Abstract
The establishment of metaphase chromosomes is an essential prerequisite of sister chromatid separation in anaphase. It involves the coordinated action of cohesin and condensin, protein complexes that mediate cohesion and condensation, respectively. In metazoans, most cohesin dissociates from chromatin at prophase, coincident with association of condensin. Whether loosening of cohesion at the onset of mitosis facilitates the compaction process, resolution of the sister chromatids, or both, remains unknown. We have found that the prophase release of cohesin is completely blocked when two mitotic kinases, aurora B and polo-like kinase (Plx1), are simultaneously depleted from Xenopus egg extracts. Condensin loading onto chromatin is not affected under this condition, and rod-shaped chromosomes are produced that show an apparently normal level of compaction. However, the resolution of sister chromatids within these chromosomes is severely compromised. This is not because of inhibition of topoisomerase II activity that is also required for the resolution process. We propose that aurora B and Plx1 cooperate to destabilize the sister chromatid linkage through distinct mechanisms that may involve phosphorylation of histone H3 and cohesin, respectively. More importantly, our results strongly suggest that cohesin release at the onset of mitosis is essential for sister chromatid resolution but not for condensin-mediated compaction.
Keywords: Cohesin, condensin, sister chromatid cohesion, chromosome condensation, histone H3 phosphorylation
In eukaryotic cells, the duplication of chromosomal DNA during S phase produces two sister DNA molecules that are topologically intertwined. In addition, a proteinaceous linkage between the sister chromatids is established during this stage and is maintained throughout G2 phase. At the onset of mitosis, loosening of cohesion accompanies the compaction of each chromatid along its longitudinal axis. The final product of this mechanistically complex process is the metaphase chromosome, in which two rod-shaped sister chromatids are juxtaposed along their entire length. The establishment of metaphase chromosome structure is an essential prerequisite of the faithful segregation of sister chromatids in anaphase. Despite its fundamental importance, very little is known about the regulatory network that contributes to the conversion of amorphous interphase chromatin into highly organized metaphase chromosomes.
Accumulating lines of evidence suggest that the cell-cycle-dependent transitions of chromosome structure can largely be explained by the dynamic behavior of two distinct structural maintenance of chromosomes (SMC) protein complexes, cohesin and condensin. Cohesin plays a central role in sister chromatid cohesion, possibly as the main constituent of the proteinaceous linkage that holds sister chromatids together (for review, see Lee and Orr-Weaver 2001; Nasmyth 2001). In metazoan cells, most cohesin complexes bound to interphase chromatin dissociate during mitotic prophase (Losada et al. 1998, 2000; Waizenegger et al. 2000; Warren et al. 2000). Although it has been proposed that this bulk release of cohesin may be required for proper chromosome condensation (e.g., Losada and Hirano 2001), no direct test for this hypothesis has been reported to date. The remaining cohesin complexes are removed from the chromosomes via separase-mediated cleavage at the onset of anaphase, leading to complete separation of the sister chromatids (Waizenegger et al. 2000; Hauf et al. 2001). On the other hand, condensin is loaded on chromosomes in prophase, at the time when condensation is initiated and cohesin-mediated linkage is destabilized (for review, see Hirano 2000). Condensin is essential for the maintenance of condensation from metaphase to anaphase and dissociates from chromosomes in telophase when decondensation becomes apparent in the daughter cell nuclei (Hirano and Mitchison 1994; Schmiesing et al. 2000).
One important question is how multiple protein kinases might regulate these changes of chromosome architecture. Three mitotic kinases, cdc2, polo, and aurora B, are of particular interest. We have shown previously that phosphorylation of condensin by cdc2–cyclin B activates its positive supercoiling and knotting activities in vitro (Kimura et al. 1998, 1999, 2001). Although this observation partially explains how chromosome condensation is initiated at the onset of mitosis, additional mechanisms are likely to dictate the mitosis-specific association of condensin with chromatin (Kimura and Hirano 2000; Steen et al. 2000). Cdc2 can also phosphorylate one of the regulatory subunits of vertebrate cohesin (SA/Scc3) in vitro, and this phosphorylation reduces the affinity of the complex for chromatin (Losada et al. 2000). However, cdc2 activity is not required for cohesin release when a pseudomitotic state is induced by addition of a phosphatase inhibitor to interphase extracts (Sumara et al. 2000). In Saccharomyces cerevisiae, Cdc5/polo kinase phosphorylates the Scc1 subunit of cohesin and thereby enhances its cleavage by separase at the onset of anaphase (Alexandru et al. 2001). The specific role of this kinase in sister chromatid separation has been revealed only recently, probably because polo plays multiple roles throughout mitosis and its mutation causes pleiotropic phenotypes (Glover et al. 1998).
A similar level of complexity has been encountered in the functional analysis of aurora B. Aurora B and its binding partner, INCENP, are referred to as chromosomal passengers on the basis of their dynamic localization during mitosis (for review, see Adams et al. 2001a). These proteins associate with chromosomes early in mitosis and are detected at their centromeric regions by metaphase. In anaphase, they redistribute to the spindle midzone and equatorial cortex and accumulate in the midbody during cytokinesis. Consistent with this dynamic behavior, mutation or depletion of aurora B causes defects in multiple mitotic events including chromosome segregation and cytokinesis. Recent studies have shown that aurora B phosphorylates the N-terminal tail of histone H3, implicating a direct contribution of this kinase to mitotic chromosome dynamics (Hsu et al. 2000; Speliotes et al. 2000; Adams et al. 2001b; Giet and Glover 2001; Murnion et al. 2001). It has long been suspected that this histone H3 modification could promote the recruitment of condensin to chromosomes (e.g., Wei et al. 1999). Nevertheless, apparently different results have been obtained among different organisms or experimental systems (Giet and Glover 2001; Morishita et al. 2001; Hagstrom et al. 2002; Kaitna et al. 2002; MacCallum et al. 2002), and the exact role of histone H3 phosphorylation in metaphase chromosome morphogenesis remains elusive (Hans and Dimitrov 2001).
In this study, we have used Xenopus egg cell-free extracts to dissect the regulatory network that leads to the establishment of metaphase chromosome structure. This in vitro system allows the assembly of chromosomes independently of other mitotic events (e.g., spindle assembly), enabling us to investigate the specific contribution of different kinases to this particular process. We have found that aurora B and polo-like kinase (Plx1) act in concert to dissociate cohesin from chromatin at the onset of mitosis. Condensin is still able to bind to chromatin when cohesin release is prevented by simultaneous depletion of both kinases and leads to the formation of rod-shaped chromosomes of apparently normal length. However, the assembled chromosomes have a profound defect in the resolution of sister chromatids.
Results
Characterization of Plx1 and aurora B in Xenopus egg extracts
We have shown previously that release of cohesin from chromatin at the onset of mitosis is not accompanied by cleavage of the Rad21/Scc1 subunit but instead correlates with phosphorylation of the SA/Scc3 subunits in Xenopus egg extracts (Losada et al. 2000). In this study, we have focused on the role of two mitotic kinases, Plx1 and aurora B, in cohesin behavior. If phosphorylation of cohesin indeed promotes its release, one would expect the responsible protein kinase(s) to act on chromatin. To test whether Plx1 and aurora B met this criterion, we investigated their chromatin-binding properties in the cell-free extracts. Sperm chromatin was first incubated with an interphase extract and then a half volume of cytostatic factor (CSF)-arrested extract was added to induce the transition to a mitotic state. Aliquots were taken from the reaction mixture at different time points, and chromatin fractions were isolated and analyzed by immunoblotting. Consistent with our previous work (Losada et al. 1998), cohesin was progressively loaded onto chromatin during interphase, and most complexes dissociated upon entry into mitosis (Fig. 1A, panel 1). Dissociation of cohesin coincided with association of condensin with chromatin (Fig. 1A, panel 2). Although Plx1 and aurora B were detectable in both interphase and mitotic chromatin (Fig. 1A, panels 3,4), their kinase activities appear to be regulated in a cell-cycle-dependent manner. Plx1 displayed a retarded migration during mitosis (Fig. 1A, panel 3, lanes 9–13), which probably reflects its mitosis-specific activation by phosphorylation (Qian et al. 1998). Although aurora B did not show a mobility shift upon mitotic entry, its binding partner INCENP did (Fig. 1A, panel 5). At the same time, phosphorylation of histone H3 at serine 10 was detected on chromatin (Fig. 1A, panel 7). Extending previous reports (Adams et al. 2000; Wheatley et al. 2001), we have found that aurora B and INCENP associate with survivin/BIR1 in a stoichiometric manner (Fig. 1B) and form a highly stable complex with a sedimentation coefficient of ∼11S (Fig. 1C). Consistently, aurora B, INCENP, and survivin/BIR1 are present at the same time in interphase and mitotic chromatin (Fig. 1A, panels 4–6).
Figure 1.
Characterization of mitotic kinases in Xenopus egg extracts. (A) Sperm chromatin was incubated with interphase low-speed supernatants (LSS) to assemble interphase nuclei; a half volume of CSF LSS was added to trigger entry into mitosis. Chromatin fractions were isolated at the times indicated (lanes 3–13) and analyzed by immunoblotting with antibodies specific to Xenopus Rad21 (XRAD21, panel 1), XCAP-H (panel 2), Plx1 (panel 3), Xenopus aurora B (XAUB, panel 4), Xenopus INCENP (XINC, panel 5), Xenopus survivin/BIR1 (XBIR1, panel 6), and histone H3 phosphorylated at serine 10 (H3-P, panel 7). An aliquot of extract (lane 1) and a sample from a mock assembly reaction without sperm chromatin (lane 2) were also included. (B) Immunoprecipitations were performed from interphase high-speed supernatants (HSS) using affinity-purified antibodies against INCENP (lanes 1,2), aurora B (lanes 3,4), and BIR1 (lanes 5,6). The corresponding antigen peptides were added at 0.4 mg/mL to demonstrate the specificity of these reactions (lanes 2,4,6). The immunoprecipitates were analyzed by immunoblotting with the antibodies indicated. Alternatively, the immunoprecipitates obtained with the anti-XINC antibody in the absence (lane 7) or presence (lane 8) of XINC peptide were analyzed by silver staining. The positions of the three polypeptides as well as the IgG heavy chains (IgG HC) and light chains (IgG LC) are indicated. (C) An aliquot of an interphase HSS was overlaid onto a 5%–20% sucrose density gradient and spun at 36,000 rpm for 14 h in a SW50.1 rotor (Beckman). Fractions were taken manually and analyzed by immunoblotting. Aurora B, INCENP, and survivin/BIR1 comigrate in the indicated 11S peak.
Dissociation of cohesin from chromatin is suppressed in the absence of Plx1 and aurora B
To test the role of Plx1 and aurora B in cohesin dissociation, the two kinases were immunodepleted from Xenopus egg extracts either individually or simultaneously (Fig. 2A). The efficiency of immunodepletion was >99% in all cases, as judged by quantitative immunoblotting. Histone H1 kinase activity in each of the depleted extracts was indistinguishable from that in a mock-depleted extract (Fig. 2B), demonstrating that normal levels of cdc2 activity can be maintained in the absence of Plx1 and/or aurora B.
Figure 2.
Immunodepletion of Plx1 and aurora B blocks cohesin dissociation at the onset of mitosis. (A) Interphase HSS were immunodepleted with control IgG (lanes 1–3; Δmock), anti-Plx1 (lane 4; ΔPlx1), a mixture of anti-XINC and anti-XAUB (lane 5; ΔXAUB), or a mixture of anti-Plx1, anti-XINC, and anti-XAUB (lane 6; ΔPlx1ΔXAUB). To estimate the efficiency of depletions, 1-μL aliquots of each extract were analyzed by immunoblotting (lanes 4–6) alongside different amounts of the mock-depleted HSS (0.01, 0.1, and 1 μL; lanes 1–3). Similar results were obtained for immunodepletion of interphase and CSF extracts (both LSS and HSS). (B) Histone H1 kinase activity was assayed in aliquots of interphase HSS (I, light gray bars) and CSF HSS (M, dark gray bars) immunodepleted as in A. (C) Sperm chromatin was incubated with the depleted interphase HSS for 2 h and then each sample was split into two aliquots. One aliquot was processed for chromatin isolation without further incubation (lanes 3–6); mitosis was induced in the other by addition of a half volume of CSF HSS (lanes 7–10). After incubation for another 2 h, chromatin fractions were isolated and analyzed by immunoblotting with the indicated antibodies (panels 1–6). An aliquot of mock-depleted interphase HSS (lane 1) and a sample from a mock assembly reaction without sperm chromatin (lane 2) were analyzed in parallel.
Sperm chromatin was first incubated with interphase extracts that had been mock-depleted or depleted of Plx1, aurora B, or Plx1 and aurora B. The assembly mixtures were then converted into a mitotic state by adding CSF-arrested extracts that had been depleted accordingly. Aliquots were taken from each sample, and chromatin fractions were analyzed by immunoblotting. We found that interphase loading of cohesin was comparable in the four extracts (Fig. 2C, panel 1, lanes 3–6). Upon mitotic entry, most cohesin dissociated from chromatin in the mock-depleted extracts (Fig. 2C, panel 1, lane 7). Whereas depletion of either Plx1 or aurora B alone resulted in a partial defect in cohesin release, simultaneous depletion of both kinases blocked the dissociation of cohesin almost completely (Fig. 2C, panel 1, lanes 8–10). Mitotic loading of condensin on chromosomes was largely unaffected in the singly or doubly depleted extracts (Fig. 2C, panel 2, lanes 7–10). As an additional control, the behavior of ISWI, a chromatin-remodeling ATPase, was analyzed in parallel. Depletion of Plx1 had little effect on the mitotic dissociation of ISWI, whereas this dissociation was greatly suppressed in the absence of aurora B, as reported previously (Fig. 2C, panel 3, lanes 7–10; MacCallum et al. 2002). Cohesin release remains under the control of aurora B even in the absence of ISWI, suggesting that this event is independent of ISWI dissociation from chromatin (data not shown). Finally, phosphorylation of histone H3 was notably diminished in the aurora B-depleted extracts, but not in the Plx1-depleted extracts (Fig. 2C, panel 6, lanes 7–10). Thus, these results suggest that aurora B cooperates with Plx1 to drive the prophase release of cohesin.
Plx1 and aurora B phosphorylate different substrates in vitro
We next tested whether Plx1 or aurora B phosphorylates cohesin in vitro. The cohesin complex was isolated from interphase extracts on protein A-agarose beads and treated with affinity-purified kinases in the presence of [γ32P]ATP. Two bands comigrating with the SA and Rad21 subunits were labeled in the bead-bound fraction incubated with Plx1 (Fig. 3A, lane 2), whereas no labeling was apparent in the fraction incubated with aurora B (Fig. 3A, lane 3). In agreement with our previous report, cdc2–cyclinB modified only the SA1 subunit (Fig. 3A, lane 1; Losada et al. 2000). Similar results were obtained in a complementary experiment in which soluble cohesin was incubated with aurora B and Plx1 fractions immobilized on protein A-agarose beads (data not shown).
Figure 3.
Plx1 and aurora B phosphorylate different substrates in vitro. (A) An immobilized fraction of interphase cohesin was mixed with a soluble fraction containing cdc2–cyclin B (lane 1), Plx1 (lane 2), or XAUB (lane 3) in the presence of [γ32P]ATP. Following incubation for 1 h, the beads were washed and the bound cohesin fractions were analyzed by SDS-PAGE followed by autoradiography. A single lane of the silver-stained gel is also shown (left). (B) Chromosomes were assembled in mock-depleted (lanes 1,3,5) and aurora B-depleted (lanes 2,4,6) mitotic HSS in the presence of [γ32P]ATP, isolated in a sucrose cushion and fractionated by SDS-PAGE. The gel was stained with silver (lanes 1,2) and incorporation of the radioactive label was detected by autoradiography (lanes 3,4). Chromosomes were also assembled in parallel reactions without [γ32P]ATP and analyzed by immunoblotting with anti-phosphoH3 antibody (lanes 5,6).
To identify potential chromosomal substrates of aurora B, mitotic chromosomes were assembled in mock-depleted and aurora B-depleted extracts containing [γ32P]ATP and analyzed by silver staining (Fig. 3B, lanes 1,2) and autoradiography (Fig. 3B, lanes 3,4). Two chromosomal polypeptides were labeled in the mock-depleted but not in the aurora B-depleted extract. One is most likely to be INCENP, which is heavily phosphorylated in mitosis but was removed from the extract upon depletion of aurora B. The other is the embryo-specific form of histone H1 (Ohsumi et al. 1993). Aurora B-dependent phosphorylation of histone H3 was evident by immunoblotting with the phospho-specific antibody (Fig. 3B, lanes 5,6), although it was obscured in the autoradiograph because of comigration of H3 with the heavily phosphorylated histone H2B (de la Barre et al. 2001). Thus, only a limited number of chromosomal polypeptides, including histone H3, are phosphorylated in an aurora B-dependent manner. The observation that Plx1 and aurora B phosphorylate different substrates in vitro, together with the finding that depletion of each protein from the extracts has an additive effect in preventing cohesin release, suggests that the two kinases contribute to this event through different pathways (see Discussion).
Rod-shaped chromosomes are assembled in the absence of Plx1 and aurora B
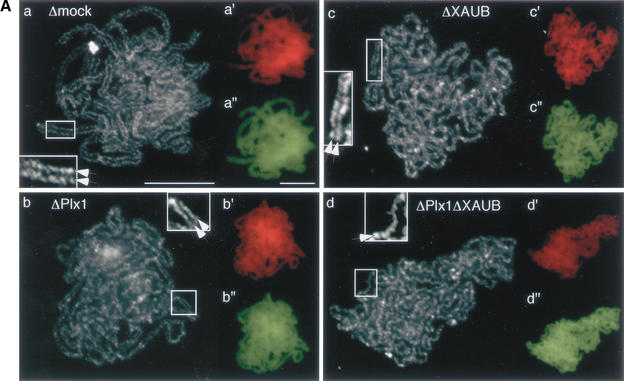
The biochemical data presented above indicated that simultaneous depletion of aurora B and Plx1 prevented the release of cohesin from chromatin but did not affect the loading of condensin (Fig. 2C). To test whether the population of condensin present on mitotic chromatin under this condition was functional, we analyzed mitotic chromosome assembly in the absence of the two kinases. Chromatin was first assembled in interphase extracts that had been mock-depleted or depleted of Plx1, aurora B, or Plx1 and aurora B, and then driven into mitosis by adding CSF-arrested extracts that had been depleted accordingly. The final products of these reactions were then fixed and examined by immunofluorescence. We found rod-shaped chromosomes in all four assembly mixtures (Fig. 4, top row). Staining of these chromosomes with an antibody against the cohesin subunit SA1 confirmed that depletion of Plx1 and aurora B had a synergistic effect on cohesin dissociation (Fig. 4, bottom two rows). These results indicate that condensin not only can bind to chromatin but also can drive chromosome compaction, despite the abnormally high levels of chromosome-bound cohesin. This finding argues against the widely accepted view that loosening of cohesion at the onset of mitosis may be a prerequisite of the action of condensin. Importantly, however, we noticed that the chromosomes obtained under the four different conditions were not completely alike: Their width decreased when the chromosomal level of cohesin became higher (Fig. 4, second row). This observation prompted us to examine, in greater detail, the structure of chromosomes assembled in the presence or absence of Plx1 and aurora B.
Figure 4.
Cohesin release is not a prerequisite of chromosome compaction. Interphase nuclei were assembled from sperm chromatin in interphase LSS that had been mock-depleted, Plx1-depleted, aurora B-depleted, or Plx1/aurora B-depleted. After 2 h, a half volume of the corresponding CSF extract was added and the mixture was incubated for another 2 h. Samples were fixed and stained with DAPI (red; top row) and anti-XSA1 antibody (green; third row). Merged images are shown in the fourth row. Chromosomal regions were selected from the DAPI images (indicated by rectangles) and their close-ups are shown in the second row (indicated by an asterisk). All images of the antibody staining were taken using the same exposure time. Bar, 10 μm.
Sister chromatid resolution is defective in the absence of Plx1 and aurora B
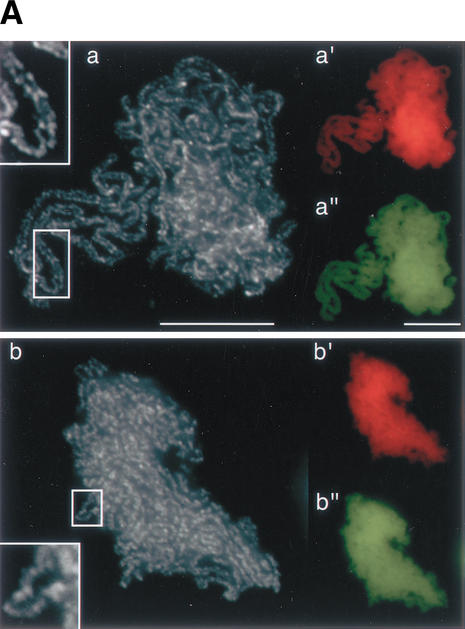
Before analyzing the degree of pairing between the sister chromatids within the assembled chromosomes, it was important to verify that chromosomal DNA was fully duplicated during interphase in the depleted extracts. To assay DNA replication, biotinylated dATP was added into the reaction mixtures and its incorporation was visualized by staining the chromosomes with fluorescein-conjugated avidin. To score the pairing between sister chromatids, we used an anti-condensin antibody that labels the chromatid axis (Losada et al. 1998). Finally, the overall morphology of chromosomes was visualized with DAPI. This triple-staining protocol was applied to chromosomes assembled in mock-depleted, Plx1-depleted, aurora B-depleted, and Plx1/aurora B-depleted extracts (Fig. 5A). Biotinylated dATP was uniformly incorporated throughout the chromosomes, demonstrating that chromosomal DNA is fully duplicated under the four different conditions (Fig. 5A, a"–d"). Sister chromatids were tightly paired but clearly resolved along the entire length of the chromosomes assembled in the mock-depleted extracts (Fig. 5A, a). In the Plx1-depleted extracts, two chromatids could be recognized in most chromosomes, yet they were less resolved than in the mock-depleted extracts (Fig. 5A, b). Similar structures were observed in the chromosomes produced in the aurora B-depleted extracts (Fig. 5A, c). Simultaneous depletion of Plx1 and aurora B had a more profound effect on sister chromatid resolution, leading to the formation of chromosomes in which two sister chromatids could no longer be discerned (Fig. 5A, d). When chromatin was first assembled in Plx1/aurora B-depleted interphase extracts and then incubated with CSF-arrested extracts containing Plx1 and aurora B, no defects in cohesin dissociation or sister chromatid resolution were observed (data not shown). This result suggests that depletion of aurora B and Plx1 only affects the mitotic behavior of cohesin but does not interfere with loading and/or establishment of cohesion during interphase.
Figure 5.
Sister chromatid resolution is defective in the absence of Plx1 and aurora B. (A) Chromosomes were assembled in mock-depleted (a,a‘,a"), Plx1-depleted (b,b‘,b"), aurora B-depleted (c,c‘,c"), and Plx1/aurora B-depleted (d,d‘,d") extracts, as described in Figure 4. Chromosomes were fixed and stained with an antibody against XCAP-E (grayscale images, a–d), DAPI (red, a‘–d‘), and FITC-conjugated avidin (yellow, a"–d"). Boxes show close-ups of chromosomal regions in which the sister chromatids (indicated by arrows) can be clearly seen. (B) Images of individual chromosomes assembled in mock-depleted (a,a‘) and Plx1/aurora B-depleted (b,b‘) extracts. For comparison, two examples of single-chromatid chromosomes assembled in the absence of DNA replication are shown (c,c‘). Chromosomes were stained with anti-XCAP-E (a–c) or FITC-conjugated avidin (a‘–c‘). (C) Quantitative analysis of chromosomes assembled in mock-depleted (left) and Plx1/aurora B-depleted (right) extracts. The distribution of fluorescence intensity (from XCAP-E staining) was measured along lines of fixed length (∼1.8 μm) drawn perpendicular to the path of the sister chromatids at regular intervals along the length of the chromosomes. Y-axis, relative fluorescence intensity of grayscale images; X-axis, distance along the drawn lines, from left to right. Bars: A, 10 μm; B,C, 5 μm.
Individual chromosomes were also observed in the assembly mixtures prepared from the depleted extracts (Fig. 5B). The average length of the chromosomes assembled in the mock-depleted and Plx1/aurora B-depleted extracts was measured to be 14.9 μm (n = 86; ς = 6.8) and 15.0 μm (n = 82; ς = 7.1), respectively. Thus, the level of axial compaction was very similar between the two sets of chromosomes. In contrast, the resolution of their sister chromatids was clearly different (Fig. 5B, a,b), and the chromosomes assembled in the Plx1/aurora B-depleted extracts morphologically resembled the “single-chromatid” chromosomes obtained in the absence of DNA replication (Fig. 5B, c). However, the incorporation of biotinylated dATP indicated that they contain fully duplicated sister DNA molecules (Fig. 5B, b‘).
An additional quantitative analysis was performed to further compare the morphology of the chromosomes assembled in the presence or absence of Plx1/aurora B. The fluorescence intensity of the condensin staining was measured along lines that had been drawn perpendicular to the sister chromatids at intervals. In the chromosome assembled in the mock-depleted extract, the fluorescence intensity was distributed in two distinct peaks corresponding to the axis of the sister chromatids (Fig. 5C, left). In contrast, a single peak of fluorescence was observed in most measurements from the chromosome assembled in the Plx1/aurora B-depleted extract (Fig. 5C, right). In this chromosome, condensin staining revealed a single ribbon-like structure twisted around itself. Such a structure could reflect an abortive condensation product in which the two sister chromatids are highly intertwined with each other.
The action of Plx1 and aurora B in sister chromatid resolution is independent of that of topoisomerase II
To better understand the mechanistic relationship between cohesin release and decatenation of the sister DNAs, we tested the contribution of topoisomerase II (topo II) to these processes. Nuclei were first assembled from sperm chromatin in interphase extracts and then converted into a mitotic state in the presence or absence of the topo II inhibitor ICRF-193 (Fig. 6A). In the presence of the drug, chromosomes tended to be clustered in a mass, and very few individual chromosomes could be found in the assembly mixture. The poor resolution of sister chromatids within these chromosomes was reminiscent of that observed in the chromosomes assembled in the absence of Plx1 and aurora B. Two results indicate, however, that the action of Plx1 and aurora B in sister chromatid resolution is independent of that of topo II. First, the decatenating activity of topo II, which was abolished by ICRF-193 (Fig. 6B, left), was not affected by depletion of the two kinases (Fig. 6B, right). Second, the inhibition of topo II activity did not prevent either cohesin dissociation or condensin loading on chromatin (Fig. 6C). Thus, we conclude that two distinct events contribute to the resolution of sister chromatids. One is the destabilization of cohesin-mediated linkage that is under the control of Plx1 and aurora B, and the other is the dissolution of DNA-mediated linkage that is catalyzed by topo II. From a mechanistic point of view, the former is likely to be a prerequisite of the latter, as a release of cohesion would help to shift the action of topo II towards decatenation of the sister DNAs.
Figure 6.
Distinct roles of topo II and Plx1/aurora B in sister chromatid resolution. (A) Sperm chromatin was incubated with interphase LSS for 2 h, and then a half volume of CSF extract without (a,a′,a") or with (b,b′b") 10 μM ICRF-193 was added. After incubation for another 2 h, chromosomes were fixed and stained as in Figure 5A. Bar, 10 μm. (B) A kinetoplast DNA decatenation assay was used to measure topo II activity in CSF extracts in the absence (lanes 2–5) or presence (lanes 6–9) of 10 μM ICRF-193, or in mock-depleted (lanes 11–14) or Plx1/aurora B-depleted (lanes 15–18) CSF extracts. Samples were taken at the indicated times, and the DNA was deproteinized and run on a 0.8% agarose gel containing 0.5 mg/mL ethidium bromide. The substrate DNA was run in lanes 1 and 10. The catenated population was retained in the wells of the gel whereas the decatenated products (∼3 kb circular DNA, relaxed or supercoiled) migrated to the positions indicated. (C) Chromatin fractions were isolated from the interphase and mitotic assembly mixtures described in A and analyzed by immunoblotting with the indicated antibodies. A sample from a mock assembly reaction (without sperm chromatin) was analyzed in parallel (lane 1).
Discussion
Establishing metaphase chromosome structure: interplay between cohesion release, DNA decatenation, and chromatid compaction
The main goal of this study was to elucidate the regulatory network that contributes to the establishment of metaphase chromosome structure. In metazoan cells, this process is accompanied by two major events. The first one is dissociation of most cohesin, which occurs independently of proteolytic cleavage of the Rad21/Scc1 subunit, and the second is association of condensin that drives the compaction of each chromatid. We have found that this prophase release of cohesin is completely blocked when aurora B and Plx1 are simultaneously depleted from Xenopus egg extracts. This observation has allowed us to address the mechanistic relationship between the loss of cohesion and the initiation of condensation. We show that condensin is able to bind to chromosomes even in the presence of abnormally high levels of cohesin. Furthermore, rod-shaped chromosomes are formed whose linear compaction is indistinguishable from that achieved in chromosomes assembled in control extracts. The resolution of sister chromatids, however, is severely compromised in the absence of Plx1 and aurora B. Neither DNA replication nor the decatenating activity of topo II is affected by depletion of the two kinases. Therefore, although it is formally possible that Plx1 and/or aurora B may affect other yet unknown factors involved in the resolution process, we think it most likely that the observed resolution defect is a direct consequence of the failure to remove cohesin from chromatin. In a recent paper, Sumara et al. (2002) reported that Plx1 promotes the prophase release of cohesin from chromatin in Xenopus egg extracts and that, in the absence of this kinase, condensin can be loaded onto chromatin containing a high level of cohesin. However, these authors did not assess the consequences that the failure of cohesin release had on metaphase chromosome structure. In this study, we have been able to assemble and analyze individual chromosomes under a condition in which cohesin dissociation is prevented. Our data strongly suggest that the prophase release of cohesin is essential to allow resolution of two sister chromatids within each chromosome, but it is not a prerequisite of the compaction process.
These results, together with our previous work (MacCallum et al. 2002), demonstrate that the chromatin loading and the action of condensin are not altered when H3 phosphorylation is drastically reduced by depletion of aurora B in Xenopus egg extracts. Plx1 is not required for condensin functions, either. Condensin binding to chromatin is only abolished in the absence of cdc2 or when a pseudomitotic state is induced by addition of okadaic acid to interphase extracts that do not contain cyclin B (A. Losada, unpubl.). All of the data support the idea that condensin is directly regulated by cdc2 in these cell-free extracts (Kimura et al. 1998). Nevertheless, regulation of condensin may vary among different organisms. For example, depletion of aurora B by RNA interference (RNAi) blocks efficient targeting of condensin in Drosophila (Giet and Glover 2001). In Caenorhabditis elegans, condensin function in prometaphase does not require aurora B (Kaitna et al. 2002), whereas its centromeric accumulation in metaphase does (Hagstrom et al. 2002). In S. cerevisiae, Ipl1/aurora kinase activity is not essential for condensin action (Lavoie et al. 2002). Thus, regulation of condensin is unlikely to be an evolutionarily conserved, essential role of aurora B.
We propose that three different kinases regulate the establishment of metaphase chromosome structure in Xenopus egg extracts (Fig. 7A). Activation of cdc2 at the onset of mitosis leads, directly or indirectly, to the activation of aurora B and Plx1, which in turn cooperate in the release of cohesin from chromatin. Cdc2 may have an additional role in preventing re-association of the released cohesin with chromatin (Losada et al. 2000). Cdc2 also phosphorylates condensin (Kimura et al. 1998), and this initiates chromosome compaction independently of cohesin dissociation. Complete resolution of the sister chromatids requires topo II-mediated decatenation of intertwined sister DNAs, and this action of topo II may be facilitated by condensin-mediated compaction (Hirano 2000). Thus, the coordinated completion of the resolution and compaction processes leads to the assembly of metaphase chromosomes that are fully competent for segregation in anaphase.
Figure 7.
(A) Regulatory networks that lead to the establishment of metaphase chromosome structure. (B) Roles of aurora B and polo-like kinase in S. cerevisiae and metazoans. For details, see the text.
Plx1 and aurora B have distinct roles in the destabilization of sister chromatid cohesion
This study provides several lines of evidence that aurora B acts in concert with Plx1 to destabilize cohesin-mediated sister chromatid linkage. First, simultaneous depletion of the two kinases is required for blocking the dissociation of cohesin completely, as judged by both biochemical and immunofluorescence analyses. Differences in the efficiency of the immunodepletion reactions and/or in the particular conditions of the dissociation assays may account for the different results obtained by Sumara et al. (2002) who reported that Plx1 alone, but not aurora B, is required for cohesin release. Second, depletion of Plx1 and aurora B also causes synergistic defects in the resolution of sister chromatids. These results argue that Plx1 and aurora B contribute to cohesin dissociation through distinct mechanisms. Consistent with this notion, only Plx1 can phosphorylate the regulatory subunits of cohesin in vitro. This phosphorylation could reduce the affinity of cohesin for chromatin, as suggested by Sumara et al. (2002), as well as by our previous results with cohesin phosphorylated by cdc2 (Losada et al. 2000). Recent structural and biochemical studies of the cohesin complex show that the non-SMC regulatory subunits bridge the two catalytic ends of the SMC heterodimer (Anderson et al. 2002; Haering et al. 2002), indicating that cohesin release may result from the opening and/or closing of the two SMC arms modulated by the non-SMC subunits.
What is the major target(s) of aurora B? We find that histone H3 and the linker histone H1 are among the few chromosomal polypeptides whose phosphorylation depends on aurora B in mitotic extracts. Although it is conceivable that aurora B phosphorylates a very low abundant chromatin component that catalyzes cohesin release, an alternative possibility is that phosphorylation of histone H3 directly contributes to the destabilization of cohesin-mediated linkage. The negative charges introduced by the phosphate groups may weaken the affinity of the histone H3 tail for DNA and enhance the structural flexibility of the chromatin fibers or, even more actively, increase the electrostatic repulsion between these fibers. Such structural changes of chromatin could, in turn, facilitate the release of cohesin complexes that had been “primed” by Plx1-mediated phosphorylation. This idea provides a novel insight into the role of histone H3 phosphorylation in mitosis and may explain why this modification is essential in Tetrahymena (Wei et al. 1999) but not in S. cerevisiae (Hsu et al. 2000; Lavoie et al. 2002). In the latter organism, the prophase release of cohesin is not observed, and cohesin dissociates in a single step at the metaphase–anaphase transition via separase-mediated cleavage of a cohesin subunit (Uhlmann et al. 1999). Accordingly, the essential role of S. cerevisiae Ipl1/aurora in chromosome segregation is to regulate kinetochore function and not to promote sister chromatid separation (Biggins et al. 1999).
Clearly, further work is required to understand the molecular mechanism by which each of the two kinases, Plx1 and aurora B, drives cohesin dissociation at the onset of mitosis. Equally challenging will be to find out how a small population of cohesin, enriched in the pericentromeric regions and essential to maintain cohesion during mitosis, is spared from this prophase release.
Dual roles of aurora B in sister chromatid resolution and kinetochore bi-orientation
Resolution of sister chromatids by metaphase is likely to be essential for their rapid and synchronous separation at the onset of anaphase in metazoans (for review, see Losada and Hirano 2001). We suggest here that the resolution process may also contribute to the assembly of properly oriented sister kinetochores. In S. cerevisiae, kinetochore bi-orientation is established immediately after DNA replication and is regulated by Ipl1/aurora (Biggins and Murray 2001; Tanaka et al. 2002; Fig. 7B, left). In higher eukaryotes, however, bipolar attachment of microtubules to chromosomes is achieved during prometaphase when the kinetochore structure is matured on progressively compact and resolved sister chromatids. In this scenario, aurora B has dual functions to ensure faithful chromosome segregation (Fig. 7B, right). The first function is to promote sister chromatid resolution and in this way facilitate the assembly of bi-oriented kinetochores during prophase. Phosphorylation of histone H3 and its centromere-specific variant CENP-A could play a direct role in this process (Zeitlin et al. 2001; Blower et al. 2002). The second is to regulate bipolar attachment of microtubules to kinetochores in prometaphase, possibly through phosphorylation of additional substrates localized at the kinetochore region, as has been shown in S. cerevisiae (Biggins et al. 1999; Kang et al. 2001; Tanaka et al. 2002). This model accounts for the complex phenotypes observed upon aurora B depletion in C. elegans and Drosophila, which include the formation of poorly resolved (“dumpy”) chromosomes, nondisjunction of sister kinetochores, and defects in metaphase chromosome alignment (Adams et al. 2001b; Kaitna et al. 2002).
The proposed dual functions of aurora B are mechanistically coupled in the mitotic chromosome cycle. In meiosis, however, the two processes occur at two different stages: Cohesion between chromosome arms is dissolved in meiosis I whereas bi-orientation of sister kinetochores occurs in meiosis II. This temporal and spatial distinction could make it easier to understand the role of aurora B in meiosis than in mitosis. In fact, a functional link between sister chromatid cohesion and histone H3 phosphorylation was first suggested by cytological observations in maize meiosis (Kaszas and Cande 2000). More recently, two studies in C. elegans have shown that aurora B functions in the release of chromosome cohesion in meiosis I (Kaitna et al. 2002; Rogers et al. 2002). Our current findings suggest that, despite apparent differences, the fundamental role of aurora B in chromosome segregation may be common in both mitosis and meiosis.
Materials and methods
Reagents
Rabbit polyclonal antisera were raised against synthetic peptides corresponding to the C-terminal amino acid sequences of Xenopus cdc2 (KSSLPDNQIRN), Plx1 (QSSKSAVAHVKASA), and XBIR1 (EYFSNQHHCSIDLDH). Polyclonal antisera were also raised against a recombinant fragment (amino acids 225–598) of Plx1. Immunization and affinity purification were performed as described previously (Hirano et al. 1997). Other antibodies used in this study were as follows: anti-XRAD21 (Losada et al. 1998); anti-XSA1 (Losada et al. 2000); anti-XCAP-E and anti-XCAP-H (Hirano et al. 1997); anti-phosphoH3 (Kimura and Hirano 2000); anti-XINC, anti-XAUB, and anti-XISWI (MacCallum et al. 2002).
Cloning of cDNAs for Xenopus BIR1
To clone cDNAs for Xenopus BIR1 (XBIR1), two degenerate PCR primers were designed on the basis of amino-acid sequence homology between the human, mouse, and Drosophila orthologs. The first primer (BR1) encodes the amino acid sequence MAEAGFI (amino acids 38–43 in human survivin) on the coding strand, and the second one (BR4) encodes the sequence GWEP D(DE)(DN) (amino acids 66–72 in human and mouse survivin; 79–85 in Drosophila CG12265) on the noncoding strand. The nucleotide sequences of the primers are as follows: BR1, 5′-GAGGAATTCATGGC(TCAG)GA(AG)GC(TCAG)GG(TCAG) TT(TC)AT-3′ (EcoRI-tag sequence is underlined; 256-fold degenerate); and BR4, 5′-GATGAATTCT(CT)(AGTC)TC(AG)T C(AGTC)GG(TC)TCCCA(AGTC)CC-3′ (EcoRI tag sequence is underlined; 512-fold degenerate). The two primers were used for RT–PCR with poly (A)+ RNA isolated from Xenopus egg extracts as a template (Hirano and Mitchison 1993). A resulting 123-bp cDNA fragment was subcloned and used as a hybridization probe to screen a Xenopus oocyte cDNA library (Stratagene). Out of 13 independent clones obtained, 8 encoded a full-length protein of 160 amino acids. The sequence is available from GenBank/EMBL/DDBJ (accession no. AY100639).
Preparation and immunodepletion of Xenopus egg extracts
Cytostatic factor (CSF)-arrested, low-speed supernatants (LSS) of Xenopus eggs were prepared in XBE2 buffer [10 mM K-Hepes (pH 7.7), 0.1 M KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA, and 50 mM sucrose] according to Murray (1991) with minor modifications (Losada et al. 1998). Interphase LSS were generated by addition of 100 μg/mL cyclohexamide and 0.4 mM CaCl2 to CSF-arrested LSS followed by incubation at 22°C for 30 min. In some experiments, LSS were further fractionated by centrifugation at 50,000 rpm at 4°C for 2 h in a Beckman TLS55 rotor to yield high-speed supernatants (HSS). Affi-Prep Protein A Support (Bio-Rad) was used for immunodepletion of LSS whereas protein A-agarose beads (GIBCO-BRL) were used for depletion of HSS. The following amounts of antibodies were bound to 25 μL of beads to deplete 50 μL of extract: for Plx1 depletion, 10 μg of anti-Plx1 (1 h × 2); for aurora B depletion, 3 μg of anti-XAUB plus 10 μg of anti-XINC (1 h), and 3 μg of anti-XINC plus 10 μg of anti-XAUB (1 h); for double depletion of Plx1 and aurora B, 10 μg of anti-Plx1 were included in the first and second incubations. Control depletions were performed using beads coated with non-immune rabbit IgG.
Biochemical analysis of chromatin and chromosomes assembled in vitro
Chromatin was assembled in vitro by incubating Xenopus sperm nuclei with interphase HSS (3000 nuclei/μL extract) or LSS (1000 nuclei/μL) that had been supplemented with an energy mix (1 mM MgATP, 10 mM creatine phosphate, and 50 μg/mL creatine kinase). After incubation at 22°C for 2 h, the extract was converted to a mitotic state by adding a half volume of CSF-arrested extract. Samples were taken and diluted fivefold (in the case of HSS) or 10-fold (in the case of LSS) with XBE2 buffer containing 0.25% Triton X-100. The chromatin fractions were isolated by centrifugation as described previously (MacCallum et al. 2002).
Morphological analysis of chromatin and chromosomes assembled in vitro
Sperm nuclei (800–1000 nuclei/μL) were incubated with freshly depleted interphase LSS supplemented with energy mix and 4 μM biotin-14-dATP at 22°C for 2 h. The extracts were then driven into mitosis by addition of a half volume of depleted CSF-arrested extract and replenished with energy mix. Incubation was then continued for another 2 h. The assembly mixtures were fixed with 10 volumes of 2% paraformaldehyde in XBE2 containing 0.25% Triton X-100 for 10 min, centrifuged onto coverslips, and processed for immunofluorescence as described previously (Losada et al. 1998). Rhodamine (TRITC)-conjugated donkey anti-rabbit secondary antibody (Jackson ImmunoResearch) was used at 6 μg/mL. Incorporation of biotin-14-dATP was detected with fluorescein (FITC)-conjugated avidin (Vector), and the DNA was counter-stained with DAPI. A Zeiss Axiophot microscope (Carl Zeiss, Inc.) equipped with a cooled charge-coupled device (CCD) camera (Photometrics Ltd.) was used to obtain grayscale images, which were later pseudocolored and merged using Adobe Photoshop. The image analysis shown in Figure 5C, as well as measurement of individual chromosome lengths, was carried out with the NIH image software (http://rsb.info.nih.gov/nih-image).
In vitro phosphorylation of cohesin
Plx1 and aurora B were purified from CSF-arrested HSS (prepared in XBE2 containing 10 mM β-glycerophosphate) by affinity chromatography. One milliliter of extract was incubated at 4°C for 2 h with 100 μL of protein A-agarose beads to which 100 μg of anti-Plx1 or 62.5 μg of anti-XINC antibody had been covalently coupled, and the solution was poured into a column. After extensive washing, the bound kinase was eluted from the column with XBE2-gly (XBE2 containing 10% glycerol instead of 50 mM sucrose) containing 0.5 mg/mL antigen peptide. The peak fractions were pooled and supplemented with 1 mM DTT. Cohesin was immunoprecipitated from 100 μL of interphase HSS using 2.5 μg of anti-XSA1 covalently coupled to 10 μL of protein A-agarose beads. After washing, the cohesin-bound beads were incubated with the kinase fractions. The reaction mixtures (15 μL) contained 0.1 mg/mL ovalbumin, 80 mM β-glycerophosphate, 0.1 mM MgATP, and 10 μCi [γ32P]ATP (6000 Ci/mmole) in XBE2-gly, and either 5 μL of ∼6 nM aurora B or 5 μL of ∼100 nM Plx1 or 0.5 μL of ∼600 nM cdc2–cyclin B (Losada et al. 2000). After incubation at room temperature for 1 h, the beads were washed with XBE2 containing 80 mM β-glycerophosphate, and the bead-bound proteins were separated by SDS-PAGE and analyzed by silver staining and autoradiography.
Other assays
Immunoprecipitation and sucrose gradient centrifugation were performed using HSS as described previously (Hirano et al. 1997). H1 kinase assay was carried out according to Murray (1991), and incorporation of [γ-32P]ATP was quantitated with an image analyzer (BAS 2000 Fuji Photofilm). DNA decatenation assay was performed exactly as described by Shamu and Murray (1992) using mock-depleted and aurora B/Plx1-depleted HSS. For comparison, the same assay was repeated in the presence or absence of the topo II inhibitor ICRF-193.
Acknowledgments
We thank A. Kumagai and W. Dunphy for a Plx1 expression plasmid and P. Budde and R. Heald for an aliquot of anti-Plx1 antibody. We are grateful to Juan Méndez and members of the Hirano laboratory for critically reading the manuscript. This work was supported by grants from the National Institutes of Health and the Human Frontier Science Program to T.H. A.L. was supported by the Robertson Research Fund and the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hirano@cshl.org; FAX (516) 367-8815.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.249202.
References
- Adams RR, Wheatley SP, Gouldsworthy AM, Kandels-Lewis SE, Carmena M, Smythe C, Gerloff DL, Earnshaw WC. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001a;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and Aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol. 2001b;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G, Uhlmann F, Mechtler K, Poupart M-A, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid cohesion in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes & Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Barre A-E, Angelov D, Molla A, Dimitrov S. The N terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J. 2001;20:6383–6393. doi: 10.1093/emboj/20.22.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila Aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover DM, Hagan IM, Tavares AAM. Polo-like kinases: A team that plays throughout mitosis. Genes & Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes & Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans F, Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters J-M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Hirano T. Chromosome cohesion, condensation and separation. Annu Rev Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hsu J-Y, Sun Z-W, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Kaitna S, Pasierbek P, Jantsch M, Loidl J, Glotzer M. The aurora B kinase, AIR-2, regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr Biol. 2002;12:798–812. doi: 10.1016/s0960-9822(02)00820-5. [DOI] [PubMed] [Google Scholar]
- Kang J-S, Cheeseman IM, Kallstrom G, Velmurugan S, Barnes G, Chan CSM. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszas E, Cande WZ. Phosphorylation of histone H3 is correlated with changes in the maintenance of sister chromatid cohesion during meiosis in maize, rather than the condensation of the chromatin. J Cell Sci. 2000;113:3217–3226. doi: 10.1242/jcs.113.18.3217. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano T. Dual roles of the 11S regulatory subcomplex in condensin functions. Proc Natl Acad Sci. 2000;97:11972–11977. doi: 10.1073/pnas.220326097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. 13S condensin actively reconfigures DNA by introducing global positive writhe: Implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Kimura K, Cuvier O, Hirano T. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J Biol Chem. 2001;276:5417–5420. doi: 10.1074/jbc.C000873200. [DOI] [PubMed] [Google Scholar]
- Lavoie BD, Hogan E, Koshland D. In vivo dissection of the chromosome condensation machinery: Reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol. 2002;156:805–815. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Orr-Weaver TL. The molecular basis of sister-chromatid cohesion. Annu Rev Cell Dev Biol. 2001;17:753–777. doi: 10.1146/annurev.cellbio.17.1.753. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano T. Shaping the metaphase chromosome: Coordination of cohesion and condensation. BioEssays. 2001;23:924–935. doi: 10.1002/bies.1133. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DE, Losada A, Kobayashi R, Hirano T. ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol Biol Cell. 2002;13:25–39. doi: 10.1091/mbc.01-09-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita J, Matsusaka T, Goshima G, Nakamura T, Tatebe H, Yanagida M. Bir1/cut17 moving from chromosome to spindle upon the loss of cohesion is required for condensation, spindle elongation and repair. Genes Cells. 2001;6:743–763. doi: 10.1046/j.1365-2443.2001.00459.x. [DOI] [PubMed] [Google Scholar]
- Murnion ME, Adams RR, Callister DM, Allis CD, Earnshaw WC, Swedlow JR. Chromatin-associated protein phosphatase regulates aurora-B and histone H3 phosphorylation. J Biol Chem. 2001;276:26656–26665. doi: 10.1074/jbc.M102288200. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Ohsumi K, Katagiri C, Kishimoto T. Chromosome condensation in Xenopus mitotic extracts without histone H1. Science. 1993;262:2033–2035. doi: 10.1126/science.8266099. [DOI] [PubMed] [Google Scholar]
- Qian YW, Erikson E, Li C, Maller JL. Activated polo-like kinase Plx1 is required at multiple points during mitosis in Xenopus laevis. Mol Cell Biol. 1998;18:4262–4271. doi: 10.1128/mcb.18.7.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E, Bishop JD, Waddle JA, Schumacher JM, Lin R. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Biol. 2002;157:219–229. doi: 10.1083/jcb.200110045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing JA, Gregson HC, Zhou S, Yokomori K. A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol Cell Biol. 2000;20:6996–7006. doi: 10.1128/mcb.20.18.6996-7006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Murray AW. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Uren A, Vaux D, Horvitz HR. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to control chromosomes and the spindle midzone. Mol Cell. 2000;6:211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- Steen RL, Cubizolles F, Le Guellec K, Collas P. A kinase-anchoring protein (AKAP)95 recruits human chromosome-associated protein (hCAP)-D2/Eg7 for chromosome condensation in mitotic extract. J Cell Biol. 2000;149:531–536. doi: 10.1083/jcb.149.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters J-M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redermann N, Nigg EA, Peters J-M. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJR, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters J-M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Warren WD, Steffensen S, Lin E, Coelho P, Loupart M-L, Cobbe N, Lee JY, McKay MJ, Orr-Weaver TL, Heck MMS, et al. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol. 2001;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]