Abstract
p27Kip1 restrains cell proliferation by binding to and inhibiting cyclin-dependent kinases. To investigate the mechanisms of p27 translational regulation, we isolated a complete p27 cDNA and identified an internal ribosomal entry site (IRES) located in its 5′UTR. The IRES allows for efficient p27 translation under conditions where cap-dependent translation is reduced. Searching for possible regulators of IRES activity we have identified the neuronal ELAV protein HuD as a specific binding factor of the p27 5′UTR. Increased expression of HuD or the ubiquitously expressed HuR protein specifically inhibits p27 translation and p27 IRES activity. Consistent with an inhibitory role of Hu proteins in p27 translation, siRNA mediated knockdown of HuR induced endogenous p27 protein levels as well as IRES-mediated reporter translation and leads to cell cycle arrest in G1.
Keywords: Cell division cycle, translation, p27 Kip1, IRES, HuD, HuR, ELAV
Cell cycle phase transitions are driven by a periodic activation and inactivation of cyclin-dependent kinases (Cdks; Morgan 1997; Ekholm and Reed 2000). Manifold antiproliferative signals act during G1 progression and interfere with Cdk activation. The Cdk inhibitory protein p27 plays a key role in regulating Cdk activity during G1 progression and in growth arrest (Hengst and Reed 1998; Sherr and Roberts 1999). Level of the inhibitor protein oscillate during the cell cycle and peak in G1 phase. p27 accumulates to high levels in quiescent cells (Hengst and Reed 1998; Sherr and Roberts 1999). Under most conditions investigated, p27 oscillations are the result of posttranscriptional mechanisms that include translational control and regulated protein stability (Pagano et al. 1995; Agrawal et al. 1996; Hengst and Reed 1996). In nonproliferating cells, enhanced polyribosomal association of the p27 mRNA was demonstrated by Millard and coworkers (1997). A region containing an U-rich element in the p27 5′UTR was identified that is necessary for efficient p27 translation in proliferating and nonproliferating cells (Millard et al. 2000). Several proteins were found to bind to this element including hnRNPs and HuR, but a direct role or mechanism for these proteins in regulating p27 expression could not be established. Hu proteins belong to a conserved family of RNA-binding factors that were originally identified in Drosophila as embryonic lethal abnormal vision (ELAV) proteins (Campos et al. 1985), and are essential for neuronal development. Four human homologs have been identified, and were implicated in regulating mRNA stability and mRNA export (Brennan and Steitz 2001). One mammalian ELAV protein, Hel-N1, was found to increase translation of neurofilament M mRNA (Antic et al. 1999), suggesting an alternative function. Hu proteins bind to AU-rich elements, normally located in the 3′UTRs of short-lived mRNAs. Among these mRNAs are several encoding cell cycle regulatory proteins like cyclins A and B (Wang et al. 2000a) and the CDK inhibitor p21 (Liu et al. 2000; Wang et al. 2000b). Binding of HuR to these elements is believed to stabilize the mRNA, therefore leading to increased protein levels. In addition to this established function we now report that the Hu/ELAV proteins HuD and HuR can decrease protein expression by inhibiting translation: expression of the CDK inhibitor p27 is repressed by HuR or HuD by reducing its internal ribosomal entry site (IRES)-dependent translation.
Results
This study aimed to identify elements that regulate p27 translation. Because translational regulation frequently involves 5′ untranslated regions (5′UTRs), we first isolated an extended 5′ noncoding sequence of the p27 transcript. Fragments of diverse length for this region have been described previously, varying in size between 152 and 479 nucleotides upstream of the initiation codon (Minami et al. 1997; Ito et al. 1999; Kamiyama et al. 1999; Millard et al. 2000). Using HeLa total RNA as a template for anchored PCR, we cloned a 5′UTR of 575 nucleotides. No products were obtained in the absence of reverse transcriptase. Longer products could not be detected with RNA isolated either from HeLa cells or from human fibroblasts (HS68; data not shown), indicating that the 575-nucleotide fragment represents the longest p27 transcript. We also isolated smaller products, most of them starting between nucleotides −469 to −473 upstream of the p27 ATG. To investigate the complexity of p27 transcripts and a possible cell cycle regulation of transcript length, we performed RNase protection assays using RNA isolated from proliferating or synchronized HeLa cells. Using a probe of 572 nucleotides complementary to the largest 5′UTR, we obtained a major protection product of about 470 nucleotides in a cluster of products of 470–400 nucleotides. Additional protected products were detected at about 570, 380, and 340 nucleotides (Fig. 1B). Almost identical patterns were obtained with RNA isolated from proliferating cells or cells arrested in G1 by lovastatin or S phase by thymidin, suggesting that p27 translation is not regulated by variations in the length of its 5′UTR and that the majority of the p27 transcripts possess an extended 5′UTR that exceeds 400 nucleotides.
Figure 1.
The p27 5′UTR. (A) Structure of the largest p27 5′UTR. The largest human p27 5′UTR is aligned to the 5′ flanking region of the mouse p27 genomic sequence (accession nos. U77915 and U10440). The upstream ORF (uORF) is marked with a line. (B) p27 mRNA contains an extended 5′UTR that is produced independently from cell cycle position. RNAse protection of 14 μg RNA isolated from proliferating (lane 4) or arrested HeLa cells (lane 5, lovastatin; lane 6, thymidin). Total RNA or 14 μg yeast tRNA hybridized to a 681-nucleotide antisense fragment containing the p27 5′UTR from nucleotides −3 to −575. Protected RNAs were detected by autoradiography. The major protection products are indicated by a clamp; additional protection products are indicated by arrows. (Lane 1) RNA marker; fragment length in nucleotides is indicated. (Lanes 2,3) Yeast tRNA (14 μg) with (lane 3) or without (lane 2) RNase treatment. (Lanes 4–6) RNA from proliferating (lane 4), lovastatin-arrested (lane 5), or thymidine-arrested (lane 6) HeLa cells hybridized and treated with RNase.
The nucleotide sequence of the human p27 5′UTR is remarkably conserved to the genomic sequence in mice, bearing an overall sequence identity of 78%. (Fig. 1A). At the 5′ end of the larger p27 transcripts is a small upstream ORF (uORF), encoding a peptide of 29 amino acids, that is also conserved in mice (Fig. 1A).
Deletion analysis of the longest 5′ leader revealed that a domain in the 5′ end of the p27 5′UTR inhibits reporter translation. This inhibition was counterbalanced in the complete 5′UTR by the presence of downstream elements (U. Göpfert and L. Hengst, unpubl.). This observation suggested that a domain in the 5′UTR might enhance translation. Among various mechanisms that enhance translation are internal ribosome entry sites (IRESs) that allow ribosomes to bind mRNA in a cap-independent manner. These elements allow bypassing of inhibitory elements such as secondary structures or uORFs that reduce or prevent ribosomal scanning.
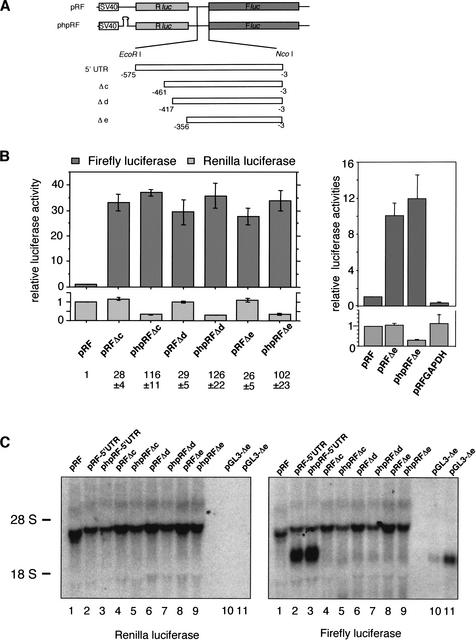
To analyze the 5′UTR for the presence of IRES elements, we employed the bicistronic reporter plasmid pRF (Stoneley et al. 1998; Coldwell et al. 2000). Three different fragments of the p27 5′UTR (starting at 356, 417, or 461 nucleotides upstream of the p27 initiation codon, respectively) representing different detectable p27 5′UTRs (cf. Fig. 1B) were inserted between the upstream Renilla and downstream firefly luciferase cistrons (Fig. 2A). Indeed, all three fragments of the p27 5′UTR dramatically increased expression of the downstream firefly luciferase, while expression of the Renilla luciferase remained unaffected (Fig. 2B, left panel). The shortest fragment used contained a 5′UTR starting 356 nucleotides upstream of the p27 start codon (Fig. 2B, left panel). We concluded that this sequence may contain an IRES element.
Figure 2.
The p27 mRNA contains an internal ribosomal entry site (IRES). (A) Bicistronic reporter plasmids. The p27 5′UTR fragments indicated (Δc from −461 to −3; Δd from −417 to −3; and Δe from −356 to −3) were inserted in the intercistronic region of pRF and phpRF. The open boxes represent the SV40 promoter; the light gray and the dark gray boxes represent the Renilla luciferase (Rluc) and the firefly luciferase (Fluc) coding sequence, respectively. phpRF contains a palindromic sequence upstream of Rluc that allows the formation of a stable hairpin in the mRNA known to reduce cap-dependent translation. (B) Relative Renilla luciferase and firefly luciferase activities. Adherent HeLa cells were transfected with bicistronic plasmids and pSV-β-Gal as a control. Both firefly (top) and Renilla (bottom) luciferase activities were normalized to β-galactosidase and expressed relative to the values obtained from pRF, which was set to 1. The ratio between the relative firefly and Renilla luciferase activities for the constructs is indicated below the diagram. (Right) A random sequence of the GAPDH mRNA fails to possess IRES activity. (C) Northern blot analysis of bicistronic mRNA expression. Total RNA (7 μg) was hybridized with a specific probe for the Renilla luciferase coding region, stripped, and rehybridized with a probe for the firefly luciferase coding region. Positions of 18S and 28S ribosomal RNAs are indicated. The generation of an additional firefly mRNA from a plasmid containing the largest 5′UTR (pRF-5′UTR and phpRF-5′UTR) may reflect promotor activity or RNA cleavage.
To ensure that the increase in firefly luciferase activity was a result of enhanced translation rather than altered mRNA stability, transcription, or the presence of monocistronic firefly luciferase mRNA, we analyzed integrity and abundance of the different transcripts by Northern blots. As shown in Figure 2C (lanes 4,6,8) this bicistronic message is expressed at comparable levels and no significant degradation products were detected. The largest p27 5′UTR (−575 to −3) could not be evaluated as its integration in the reporter gave rise to a large amount of monocistronic firefly luciferase mRNA, possible due to a (cryptic) promoter activity (Fig. 2C, lanes 2,3). To exclude alternative translational mechanisms like reinitiation, a second plasmid was used where a palindrome was inserted upstream of the Renilla luciferase gene. This palindrome allows the formation of a stable RNA hairpin that inhibits translation of the Renilla luciferase (Stoneley et al. 1998; Coldwell et al. 2000). If the synthesis of firefly luciferase were due to the reinitiation of ribosomes, then a comparable reduction in the synthesis of Renilla and firefly luciferase would be expected. However, the hairpin only reduced expression of the upstream Renilla luciferase cistron, but had no effect on the induction of firefly luciferase expression in the presence of p27 5′UTR fragments (Fig. 2B). This argues against reinitiation as a possible mechanism. Because the presence of detectable levels of truncated mRNAs could be excluded by Northern blot (Fig. 2C, lanes 5,7,9), the strong increase in translation of the downstream ORF by sequences of the p27 5′UTR implies that a potent IRES is present in this region.
Finally, a nonrelated sequence from the GAPDH cDNA (356 nucleotides) failed to enhance firefly luciferase activity, confirming lack of IRES activity in random sequences inserted into the intercistronic region of the bicistronic messenger (Fig. 2B, right panel).
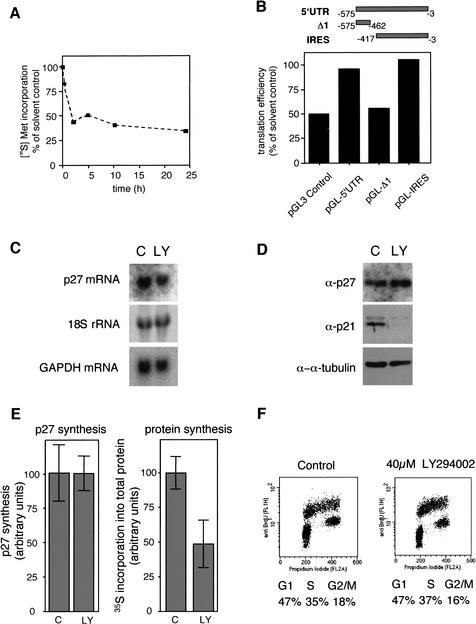
To verify IRES functions under physiologic conditions, we made use of the observation that inactivation of the PI3-kinase/mTOR pathway leads to hypophosphorylation of eIF4E-BP1 and to inhibition of cap-dependent translation (Gingras et al. 1999; Raught et al. 2000). General inhibition of protein synthesis by the PI3-kinase inhibitor LY294002 in HeLa cells was measured by pulse-labeling with (35S) methionine (Fig. 3A). Overall protein synthesis rates were reduced to ∼50%. Although reporter translation of control vectors was also reduced to ∼50%, insertion of the p27 5′UTR leads to complete resistance to LY294002 repression of translation. Analysis of shorter fragments indicated that this resistance was mediated by the IRES element of the 5′UTR (Fig. 3B). Importantly, these findings can be recapitulated for endogenous p27. Inhibition of general protein synthesis even induced a slight increase in p27 protein (Fig. 3D), while p27 mRNA levels remained unchanged (Fig. 3C). Further, synthesis rate of p27 remained identical upon addition of the drug (Fig. 3E), while overall protein synthesis was reduced to ∼50% of control treated cells (Fig. 3E, right panel). We observed no significant alteration of the stability of p27 in pulse-chase experiments (data not shown). Finally, cell cycle distribution of HeLa cells was not affected by LY294002 treatment (Fig. 3F), indicating that changes in endogenous p27 are not due to cell cycle synchronization effects. Taken together, these findings demonstrate internal initiation of translation for both, the endogenous p27 and reporter under control of the p27 5′UTR.
Figure 3.
The IRES domain of the p27 5′UTR confers resistance to translational repression by PI3-kinase inhibitor LY294002. (A) Protein synthesis rates of HeLa cells after addition of PI3-kinase inhibitor LY294002. Cells treated with LY294002 or solvent (ethanol) for 0 min, 10 min, 2 h, 5 h, or 24 h were labeled with (35S)-methionine for an additional 30 min. The incorporation of radioactivity into proteins in LY294002 treated cells was plotted as a percentage of the incorporation in solvent-treated cells. (B) Analysis of PI3-kinase inhibition on the translation of monocistronic mRNAs with p27 5′UTR leaders. Transfected HeLa cells were incubated for 24 h with LY294002 or solvent. Luciferase acivities were normalized to luciferase mRNA levels determined in Northern blots. (C,D) Endogenous p27 protein and mRNA levels are unaltered in LY294002-treated cells. HeLa cells were incubated with LY294002 (LY) or the solvent (C). p27 mRNA levels (top) were analyzed by Northern blot of total RNA with probes specific for p27 mRNA and GAPDH mRNA (bottom). The 18S small ribosomal RNA serves as an additional loading control. (D) Protein levels of p27 (top), p21 (middle), or α-tubulin (bottom) were detected by immunoblotting using monoclonal antibodies. (E) Synthesis rates of p27 are unaltered in the presence of LY294002. After 23-h incubation with LY294002 or solvent (control), adherent HeLa cells were pulse-labeled with (35S) methionine and (35S) cysteine for 1 h. Extracts adjusted for equal amounts of protein were boiled and p27 was immunoprecipitated. p27 synthesis was quantified after SDS-PAGE using a PhosphorImager. (Left) The average of six experiments is represented. (Right) Incorporation of radioactivity in total protein was determined by TCA-precipitation of protein extracts of identical protein concentrations from solvent (C) or LY294002 (LY)-treated cells and liquid scintillation counting. Three experiments are represented. (F) Flow cytometry analysis of cell cycle distribution of adherent HeLa cells after 24 h treatment with LY294002 or solvent.
Recently, Miskimins et al. (2001) reported the identification of an IRES element of 217 nucleotides in the murine p27 5′UTR. This IRES renders p27 translation cap-independent under conditions of elevated cyclic AMP levels (Miskimins et al. 2001). As is demonstrated below (Fig. 4D), the complete human IRES element is, however, significantly larger.
Figure 4.
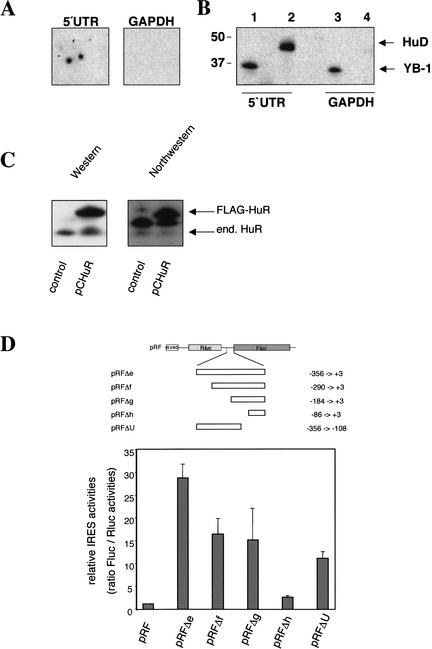
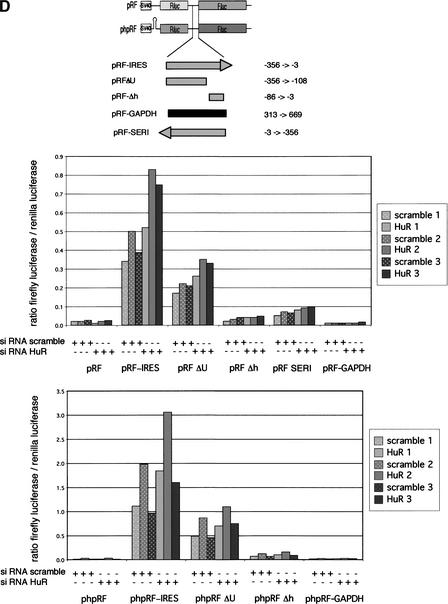
HuD and HuR proteins bind to the p27 5′UTR. (A) Identification of HuD as a factor binding to the p27 5′UTR. A protein array of 37,200 colonies expressing human cDNAs was hybridized with a radioactive-labeled p27 5′UTR. Each colony was spotted twice onto a nylon membrane in a defined orientation. The autoradiography shows an area of the filter that contains the HuD cDNA-expressing cells (left). Specificity of HuD binding was demonstrated by stripping and second hybridization with a 32P-labeled GAPDH mRNA. (B) Recombinant HuD protein was UV cross-linked to either the p27 5′UTR or the GAPDH mRNA. As a control, the RNA binding YB-1 protein was analyzed. Protein–RNA complexes were resolved by SDS-PAGE and the dried gels exposed to a film. While YB-1 binds to the p27 5′UTR and to the GAPDH mRNA, HuD specifically binds to the p27 5′ leader. (C) HuR protein binds to the p27 5′UTR. FLAG-tagged HuR was overexpressed in HeLa cells and protein extracts were separated by SDS-PAGE and transferred to a PVDF membrane. The membrane was first processed for Northwestern analysis using a radioactive-labeled fragment of the p27 5′UTR and thereafter for immunodetection of HuR-proteins with an HuR-specific antibody. FLAG-tagged fusion and endogenous HuR proteins are indicated by arrows. The additional band in the Northwestern analysis between the endogenous and FLAG-tagged HuR represents a yet uncharacterized nonspecific RNA-binding protein present in HeLa cell extracts. (D) A U-rich element known to bind HuR lacks significant IRES activity. 293T cells were transfected with plasmids expressing bicistronic mRNA that contains fragments of the p27 5′UTR in the intercistronic region, as indicated above the bar graph. Firefly and Renilla luciferase activities were determined, and the ratio between firefly and Renilla luciferase was calculated. The ratio for the empty vector pRF was set to 1.
To identify proteins that might regulate p27 IRES activity, we hybridized a cDNA expression array of human brain tissue (menstrual age 14.8 and 15.8) with a radioactive RNA probe of the complete p27 5′UTR. One of the strongest signals was obtained from the brain specific ELAV protein HuD (Fig. 4A). The corresponding HuD cDNA contained the coding region for HuD, lacking only three initial codons. Specificity of HuD binding to the p27 5′UTR was determined in solution by UV cross-linking experiments. The HuD protein binds specifically to p27 5′UTR RNA, as no interaction was detected with GAPDH mRNA of comparable length (Fig. 4B). Another member of the ELAV family, HuR (HuA), was previously identified as a factor binding to a p27 5′UTR starting 153 nucleotides upstream of the p27 start codon (Millard et al. 2000). This sequence overlaps with the predicted IRES element. Because HuD expression is restricted to neuronal cells, we speculated that the ubiquitously expressed and closely related HuR protein might fulfill HuD functions in other cell types. As for HuD, also endogenous or overexpressed HuR protein binds to the extended p27 5′UTR in Northwestern assays (Fig. 4C). The binding site of HuR has previously been mapped to an U-rich element in a truncated p27 5′UTR; however, a function in regulating p27 level could not be determined (Millard et al. 2000).
To further characterize the IRES element and to investigate if the described HuR binding site (nucleotides −75 to −33) was part of the IRES, we analyzed deletion mutants of the 5′UTR (Fig. 4D, upper panel). Truncation of the IRES element from 356 to 290 or 184 nucleotides diminished IRES activity to <60%, indicating that the region between nucleotides −356 and −290 is required for maximal IRES activity (Fig. 4D, lower panel). A smaller fragment containing the characterized HuR binding domain had only minimal IRES activity (10% of the entire IRES domain), but deletion of this U-rich HuR binding element also weakened the strength of the IRES (Fig. 4D). The characterized HuR binding region therefore contributes to, but is not essential for IRES activity of the p27 5′UTR.
Because one mammalian ELAV protein, Hel-N1 (HuB) was found to increase translation of neurofilament M and GLUT1 mRNAs (Jain et al. 1997), we wished to investigate if HuD or HuR might directly alter p27 translation. For this, we overexpressed HuD and HuR and analyzed a luciferase reporter containing the complete p27 5′UTR as leader. Surprisingly, overexpression of both, HuD or HuR did not increase, but inhibited reporter translation (Fig. 5A). To investigate whether this repression was due to interference with the IRES activity, we investigated bicistronic mRNA translation. A dramatic reduction of IRES activity was detected after HuD or HuR overexpression (to between 20% and 35% of the vector control, Fig. 5C). This strong repression was also observed for a bicistronic plasmid that contained a stable hairpin to reduce cap-dependent translation (Fig. 5C). These data support a new and unexpected role for Hu/ELAV proteins in inhibiting IRES-dependent translation.
Figure 5.
HuD and HuR repress p27 translation. 293T cells were cotransfected with luciferase reporter plasmids, expression vectors for HuD or HuR, and pSV-β-Gal. Twenty-four hours after transfection the relative luciferase activity and β-galactosidase activities were determined. (A) Cells expressing firefly luciferase with or without the p27 5′UTR (pGL3 or pGL5′UTR) or the IRES domain of the p27 5′UTR (pGL-IRES) were cotransfected with HuD, HuR, or empty-vector DNA. Normalized luciferase translation rates are expressed relative to that of pGL3 (set to 1). (B) FLAG-tagged HuR and HuD were expressed in 293T cells. Expression level of both proteins were compared by immunoblotting with an anti FLAG antibody. (C) HuR and HuD inhibit the p27-IRES activity. pRF, pRF-IRES, and phpRF-IRES were cotransfected with expression vectors for HuD or HuR. IRES activities were determined as ratios of firefly to Renilla luciferase activities relative to the empty vector. Representative results of at least three independent experiments are shown in A and C.
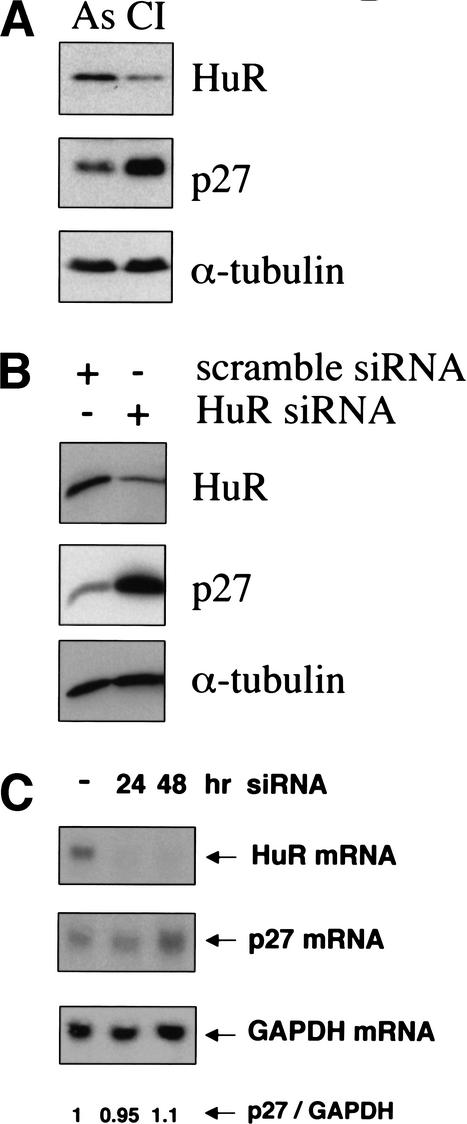
We had previously observed that p27 translation is increased in cells that are quiescent due to contact inhibition (Hengst and Reed 1996). To investigate if HuR might be involved in increasing p27 under these conditions, we grew human diploid fibroblasts into contact inhibition and compared p27 and HuR protein level. As expected, p27 protein increased in density-arrested cells (Fig. 6A). At the same time HuR protein level declined, consistent with the hypothesis that reduced HuR level may increase p27 translation.
Figure 6.
(A) Reduced HuR correlates with induced p27 in density-arrested fibroblasts. Human diploid fibroblasts were grown into contact inhibition, and HuR and p27 levels were compared by immunoblotting to those of proliferating fibroblasts. (B–D) Knockdown of HuR expression by RNA interference induces p27 protein. HeLa cells were transfected with siRNAs targeting HuR mRNA (siHuR). (B) Western blot analysis of HuR RNAi in HeLa cells. Twenty-five micrograms of total cell extract from either control-transfected (siRNA) or siHuR-transfected cells were isolated 24 h after transfection, and protein extracts were separated by SDS-PAGE and analyzed for p27, HuR, or α-tubulin. (C) Northern analysis of HuR RNAi in HeLa cells. Ten micrograms of total RNA were separated and immobilized on a nylon membrane. The filter was sequentially hybridized with 32P-labeled probes specific for p27, HuR, and GAPDH mRNAs. Relative hybridization signals for p27 and GAPDH were determined using a PhosphorImager, and the calculated relative ratio of these mRNAs is given below the blot. (D) Down-regulation of HuR expression induces p27-IRES activity. HeLa cells were transfected with the bicistronic constructs pRF, pRF-IRES, and phpRF-IRES, together with siHuR and pSV-β-Gal. Cells were harvested 24 h after transfection and extracts analyzed for Renilla and firefly luciferase expression. As a reference a scrambled siRNA was cotransfected (control). IRES activities were determined as ratios of firefly to Renilla luciferase activities relative to the empty vector. Three sets of independent experiments are shown. Samples of one experiment are shown in the same gray tone (e.g., experiment 1: scramble 1 and HuR 1).
To directly investigate the regulation of p27 by HuR in the absence of overexpression, we employed mRNA “knockdown” by RNA interference (Elbashir et al. 2001). Transfection of siRNA oligonucleotides directed against the HuR mRNA efficiently reduced HuR mRNA and protein (Fig. 6B,C). Reduction of HuR protein by siRNA (similar to level observed in density-arrested cells; Fig. 6, cf. A and B) dramatically induced p27 protein (Fig. 6B). The increase in p27 protein was already detected 24 h after transfection, and p27 levels remained at this level for at least 96 h (data not shown).
HuR enhances stability and mediates nuclear export of specific mRNAs (Brennan and Steitz 2001). If this were also the case for p27, reduced p27 mRNA and protein levels would be expected in siRNA treated cells. Instead, we observed an increase of p27 protein, while p27 mRNA level remained unchanged at 24 h and increased even slightly (10%) at 48 h (Fig. 6C).
We next asked whether increased p27 levels upon HuR depletion were due to a direct effect on translation. For this, we used the bicistronic reporter constructs with or without hairpin, because these plasmids allow distinguishing direct interference with IRES-dependent translation from indirect effects, for example, through mRNA localization. Alterations in mRNA export or stability would affect both cistrons of the bicistronic messenger, and the ratio between firefly and renilla luciferases would remain constant. In contrast, mechanism modulating specific cap-dependent versus IRES-dependent translation should affect this ratio. We cotransfected the constructs with HuR specific siRNA or a control siRNA of identical length, determined the activity of both luciferases, and calculated the ratio. In comparison to the control siRNA, “knockdown” of HuR clearly increased IRES-dependent translation in both types of the constructs (Fig. 6D). The increase in internal initiation rate was in average 1.66-fold for the bicistonic vector pRF (n = 8, p < 0.01) and 1.47-fold for the haipin containing construct (n = 8, p < 0.01).
We next wanted to test whether this effect was mediated via the mapped HuR binding sites. For this, we introduced vectors lacking the U-rich element that was shown to bind HuR (pRF-ΔU and phpRF-ΔU). This 5′UTR still showed response to HuR knockdown (pRF-ΔU: n = 8, 1.44-fold increase and phpRF-ΔU: n = 10, 1.29-fold increase); however, in comparison with the entire IRES, this response was clearly reduced (Fig. 6D). Although we only observed a partial loss of regulation, this was statistically highly significant (bicistronic constructs pRF: n = 10, p < 0.01; hairpin constructs phpRF: n = 8, p < 0.01). This suggests that the mapped HuR binding element contributes to but is not the only HuR responsive element in the p27 5′UTR. Taken together, these findings suggest a new and unexpected role for HuR in reducing p27 protein level.
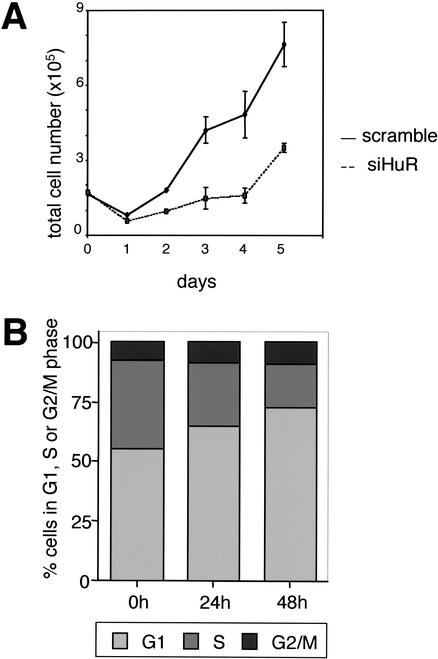
If HuR regulates p27 level, siRNA mediated knockdown might lead to a cell cycle delay/arrest in G1. We, therefore, investigated proliferation and cell cycle phase distribution in cells treated with HuR siRNA or control siRNA. Indeed, HuR knockdown significantly reduced the doubling time of HeLa cells (Fig. 7A). Concomitantly, within 48 h incubation with siRNA, G1-phase cells increased from 56% to 73% while S-phase cells declined from 37% to 18% (Fig. 7B). This supports a key role of HuR in regulating cell proliferation, in part by inhibiting p27 translation.
Figure 7.
siRNA-mediated knockdown of HuR induces cell cycle arrest in G1. (A) Growth curve of HeLa cells after HuR knockdown. Cells were transfected with siRNA oligonucleotides on day 0 and day 1, and cell numbers were determined. (B) Cell cycle distribution of HeLa cells transfected with siRNA directed against HuR. Cell cycle phase distribution using BrdU and propidium iodide was determined by flow cytometry.
Discussion
We have characterized a potent IRES element in the 5′ leader of the human p27 mRNA. A p27 IRES in a mouse macrophage cDNA has been independently identified by Miskiminis et al. (2001). The mapped IRES covered their complete 5′UTR of 217 nucleotides, and may well be truncated. The region conferring maximal IRES activity of human p27 is significantly larger and includes 356 nucleotides upstream of the ATG. Truncation of this fragment to 290 nucleotides already severely impaired IRES activity (Fig. 4D). Because human cDNA and the genomic mouse sequences are well conserved in their 5′ region, it seems likely that longer 5′UTRs with more potent IRES elements exist for the mouse as well.
Our attempts to map the IRES domain allowed a truncation from 461 to 356 nucleotides without a significant alteration in IRES activity (Fig. 2B). Together with our RNase protection analysis (Fig. 1B), these data demonstrate that the vast majority of human p27 mRNAs contains the functional IRES element. Further truncations reduced but did not abrogate the IRES activity, suggesting that the p27 IRES domain is composed of different sequence elements that can function independently from each other. A similar behavior was reported for the shorter murine p27 IRES (Miskimins et al. 2001), and has also been reported for the IRES of the c-sis or the eIF4G mRNAs (Gan et al. 1998; Sella et al. 1999).
What is the physiologic role of the IRES? Our analysis of the effect of PI3 kinase inhibition on p27 translation suggests that p27 translation can be mediated in large parts via its IRES element. We would like to speculate that the IRES serves to sustain p27 synthesis under unfavorable growth conditions, when overall cap-dependent protein synthesis is reduced and persisting p27 expression is required, for example, in the case of some viral infections (Pe'ery and Mathews 2000). In addition, maintenance of translation under stress conditions or during quiescence may relay on IRES elements. For example, VEGF is thought to be translated by internal ribosomal entry under hypoxia, when overall protein synthesis is compromised (Stein et al. 1998). Interestingly hypoxia has also been shown to induce p27 protein levels (Krtolica et al. 1998, 1999; Gardner et al. 2001). Efficient translation of the Cdk inhibitor under these conditions may therefore depend on its IRES element.
It should be interesting to analyze the integrity of the p27 5′UTRs isolated from tumor samples or cancer cell lines displaying reduced p27 protein levels. Mutations or deletions in the IRES sequence could well account for reduction in p27 levels. Gain of function mutations within the IRES of the protooncogene c-myc were found at high rate in bone marrow samples from patients with multiple myeloma (Chappell et al. 2000).
To understand a possible regulation of the p27 IRES, we searched for proteins that bound to the p27 5′UTR and identified HuD on protein arrays. The RNA binding protein HuD is expressed in neuronal tissue; however, a related member of the human Hu/Elav protein family is ubiquitously expressed. HuR was previously identified as protein binding to a U-rich element in the p27 5′UTR (Millard et al. 2000). By overexpression and siRNA mediated HuR knockdown we could demonstrate that HuR represses internal initiation of p27 translation.
One binding site of HuR was mapped to an U-rich element in proximity to the p27 initiation codon. The HuR binding domain is part of the IRES element since its deletion reduces IRES activity. Deletion of the HuR binding element partially reduced the IRES induction by HuR knockdown but failed to render internal initiation insensitive to HuR. The residual degree of regulation by HuR might be explained by additional HuR binding sites in the p27 5′UTR. It was recently described that HuR binding sites do not need to be AU rich elements (ARE), but that in addition UC-rich sequences can serve as binding motives (Yeap et al. 2002). It remains to be investigated if HuR by itself or in association with other proteins can bind to additional sites in the p27 5′UTR and whether this binding accounts for the remaining regulation of the p27 IRES by HuR. Alternatively, HuR may also act on p27 IRES by some indirect mechanism, for example, by influencing the expression level of factors involved in p27 translation.
Interestingly, HuR seems to fulfill a very different role for the closely related CDK inhibitor p21. Here, HuR binds to an ARE in the 3′UTR of the p21 mRNA (Joseph et al. 1998) and stabilizes the transcript after UV damage (Wang et al. 2000b). Similarly, all other RNA interactions described for HuR this far lead to an increase in protein levels by conferring mRNA stability to the bound mRNA. In each of these cases, HuR binds to the 3′UTR of the respective message. It will be interesting to see whether other mRNAs with HuR binding sites in their 5′UTR exist.
Is an inhibitory role of HuR consistent with the regulated translation of p27 in growth arrest or during the cell cycle? We found a decline of HuR level in contact inhibited human diploid fibroblasts. This decline is paralleled by an increase in p27 protein. Further, murine HuR (mHuA) is down-regulated in quiescent cells and induced upon serum stimulation (Atasoy et al. 1998), and human HuR expression decreases during senescence (Wang et al. 2001). High expression of HuR correlates therefore with growth conditions where reduced p27 translation have been described (Pagano et al. 1995; Agrawal et al. 1996; Hengst and Reed 1996).
Translation of p27 is also regulated during the cell cycle with a decrease at the G1/S transition (Hengst and Reed 1996). HuR is predominantly nuclear in early and mid G1 phase and increasingly cytoplasmic during late G1, S phase, and G2/M. (Wang et al. 2000a). This change in localization would be consistent with a role of cytoplamic HuR in inhibiting translation of p27.
Interestingly, HuR appears to control cell cycle progression at several levels. Besides reducing p27 translation, it can enhance stability of cyclin A and B mRNA (Wang et al. 2000a) as well as of additional growth regulatory mRNAs (Brennan et al. 2000; Gallouzi and Steitz 2001; Gallouzi et al. 2001). By simultaneously reducing levels of cell cycle inhibitors and enhancing levels of cell cycle promoting factors, HuR may be a key factor in controlling cell proliferation. Consistent with this idea, siRNA mediated knockdown of HuR in HeLa cells leads to a strong reduction in growth rate and concomitant increase in G1 cells (Fig. 7).
It has been previously reported that RKO cell lines that stably express HuR antisense mRNA increase in S/G2/M phases (Wang et al. 2000a). It remains to be determined if this difference is cell type specific, or if selection of stable cell lines may lead to elimination of cells that arrest in G1 and cease proliferation.
In conclusion, our data describe HuR as a negative regulator for p27 translation. Together with its known function in positively regulating cyclins A and B, careful regulation of HuR level and localization must be of the essence for proper growth control. Interestingly members of the Hu/ELAV family have already been found (over)expressed in tumors, for example, HuR in malignant brain tumors or colon cancer (Dixon et al. 2001; Nabors et al. 2001).
Materials and methods
Tissue culture
HeLa, 293T cells, and human foreskin fibroblasts (Clonetics) were grown in Dulbecco modified eagles medium (DMEM) with 10% fetal bovine serum. For siRNA experiments, cells were transferred to Opti-MEM1 (Gibco). Where indicated, LY294002 (Biomol) was added to medium to a final concentration of 40 μM. Cell cycle phase distributions were determined by flow cytometry using propidium iodine and BrdU pulse labeling. Cells incorporating BrdU were determined using a FITC-coupled anti-BrdU antibody (BD Biosciences). Transfections were performed with SuperFect reagent (Qiagen), Oligofectamine, or Lipofectamine 2000 (both Invitrogen) according to the manufacturer's instructions. Typically, cells growing on 60-mm Petri dishes were transfected with 0.2 μg reporter plasmid and 1.8 μg pSV-β-Gal vector (Promega) when SuperFect reagent was used or 0.4 μg reporter plasmid together with 7.6 μg pSV-β-Gal vector (Promega) in the case of Lipofectamine 2000 transfection, 0.4 μg reporter plasmid, 2.1 μg siRNA, and 5.5 μg pSV-β-Gal vector were transfected. Cells were analyzed 24 to 48 h after transfection.
For determining the doubling time of HeLa cells during RNA interference, two 60-mm Petri dishes per siRNA were transfected with the respective siRNAs (day 0); 6 h after this first transfection, cells were split to six well plates and 16 h later transfected a second time with siRNAs (day 1). siRNA complexes were left on the cells for 5 d, and each day cells were harvested for determining cell numbers and efficiency of RNA interference.
DNA manipulations
Detailed information about DNA cloning procedures can be obtained from the authors upon request. The p27 5′UTR from −575 to −108 was isolated by anchored PCR (5′RACE Kit, Life Technologies) from Hela total RNA. Fragments of the p27 5′UTR were generated by PCR and cloned into reporter plasmids as indicated. For in vitro transcription, linearized and gel-purified template DNAs (Easy Pure, Biozym) were used. Bicistronic reporter plasmids pRF and phpRF (Stoneley et al. 1998; Coldwell et al. 2000) were a generous gift of A.E. Willis (Leicester). Fragments of the p27 5′UTR, obtained by PCR amplification of the respective regions in p27 5′UTR, were inserted into the EcoRI and NcoI sites of these vectors.
The HuD coding region of the EST clone MPMGp800E17542Q102 (RZPD, GmbH, Berlin) and the coding region for HuR (clone IMAGp998J057162Q2) were isolated by PCR and inserted into the expression plasmid pCruzOctA (Santa Cruz). The GAPDH cDNA fragment (356 nucleotides from 313 to 669) was amplified by PCR from a cDNA.
Reporter analysis
Cells transfected with monocistronic reporter plasmids were lysed in Reporter Lysis Buffer (Promega), and firefly luciferase expression was determined using the Luciferase Assay System (Promega). For bicistronic reporter plasmids, cells were lysed in Passive Lysis Buffer and both firefly (Photinus pyralis) and sea pansy (Renilla reniformis) luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega). β-Galactosidase activity was determined in the same lysate using the β-galactosidase Enzyme Assay System (Promega).
RNA analysis
Total RNA was isolated with the RNeasy Mini Kit (Qiagen). Denatured RNA was separated through 1% agarose gels containing 440 mM formaldehyde, transferred to a positively charged membrane (HybondN+; AP biotech), cross-linked by UV irradiation and hybridized at 65°C in 1 M NaCl, 10% dextran sulfate, 1% SDS, and 0.01% salmon sperm DNA. Radioactive probes were generated by random prime labeling (Promega) of isolated DNA fragments or by PCR amplification. Blots were analyzed by autoradiography and by using a PhosphorImager.
RNAse protection analysis was performed using the Kit from Roche according to the manufacturers instructions. Per sample, 14μg total RNA was hybridized with an antisense RNA probe (1.5 × 105 cpm/sample) of 681 nucleotides including the p27 5′UTR (nucleotides −3 to −575) and flanking vector sequences. After digestion with RNase A (3.5 U) and RNase T1 (25 U), RNase was degraded with proteinase K and proteins were removed by phenol/chloroform extraction. RNAs were precipitated and protected products were separated on a denaturating polyacrylamide gel and detected by autoradiography.
RNA interference (RNAi)
RNA oligonucleotides were synthesized by Dharmacon Research. Annealing and RNA interference analysis was described by Elbashir et al. (2001). Best results for HuR knockdown were obtained by combining two siRNA pairs (HuRI, 5′-GUUGAAU CUGCAAAACUUAdTdT-3′/5′-UAAGUUUUGCAGAUUCA ACdTdT-3′; and HuRII, 5′-UGUGAAAGUGAUCCGCGAC dTdT-3′/5′-GUCGCGGAUCACUUUCACAdTdT-3′). As a control premade “scramble” dsRNA, available at Dharmacon Research, with the sequences 5′-GCGCGCUUUGUAGGAUUC GdTdT-3′/5′-CGAAUCCUACAAAGCGCGAdTdT-3′ was used.This sequence lacks complementary sequences in the human genome. dsRNA was transfected into Hela cells with Oligofectamine (Invitrogen) according to the manufacturer's recommendation. Cotransfections of reporter-DNA with siRNA was done by using Lipofectamine 2000 (Invitrogen) as transfection reagent.
Protein analysis
Western blots were performed using standard procedures. Monoclonal mouse antibodies against p27 (clone 57, Transduction Laboratories), HuR (19F12, Molecular Probes), FLAG-epitope and anti-α-tubulin (M2 and clone DM 1A, Sigma) were used. To determine protein synthesis rates and stabilities, pulse-chase experiments of HeLa cells were performed as described (Hengst and Reed 1996). To measure the total uptake of radioactivity, 50-μL aliquots of the cell suspension were spotted directly onto Whatman GF/C filters. The filters were dried and the radioactivity measured by liquid scintillation counting. To quantify the radioactivity that was incorporated into proteins, 50-μL cell suspension was subjected to trichloroacetic acid (TCA) precipitation. The samples were filtered through Whatman GF/C filters. The filters were washed twice with ice-cold 10% TCA and ethanol, dried, and analyzed by liquid scintillation counting. The percentage of incorporated radioactivity was calculated from LY294002 and solvent treated cells.
RNA–protein interactions
Protein expression arrays were obtained from the Resource Center/Primary database (RZPD, Berlin, Germany). The arrays contained 37,200 colonies expressing hexahistidine tagged human cDNAs of a human brain tissue library, menstrual age 14.8 and 15.8. For RNA binding studies, RNA was synthesized by SP6 bacteriophage polymerase (Roche) and labeled by incorporation of α32P-CTP according to the manufacturers recommendation. The integrity of the riboprobes was tested by denaturing RNA-gel electrophoresis.
For Northwestern screening, filter immobilized proteins were renatured overnight at 4°C under continuos shaking in Northwestern binding buffer (NWBB: 10 mM Tris-HCl at pH 7.5, 1 mM EDTA, 50 mM NaCl, 0.1% Triton X-100, 0.02% BSA, 0.02% Ficoll 400, and 0.02% PVP. Filters were then transferred into 200 mL of fresh NWBB and prehybridized in the presence of 0.5 μg/μL heparin and 0.5 μg/μL yeast tRNA for 15 min at room temperature. Then 2 × 107 cpm of gel-purified full-length 5′UTR transcript (105 cpm 32P-labeled transcript per milliliter) was added and further hybridized for 1 h. Before extensive washing of the filters in NWBB, an aliquot of the hybridization solution was propanol precipitated and resolved by denaturing RNA gel electrophoresis for controlling integrity of the transcript during the hybridization procedure.
Alternatively, protein of 293T cells overexpressing FLAG-tagged HuR or HuD were separated by SDS-PAGE and transferred to the PVDF membrane (Hybond P, Amersham Pharmacia) and analyzed as described above.
RNA/protein UV cross-linking
Recombinant His-tagged HuD was incubated on ice for 10 min in 0.5 μg/μL Heparin (Calbiochem) and 0.25 μg/μL yeast tRNA (Calbiochem) dissolved in 40 μL RNA binding buffer (10 mM HEPES at pH 7.9, 100 mM KCl, 4 mM MgCl2, 1 mM EDTA, and 10% Glycerol); 5 × 104 cpm of 32P-labeled transcript was added. After 20 min incubation on ice, the binding reaction was UV-irradiated with 10 mJ in a UV Stratalinker 1800 (Stratagene). After treatment with RNase A (0.2 μg/μL) and RNase T1 (40 units) for 30 min at 37°C, cross-linked reaction mixtures were resolved by 10% SDS-PAGE and visualized by autoradiography.
Statistics
Statistic analysis was performed using the Sigma Stat 2.0 software (Jandel Scientific). For samples lacking proven normal distribution, p-values were calculated using Wilcoxan signed rank algorithm. (Lower p-values obtained using a Sudent's t-test assuming normal distribution of the sample values).
Acknowledgments
We thank Ansgar Resch for assistance with the statistical analysis and Irina Sperling for expert technical support. We especially thank Anne E. Willis for the bicistronic vectors pRF and phpRF, and members of the laboratory, Frauke Melchior and Sowmya Swaminathan, for stimulating discussions and critical reading of the manuscript. This work was funded by grants of the Max-Planck Society; the government of Oberbayern; and Roche, Penzberg.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hengst@biochem.mpg.de; FAX 49-89-8578-2361.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.248902.
References
- Agrawal D, Hauser P, McPherson F, Dong F, Garcia A, Pledger WJ. Repression of p27kip1 synthesis by platelet-derived growth factor in BALB/c 3T3 cells. Mol Cell Biol. 1996;16:4327–4336. doi: 10.1128/mcb.16.8.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antic D, Lu N, Keene JD. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes & Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AR, Grossman D, White K. Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects. A developmental-genetic analysis. J Neurogenet. 1985;2:197–218. doi: 10.3109/01677068509100150. [DOI] [PubMed] [Google Scholar]
- Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, Ralston SH, Helfrich MH, Willis AE. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: A novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekholm SV, Reed SI. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol. 2000;12:676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Steitz JA. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science. 2001;294:1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Steitz JA. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7:1348–1361. doi: 10.1017/s1355838201016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, LaCelle M, Rhoads RE. Functional characterization of the internal ribosome entry site of eIF4G mRNA. J Biol Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- Gardner LB, Li Q, Park MS, Flanagan WM, Semenza GL, Dang CV. Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem. 2001;276:7919–7926. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- Gingras A-C, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Hengst L, Reed SI. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- ————— Inhibitors of the Cip/Kip family. Curr Top Microbiol Immunol. 1998;227:25–41. doi: 10.1007/978-3-642-71941-7_2. [DOI] [PubMed] [Google Scholar]
- Ito E, Iwahashi Y, Yanagisawa Y, Suzuki Y, Sugano S, Yuasa Y, Maruyama K. Two short sequences have positive effects on the human p27Kip1 gene transcription. Gene. 1999;228:93–100. doi: 10.1016/s0378-1119(99)00022-0. [DOI] [PubMed] [Google Scholar]
- Jain RG, Andrews LG, McGowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Orlian M, Furneaux H. p21(waf1) mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J Biol Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- Kamiyama J, Inoue T, Ohtani-Fujita N, Minami S, Yamagishi H, Sakai T. The ubiquitous transcription factor NF-Y positively regulates the transcription of human p27Kip1 through a CCAAT box located in the 5-upstream region of the p27Kip1 gene. FEBS Lett. 1999;455:281–285. doi: 10.1016/s0014-5793(99)00899-6. [DOI] [PubMed] [Google Scholar]
- Krtolica A, Krucher NA, Ludlow JW. Hypoxia-induced pRB hypophosphorylation results from downregulation of CDK and upregulation of PP1 activities. Oncogene. 1998;17:2295–2304. doi: 10.1038/sj.onc.1202159. [DOI] [PubMed] [Google Scholar]
- ————— Molecular analysis of selected cell cycle regulatory proteins during aerobic and hypoxic maintenance of human ovarian carcinoma cells. Br J Cancer. 1999;80:1875–1883. doi: 10.1038/sj.bjc.6690615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Shen X, Nguyen VA, Kunos G, Gao B. α(1) adrenergic agonist induction of p21(waf1/cip1) mRNA stability in transfected HepG2 cells correlates with the increased binding of an AU-rich element binding factor. J Biol Chem. 2000;275:11846–11851. doi: 10.1074/jbc.275.16.11846. [DOI] [PubMed] [Google Scholar]
- Millard SS, Yan JS, Nguyen H, Pagano M, Kiyokawa H, Koff A. Enhanced ribosomal association of p27(Kip1) mRNA is a mechanism contributing to accumulation during growth arrest. J Biol Chem. 1997;272:7093–7098. doi: 10.1074/jbc.272.11.7093. [DOI] [PubMed] [Google Scholar]
- Millard SS, Vidal A, Markus M, Koff A. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol Cell Biol. 2000;20:5947–5959. doi: 10.1128/mcb.20.16.5947-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami S, Ohtani-Fujita N, Igata E, Tamaki T, Sakai T. Molecular cloning and characterization of the human p27Kip1 gene promoter. FEBS Lett. 1997;411:1–6. doi: 10.1016/s0014-5793(97)00660-1. [DOI] [PubMed] [Google Scholar]
- Miskimins WK, Wang G, Hawkinson M, Miskimins R. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol Cell Biol. 2001;21:4960–4967. doi: 10.1128/MCB.21.15.4960-4967.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. [PubMed] [Google Scholar]
- Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- Pe'ery T, Mathews MB. Viral translational strategies and host defense mechanisms. In: Mathews MB, editor. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 371–424. [Google Scholar]
- Raught B, Gingras A-C, Sonenberg N. Regulation of ribosomal recruitment in eukaryotes. In: Mathews MB, editor. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 245–293. [Google Scholar]
- Sella O, Gerlitz G, Le SY, Elroy-Stein O. Differentiation-induced internal translation of c-sis mRNA: Analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol Cell Biol. 1999;19:5429–5440. doi: 10.1128/mcb.19.8.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes & Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000a;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000b;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M. Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol. 2001;21:5889–5898. doi: 10.1128/MCB.21.17.5889-5898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap BB, Voon DC, Vivian JP, McCulloch RK, Thomson AM, Giles KM, Czyzyk-Krzeska MF, Furneaux H, Wilce MC, Wilce JA, et al. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J Biol Chem. 2002;277:27183–27192. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]