is absent from sxi1α deletion strains (Fig. 8A, third row, lanes 1,2), and in a/α cells this induction in wild-type cells is even greater (Fig. 8A, third row, lanes 3,4). This suggests that a factor from a cells induces the expression of SXI1α following the fusion of a and α cells, and Sxi1α can then directly or indirectly regulate pheromone genes and other as-yet-unidentified target genes.
Discussion
We have described here the discovery of a novel region of the C. neoformans MAT locus that encodes the cell-type regulator Sxi1α. A deletion of the previously identified ∼50-kb region of the MATα allele in C. neoformans had an unexpected phenotype because it did not perturb cell fate. That is, a/α cell identity was not lost with the deletion of MATα. This result prompted us to investigate the mechanisms by which sexual differentiation is controlled. Using homozygous a/a and α/α diploid strains in filamentation assays, we ruled out the hypothesis that ploidy is the sole regulator of diploid filamentation and showed that both a and α information are necessary for sexual differentiation. This discovery led us to consider that cell-type-specific components related to those in other fungi could be responsible for directing cell identity and sexual development. We identified the homeodomain-encoding gene SXI1α and discovered that it is both necessary and sufficient for specifying α identity and for promoting proper development during mating and diploid filamentation. Sxi1α is a formal repressor of the a and α pheromone genes, and levels of SXI1α transcript are increased after fusion with a mating partner. These data support a model in which Sxi1α functions as a transcriptional regulator responsible for signaling cell identity and allowing progression through the sexual cycle.
Sxi1α acts after cell fusion
The experiments presented here show how a single mating-type-specific protein can profoundly influence the process of sexual development. Our data provide insight into when and how Sxi1α exerts its influence and lead us to propose that Sxi1α is important for signaling the dikaryotic state (Fig. 8B). This unique time in development is one in which each cell in a filament contains two nuclei (one from a and one from α) whose replication and transport into the next cell must be carefully orchestrated to maintain the process of development and allow spore formation. In this model, Sxi1α could signal dikaryon formation after cell fusion by repressing haploid-specific genes that are no longer necessary for mating.
The idea that the primary role for Sxi1α is after mating partners fuse is supported by several observations. First, the frequency of dikaryon formation during mating is not altered in an sxi1α deletion strain, indicating that Sxi1α is not required for signaling to a mating partner or promoting the process of cell fusion. Second, diploid strains do not filament properly in the absence of Sxi1α. Stable diploid strains have already undergone cell and nuclear fusion, and thus, the filamentation defect seen in an a/α sxi1α deletion strain must be attributable to a block after fusion. These data could be interpreted to suggest that Sxi1α is simply an inducer of filamentation; however, this is not the case. sxi1α deletion strains do not have a defect in haploid fruiting. Under low-nitrogen, desiccating conditions, haploid α sxi1α strains form filaments, basidia, and spores like wild-type α cells. In addition, overexpression of SXI1α in α cells does not result in diploid filament formation as it does in a cells, indicating that the role of Sxi1α is to mediate sexual development and not simply to induce filament formation.
An a-specific factor is required for sexual development
How Sxi1α induces sexual development is not yet clear. The process requires Sxi1α and at least one a-specific component, and this is reminiscent of a classic homeodomain heterodimeric complex. A well-studied example of this form of regulation is that of the a1–α2 heterodimer in S. cerevisiae. In this ascomycete, the a1 protein from a cells associates with the α2 protein from α cells to form a novel activity in diploid cells that represses haploid-specific genes and allows diploid cells to undergo meiosis and sporulation (Johnson 1995). This pattern of regulation is also seen in basidiomycetes like Ustilago maydis, where the bE1 and bW1 proteins function as a heterodimer in the diploid state to repress genes and promote sexual development (Kamper et al. 1995; Brachmann et al. 2001). Although we favor a model in which Sxi1α forms a heterodimeric regulatory complex with an a-specific factor, the sequence of the complete MATa allele (Lengeler et al. 2002) has not revealed any strong candidates for the homeodomain, HMG box, or α-domain proteins that are often present in the MAT loci of other fungi (Coppin et al. 1997). In fact, only a single a-specific gene, NCP1a, has been identified in the MATa sequence, and its predicted product has no similarity to any known sequence or structural motifs. Deletion of NCP1a does not lead to a detectable mating phenotype, excluding this gene as encoding the Sxi1α partner (C.M. Hull, K. Forrester, and J. Heitman, unpubl.). In the absence of a clear Sxi1α binding partner but a clear dependence of sexual differentiation on an a-specific factor, an alternate model is one in which one of the a alleles in MATa has a disparate function from its partner allele in MATα. The product of the a-specific allele could interact with or modify Sxi1α to direct its function in the dikaryon.
SXI1α is induced during mating
The expression pattern of SXI1α also distinguishes this cell fate determinant from its homeodomain counterparts in other fungi. Unlike many other homeodomain regulators of cell identity, SXI1α is not expressed at detectable levels in haploid cells. Transcript levels are induced in a mating between a and α cells and are significantly induced in a/α diploid cells. This pattern is consistent with a model in which activation of SXI1α is dependent on an a-specific factor that acts on the SXI1α gene after cell fusion (Fig. 8B, factor X). Expression of SXI1α is even higher in the diploid state because there is constitutive induction of its transcription in the diploid nucleus. The expression of SXI1α is also dependent on an additional α-specific factor (Fig. 8B, factor Y). Because SXI1α is not expressed in a cells under the control of its own promoter, and because Sxi1α activity is preserved in the a/Δ strain, we conclude that another component from MATα (that resides outside the deleted ∼50-kb region) is necessary to activate SXI1α.
C. neoformans has an unusual MAT locus
Although clearly classed with the basidiomycete phylum of fungi, C. neoformans is very different from its basidiomycete relatives. It is the only human pathogen in the basidiomycete phylum (although there are many plant pathogens), and it does not have the classic MAT locus architecture that has been defined for model basidiomycetes. Well-studied basidiomycetes like U. maydis, Coprinus cinereus, and Schizophyllum commune all have two MAT loci in which one locus encodes pheromones and pheromone receptors, and the other encodes homeodomain DNA-binding proteins (Casselton and Olesnicky 1998). This structure has not been conserved in C. neoformans. The work presented here predicts the existence of a second, linked, MATα region containing SXI1α or the inclusion of SXI1α in a locus larger than the ∼50-kb region identified previously. Recent evidence to be presented elsewhere demonstrates that the MAT locus of C. neoformans is in fact much larger than previously described (Lengeler et al. 2002). Large-scale BAC mapping and sequence analyses have shown that the MAT locus is ∼105 kb in size, contains genes never seen before in MAT loci, and encodes pheromones, pheromone receptors, and now, the homeodomain regulatory protein, Sxi1α.
Although the C. neoformans MAT locus architecture differs from that of its basidiomycete relatives, the identification of a homeodomain regulator of its sexual cycle is entirely consistent with established paradigms by which fungal sexual development is governed. Nearly all fungi in which a sexual cycle has been identified (and some where one has not) contain a presumptive MAT locus with regulatory proteins. In sexual fungi, homeodomain, α-domain, and HMG-box proteins control mating, cell and nuclear fusion, meiosis, and spore formation (Coppin et al. 1997; Turgeon 1998). Thus, the absence of clear MAT DNA-binding proteins in C. neoformans had been somewhat mysterious. The classic DNA-binding proteins were not present in the known MATα region, and identifying such proteins in the C. neoformans genome sequence had not been successful previously. No similarities between the C. neoformans genome and other fungal MAT DNA-binding proteins resulted in significant scores by BLAST search analysis.
Our approach to identify SXI1α and other putative homeodomain proteins was based on direct comparisons of sequences with weak BLAST scores with known homeodomain structural motifs. This approach enabled us to identify candidates that contain residues conserved in the DNA-binding helices of many homeodomain proteins and map well to the S. cerevisiae a1–α2 crystal structure (Li et al. 1995). Given the weak identity between fungal homeodomains and other fungal DNA-binding regulators (fungal α-domain proteins share quite low similarity at the amino acid level), this weak primary sequence similarity was not surprising. However, a broader comparison of the predicted Sxi1α homeodomain region with other homeodomain proteins from plants and animals resulted in a clear relationship. In fact, the Sxi1α homeodomain is more similar to the nonfungal homeodomain protein Gsp1 from the green algae Chlamydomonas reinhardtii and the mouse homeodomain protein Meis2 than it is to those of its basidiomycete relatives C. cinereus and U. maydis.
Although many features of Sxi1α are unique, it still belongs to a class of gene-specific regulators of cell identity that includes S. cerevisiae MATa1, C. reinhardtii GSP1, and the M. musculus Sry. One hallmark characteristic of all of these regulators is that they alter (or establish) the identity of a cell. Expression of MATa1 in haploid α cells results in the repression of haploid-specific genes and creates a meiosis-competent state; expression of GSP1 in gamete cells activates the zygote developmental program; expression of Sry converts XX animals to the XY fate, and the expression of SXI1α in haploid a cells activates a diploid sexual development program. In each case the fate of the cell is dependent on the presence of a single, dominant factor that drastically alters the expression patterns of cell-type-specific genes. Discovering additional targets of Sxi1α will reveal the extent to which transcription profiles must be altered to achieve the dikaryotic state. With the discovery of Sxi1α, we have answered the question as to whether such cell-type-specific regulators exist in C. neoformans, and this advance allows us to now move on to elucidate the mechanism by which Sxi1α and regulators like it establish cell fate and control sexual differentiation. The discovery of the central role that Sxi1α plays in establishing cell fate also provides a molecular foundation from which to explore the known link between the α allele of the MAT locus and virulence.
Materials and methods
Constructing matα and sxi1α deletion strains
To create the matα deletion strain, a PCR overlap approach (Ho et al. 1989; Horton et al. 1989; Davidson et al. 2002) was used to make a disruption fragment. The left end of the locus was amplified with primers JOHE3186 (AGGTACGGAATCATTTCT CAT) and JOHE4439 (CCACCTCCTGGAGGCAAGATGCC TAACGACAGCCAG). The right end of the locus was amplified with primers JOHE5076 (GGATCCACTAGTTCTAGAAAC GCC) and JOHE5700 (GGTCGAGCAACTTCGCTCTGCTGT TGTGAGCGGCGT). The URA5 selectable marker was amplified with primers JOHE4440 (CTGGCTGTCGTTAGGCATCT TGCCTCCAGGAGGTGG) and JOHE5699 (ACGCCGCTCA CAACAGCAGAGCGAAGTTGCTCGACC). The amplified products were gel purified and extracted together. Primers JOHE3186 and JOHE5076 were then used in a PCR to overlap the three templates into a 4.8-kb matα∷URA5 deletion construct containing the left and right portions of the known MATα region flanking the URA5 gene. The linear disruption allele was then introduced into the serotype D ura5/ura5 diploid strain RAS008 by biolistic transformation as described by Toffaletti et al. (1993). Eighty transformants were screened by PCR for the absence of signal with STE12α-specific primers. Southern analysis was performed using probes for several genes in the locus (left end, STE11α, ZNF1α, PRT1α) to confirm the deletion in one transformant (a/matαΔ, RDC42-5).
Similarly, two different PCR-generated disruption constructs were created to delete most of the SXI1α open reading frame. Portions of the upstream and downstream regions of SXI1α flanking URA5 were created using the following primers: left flank, JOHE7032 (GTTGTTGCTTAAATCGATGC) and JOHE7034 (GGTCGAGCAACTTCGCTCCTAGCAAAAGTG ACTCTATTC); right flank, JOHE7035 (CCACCTCCTGGAG GCAAGCGTGTTAATACAGATAAACC) and JOHE7037 (CC ATTGGAGGAAGCTGTGGGCTG); URA5 marker, JOHE7033 (GAATAGAGTCACTTTTGCTAGGAGCGAAGTTGCTCGA CC) and JOHE7036 (GGTTTATCTGTATTAACACGCTT GCCTCCAGGAGGTGG). Portions of the upstream and downstream regions of SXI1α flanking the dominant selectable marker NAT1 were created using the following primers: left flank, JOHE7032 (GTTGTTGCTTAAATCGATGC) and JOHE7134 (CAGCTCACATCCTCGCAGCCTAGCAAAAG TGACTCTATTC); right flank, JOHE7070 (CATCTCTTCTA TAAGCTTCGTGTTAATACAGATAAACC) and JOHE7037 (CCATTGGAGGAAGCTGTGGGCTG); NAT1 marker, JOHE7068 (GAATAGAGTCACTTTTGCTAGGCTGCGAGG ATGTGAGCTG) and JOHE7071 (GGTTTATCTGTATTAA CACGAAGCTTATAGAAGAGATG). The sxi1α∷URA5 PCR-generated overlap fragment was transformed using biolistic transformation into the ura5 haploid strain JEC34 and the ura5/ura5 diploid strain RAS009. Recovered transformants were screened by PCR and Southern blots and resulted in the haploid and diploid deletion strains CHY610 and CHY615. The sxi1α∷NAT1 PCR fragment was transformed into the previously constructed and characterized matα deletion strain RDC42-5 to create the matαΔ sxi1αΔ diploid strain CHY625. Transformations were carried out by biolistics on YPD plates containing 1 M sorbitol. Transformants were allowed to recover at 30°C for 4–6 h, scraped off the transformation plates, and plated on YPD plates containing 100 μg/mL nourseothricin.
PCR amplification
All PCR amplifications were performed using the ExTaq PCR system (Intergen). Primers were used at a concentration of 0.5 μM, and templates for PCR reactions were titrated and evaluated empirically for each product. PCR overlap conditions used were as described previously (Davidson et al. 2002). For diagnostic PCR to distinguish a/a and α/α cells, primers to the STE20a and STE20α genes were used as in Sia et al. (2000) on genomic DNA templates. PCR to confirm the matα deletion strain was carried out with the following primer sets, and a subset of these primers was also used to generate probes used in Southern and Northern analysis: control primers to the CPK1 gene, JOHE1703 (AAGGATCCATATGACAATCGACCAAA GCCAAATC) and JOHE1704 (AACTGCAGGCACCTCATCG TAAATCATTCCT); primers A, JOHE3186 (AGGTACGGAAT CATTTCTCAT); B, JOHE5548 (GCAGGAAACTCCCCT TCT); C, JOHE6332 (TGGATCATGACGATCGGACAC); D, JOHE5910 (CAACTATGTGGCAACAACATGG); STE12α, JOHE4545 (ACAACCTGGCTTAGAAGA) and JOHE4546 (CC TCTCGAAACCATTTCT); STE20α, JOHE3069 (GATTTATC TCAGCAGCCACG) and JOHE3070 (AAATCGGCTACGG GACGTC); STE11α, JOHE5306 (GGTCGAGCAACTTCGCT CATTTACAGGGCTGTCCTG) and JOHE5391 (GCTCGT TCTCCCCTGTAC); MFα, JOHE1204 (TTTTACGCTTTTTG CAGATTCCGCCAAA) and JOHE3242 (GACCACTGTTT CTTTCGTTCT); MFa, JOHE6683 (TTCTTCGGCAGCCTCA CTAT) and JOHE6684 (GAAAAGAGGTACGAGTAGAT); ACT1, JOHE6307 (CTGTCTTCCCTTCTATTGTTGGTCG) and JOHE6308 (CACTGTACTTTCGCTCGGGAGG). Primers to the SXI1α gene used to show α-specific segregation and to generate a probe for Southern and chromoblot hybridizations were JOHE6510 (CGTCCGTCTCTGGTTTCTTG) and JOHE6511 (GGCGATAATTAGTCAGATCA).
Strain manipulations and media
All strains used were of the serotype D background and are described in the relevant procedures. All were handled using standard techniques and media as described by Alspaugh et al. (1997) and Guthrie and Fink (1991). Mating and self-filamentation assays were conducted on V8 medium at room temperature in the dark for 2–5 d. Filamentation was evaluated by observing the periphery of test spots on V8 medium. The mating tester strains used were JEC20 (a) and JEC21 (α). Diploid strains were all prototrophic, with the exception of the a/0 strain (RDC42-5). This strain was created by selecting against the URA5 gene on 5-FOA, which led to either the loss of the α chromosome creating a 2n − 1 strain, or a mitotic cross-over event, resulting in a functional a/a strain. The a/0 strain was tested for filamentation on V8 medium supplemented with uracil.
Assisted mating reactions
Matings to create a/a and α/α homozygous diploid strains were carried out as follows: For a/a strains, JEC171 (a ade2 lys2) and JEC53 (a ura5 lys1) were grown on YPD medium and resuspended in water in approximately equal amounts with (assisted mating) and without (unassisted mating) JEC157 (α ura5 ade2 lys1). For α/α strains, JEC170 (α ade2 lys2) and JEC52 (α ura5 lys1) were grown on YPD medium and resuspended in water in approximately equal amounts with and without JEC169 (a ade2 ura5 lys1). Mixtures were spotted onto V8 medium and incubated in the dark at room temperature for 4–7 d. Mating mixes were then scraped off the V8 medium; resuspended in water; plated to selective plates lacking uracil, adenine, and lysine; and grown at 30°C for 3 d. No prototrophs were recovered from the unassisted mating mixes. Prototrophs from the assisted mating were tested for DNA content by FACS. FACS analysis was carried out according to Sia et al. (2000). The control α haploid strain was JEC21. The control a/α diploid strain was RAS009. Test strains were a/a CHY600 and α/α CHY601. Positive diploid strains by FACS were tested for a and α information by PCR (see PCR section).
Ectopic expression experiments
The predicted SXI1α ORF was generated via PCR using primers JOHE8206 (CGAGGATCCATGCTTTCGACCTGTGAT) and JOHE8523 (CGGGATCCCTTTAGTTTAAGTCGGGGATCA) and a cloned genomic fragment as template. The resulting PCR product was cloned and sequenced to verify a wild-type clone (pCH254). The SXI1α ORF was liberated from pCH254 with BamHI, and cloned into the BamHI site of the telomeric, GPD1-containing plasmid pRCD85 to create pCH258. pCH258 was digested with the meganuclease I-SceI to expose its telomeric ends and then transformed by electroporation into JEC34 (a ura5), JEC43 (α ura5), and CHY618 (α sxi1α∷NAT1 ura5) to create the strains CHY656, CHY657, and CHY658, respectively. pCH258 was also transformed into a 5-FOA-resistant derivative of CHY625 (a/Δ sxi1αΔ) and found to restore filamentation behavior to that of the a/Δ strain RDC42-5. SXI11α overexpression strains were evaluated for self-filamentation and mating on V8 plates at room temperature for 2–5 d in the dark.
Southern blot analysis
Genomic DNA was isolated from strains using the method of Pitkin et al. (1996). Twenty micrograms of genomic DNA was digested with the indicated enzymes and electrophoresed on 0.8% TBE agarose gels. Transfer, hybridization, and autoradiography were performed as previously described (Ausubel et al. 1992) and by using the Alk-Phos Direct Detection System (Amersham Pharmacia Biotech). PCR-generated fragments were used as probes for Southern blot hybridizations: left end, JOHE3186 (AGGTACGGAATCATTTCTCAT) and JOHE4439 (CCACCTCCTGGAGGCAAGATGCCTAACGA CAGCCAG); PRT1α, JOHE6526 (AAATCTTCCATCGGTC TCAG) and JOHE6527 (ACAATTCCTCAGTCTCATCG); ZNF1α, JOHE6149 (CTTTTGCCCTCGTACTCCGCACC) and JOHE6150 (CCGGATTGCCATGCACAGGACGC); see PCR section for remaining primer sequences. Chromoblot was a gift from Klaus Lengeler (Duke University, Durham, NC) and was hybridized in parallel under the same conditions as the Southern blots.
Northern blot analysis
RNA was prepared from C. neoformans cells using a hot phenol extraction protocol for S. cerevisiae (Ausubel et al. 1992). Northern blots were carried out according to standard protocols (Ausubel et al. 1992) with 10 μg of total RNA used for each sample. The MFα, MFa, SXI1α, and ACT1 probes were generated by PCR as described in the PCR section, and radiolabeled probes (Rediprime II kit from Amersham Pharmacia Biotech) were used in hybridization reactions as described previously (Church and Gilbert 1984) at 65°C.
Sequence manipulations
Sequence comparisons were conducted using the BLAST algorithm (Altschul et al. 1997) against the Stanford C. neoformans genome sequence [version 5, C. neoformans Genome Project, Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/C.neoformans/index.html), funded by the NIAID/NIH under cooperative agreement U01 AI47087, and The Institute for Genomic Research, funded by the NIAID/NIH under cooperative agreement U01 AI48594]. Sequence analyses were conducted and phylogenetic trees and alignments were generated using SeqWeb Version 2 (Accelrys). Sequence alignment was displayed using SeqVu Version 1.1 (The Garvan Institute of Medical Research). The SXI1α sequence can be obtained from GenBank using accession no. AY162324.
Acknowledgments
The authors thank R. Brazas, R. Wharton, D. Lew, B. Capel, and J. McCusker for helpful comments and discussions; M.-J. Boily, K. Forrester, and C. Arndt for technical assistance; and other members of the Heitman Laboratory, especially K. Lengeler, for their support. This work was supported by an NIAID RO1 grant AI50113 to J.H. and an NIAID program project grant AI44975 to the Duke University Mycology Research Unit. C.M.H. is supported by a Damon Runyon Cancer Research Fund Fellowship, DRG-1694. J.H. is a Burroughs-Wellcome Scholar in Molecular Pathogenic Mycology and an Associate Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL heitm001@duke.edu; FAX (919) 684-5458.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1041402.
References
- Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes & Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspaugh JA, Davidson RC, Heitman J. Morphogenesis of Cryptococcus neoformans. Contrib Microbiol. 2000;5:217–238. doi: 10.1159/000060352. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York: Greene Publishing Associates and Wiley-Interscience; 1992. [Google Scholar]
- Banuett F, Herskowitz I. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci. 1989;86:5878–5882. doi: 10.1073/pnas.86.15.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A, Weinzierl G, Kamper J, Kahmann R. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol Microbiol. 2001;42:1047–1063. doi: 10.1046/j.1365-2958.2001.02699.x. [DOI] [PubMed] [Google Scholar]
- Burglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B. Sex in the 90s: SRY and the switch to the male pathway. Annu Rev Physiol. 1998;60:497–523. doi: 10.1146/annurev.physiol.60.1.497. [DOI] [PubMed] [Google Scholar]
- Casadevall C, Perfect JR. Cryptococcus neoformans. 1st ed. Washington, DC.: ASM Press; 1998. [Google Scholar]
- Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Wickes BL, Miller GF, Penoyer LA, Kwon-Chung KJ. Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J Exp Med. 2000;191:871–882. doi: 10.1084/jem.191.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DL, Woodlee GL, McClelland CM, Seymour TS, Wickes BL. The Cryptococcus neoformans STE11α gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Mol Microbiol. 2001;40:200–213. doi: 10.1046/j.1365-2958.2001.02375.x. [DOI] [PubMed] [Google Scholar]
- Coppin E, Debuchy R, Arnaise S, Picard M. Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev. 1997;61:411–428. doi: 10.1128/mmbr.61.4.411-428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Moore TD, Odom AR, Heitman J. Characterization of the MFα pheromone of the human fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2000;38:1017–1026. doi: 10.1046/j.1365-2958.2000.02213.x. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Blankenship JR, Kraus PR, de Jesus-Berrios M, Hull CM, D'Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. San Diego: Academic Press; 1991. [Google Scholar]
- Haqq CM, Donahoe PK. Regulation of sexual dimorphism in mammals. Physiol Rev. 1998;78:1–33. doi: 10.1152/physrev.1998.78.1.1. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980;85:811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I, Rine J, Strathern J. Mating-type determination and mating-type interconversion in Saccharomyces cerevisiae. In: Jones EW, et al., editors. Molecular and cellular biology of the yeast Saccharomyces: Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 583–656. [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- Johnson AD. Molecular mechanisms of cell-type determination in budding yeast. Curr Opin Genet Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- Kamper J, Reichmann M, Romeis T, Bolker M, Kahmann R. Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell. 1995;81:73–83. doi: 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- Karos M, Chang YC, McClelland CM, Clarke DL, Fu J, Wickes BL, Kwon-Chung KJ. Mapping of the Cryptococcus neoformans MATα locus: Presence of mating type-specific mitogen-activated protein kinase cascade homologs. J Bacteriol. 2000;182:6222–6227. doi: 10.1128/jb.182.21.6222-6227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Staben C. Mating type in filamentous fungi. Annu Rev Genet. 1997;31:245–276. doi: 10.1146/annurev.genet.31.1.245. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Davidson RC, D'Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, Heitman J. Mating type locus of Cryptococcus neoformans: A step in evolution of sex chromosomes. Eukaryot Cell. 2002;1:704–718. doi: 10.1128/EC.1.5.704-718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Stark MR, Johnson AD, Wolberger C. Crystal structure of the MATa1/MATα2 homeodomain heterodimer bound to DNA. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- Moore TD, Edman JC. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol. 1993;13:1962–1970. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst SM, Meneely PM. Sex determination and dosage compensation: Lessons from flies and worms. Science. 1994;264:924–932. doi: 10.1126/science.8178152. [DOI] [PubMed] [Google Scholar]
- Pitkin JW, Panaccione DG, Walton JD. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology. 1996;142:1557–1565. doi: 10.1099/13500872-142-6-1557. [DOI] [PubMed] [Google Scholar]
- Sia RA, Lengeler KB, Heitman J. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol. 2000;29:153–163. doi: 10.1006/fgbi.2000.1192. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon BG. Application of mating type gene technology to problems in fungal biology. Annu Rev Phytopathol. 1998;36:115–137. doi: 10.1146/annurev.phyto.36.1.115. [DOI] [PubMed] [Google Scholar]
- Wickes BL, Mayorga ME, Edman U, Edman JC. Dimorphism and haploid fruiting in Cryptococcus neoformans: Association with the α-mating type. Proc Natl Acad Sci. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue C, Cavallo LM, Alspaugh JA, Wang P, Cox GM, Perfect JR, Heitman J. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Lu M, Singh R, Snell WJ. Ectopic expression of a Chlamydomonas mt+-specific homeodomain protein in mt− gametes initiates zygote development without gamete fusion. Genes & Dev. 2001;15:2767–2777. doi: 10.1101/gad.919501. [DOI] [PMC free article] [PubMed] [Google Scholar]