Abstract
The plant hormone auxin regulates many aspects of plant growth and development. Although several auxin biosynthetic pathways have been proposed, none of these pathways has been precisely defined at the molecular level. Here we provide in planta evidence that the two Arabidopsis cytochrome P450s, CYP79B2 and CYP79B3, which convert tryptophan (Trp) to indole-3-acetaldoxime (IAOx) in vitro, are critical enzymes in auxin biosynthesis in vivo. IAOx is thus implicated as an important intermediate in auxin biosynthesis. Plants overexpressing CYP79B2 contain elevated levels of free auxin and display auxin overproduction phenotypes. Conversely, cyp79B2 cyp79B3 double mutants have reduced levels of IAA and show growth defects consistent with partial auxin deficiency. Together with previous work on YUCCA, a flavin monooxygenase also implicated in IAOx production, and nitrilases that convert indole-3-acetonitrile to auxin, this work provides a framework for further dissecting auxin biosynthetic pathways and their regulation.
Keywords: Auxin, indole-3-acetic acid, tryptophan, glucosinolates, cytochrome P450, flavin monooxygenase
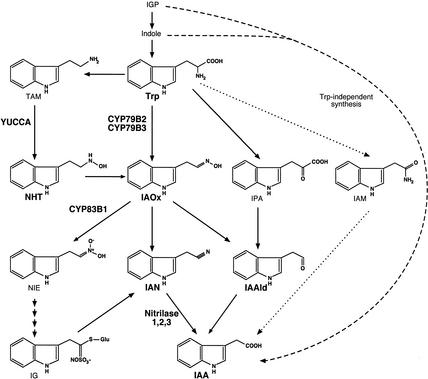
Indole-3-acetic acid (IAA), the predominant naturally occurring auxin, is implicated in almost every aspect of plant growth and development, including cell division, cell elongation, cell differentiation, tropism, flower development, and vascular system patterning. Although much progress has been made in defining IAA signaling pathways, the biosynthesis of IAA and its regulation by environmental and developmental signals remain poorly understood, although several biosynthetic schemes have been proposed (Fig. 1; Normanly and Bartel 1999; Bartel et al. 2001; Ljung et al. 2001b).
Figure 1.
Proposed pathways for IAA biosynthesis. Proposed Trp-dependent pathways are shown, and key intermediates and enzymes discussed in the text are in bold. Trp-independent IAA biosynthesis is indicated by the dashed arrow, and Agrobacterium pathway for IAA biosynthesis is indicated by the dotted arrow. IAA, indole-3-acetic acid; IAAld, indole-3-acetaldehyde; IAM, indole-3-acetamide; IAN, indole-3-acetonitrile; IAOx, indole-3-acetaldehyde; IG, indole glucosinolate; IGP, indole-3-glycerol phosphate; IPA, indole-3-pyruvic acid; NHT, N-hydroxyl tryptamine; NIE, 1-aci-nitro-2-indolyl-ethane; TAM, tryptamine; Trp, tryptophan.
Genetic dissection of IAA biosynthesis has proven difficult, and no IAA-deficient mutants have been identified. Many of the difficulties in dissecting IAA biosynthesis by using genetics have been attributed to redundancy between and within these various proposed IAA biosynthesis pathways (Fig. 1). Although labeling studies indicate that both tryptophan (Trp)-dependent and Trp-independent IAA biosynthesis pathways function in plants, it is not clear whether all proposed Trp-dependent pathways exist in all plant species and whether these pathways are redundant with respect to their in planta functions.
Relative to the number of proposed pathways, few IAA biosynthesis genes have been identified and characterized. Among the genes proposed to function in IAA biosynthesis, only the YUCCA gene has been implicated in IAA biosynthesis both genetically and biochemically (Zhao et al. 2001). YUCCA encodes a flavin monooxygenase that catalyzes the N-hydroxylation of tryptamine, a proposed step in IAA biosynthesis (Fig. 1). The gain-of-function yucca mutant displays phenotypes strongly indicative of IAA overproduction; namely, light-grown yucca mutants have elongated hypocotyls and epinastic cotyledons, whereas dark grown yucca plants have short hypocotyls and lack an apical hook. This mutant was shown to have elevated endogenous levels of free IAA resulting from increased transcription of the YUCCA gene. In addition, the mutant is resistant to toxic Trp analogs such as 5-methyltryptophan (5MT), further indicating that YUCCA is a key component of Trp-dependent IAA biosynthesis. YUCCA has 10 homologs in the Arabidopsis genome, some of which have been shown to be functionally redundant with YUCCA. For example, overexpression of either YUCCA or YUCCA2 in Arabidopsis leads to IAA overproduction phenotypes, but loss-of-function mutants of yucca and yucca2 fail to display any developmental phenotypes, consistent with genetic redundancy in the biosynthetic pathway as had been proposed (Zhao et al. 2001).
Nitrilases that can convert indole-3-acetonitrile (IAN) to IAA are also proposed to function in IAA synthesis (Fig. 1). Unlike YUCCA, nitrilase activity appears not to be rate-limiting in IAA biosynthesis because gain-of-function nitrilase mutants fail to display any developmental phenotypes (Normanly et al. 1997). Four nitrilases genes, of which three encode enzymes capable of IAN metabolism, have been identified in Arabidopsis (Bartel and Fink 1994; Piotrowski et al. 2001). The nitrilase genes may also have overlapping functions because a NIT1 loss-of-function mutant does not have developmental phenotypes. Nitrilases and YUCCA have been proposed to function in the same IAA biosynthetic pathway, with YUCCA catalyzing the rate-limiting step upstream of the nitrilases (Fig. 1; Zhao et al. 2001). However, it is not known how N-hydroxyl tryptamine (NHT) generated by YUCCA is converted to IAN and then finally converted to IAA. It is also not clear whether IAN is a necessary downstream intermediate in the YUCCA pathway or whether other intermediates, such as indole-3-acetaldehyde (IAAld), could also play a role in IAA biosynthesis. Nevertheless, it is apparent that other intermediates are needed between NHT and IAN or IAAld in the proposed YUCCA pathway. One possibility is that NHT is converted to indole-3-acetaldoxime (IAOx), which is then converted to IAN or IAAld (Fig. 1). Therefore, determination that IAOx is a precursor for IAA would provide crucial evidence in support of the proposed IAA biosynthetic pathway mediated by YUCCA.
The Arabidopsis cytochrome P450 enzymes encoded by CYP79B2 and CYP79B3 have previously been implicated in Trp metabolism and indolic glucosinolate (IG) biosynthesis (Hull et al. 2000; Mikkelsen et al. 2000). Both enzymes catalyze the conversion of Trp to IAOx in vitro, suggesting that they could partake in IAA biosynthesis if IAOx, indeed, is an important intermediate for IAA biosynthesis. There is some indirect evidence that IAOx is a key intermediate in IAA biosynthesis. The second step in IG formation is the conversion of the IAOx to 1-aci-nitro-2-indolyl-ethane catalyzed in Arabidopsis primarily by CYP83B1 (Bak et al. 2001). The sur2 and rnt-1 mutants are loss-of-function alleles of CYP83B1, and these plants have a 50% reduction in IGs and display long hypocotyls and epinastic cotyledons caused by IAA overproduction (Barlier et al. 2000; Bak et al. 2001). The blockage of IG synthesis, and concomitant increase in the IAOx levels, presumably leads to elevated levels of IAA in cyp83B1 mutants. However, there is no direct in planta evidence supporting the hypothesis that IAOx is a key IAA biosynthesis intermediate. In fact, it is generally believed that the primary function of these CYP79B2 and CYP79B3 is to promote glucosinolate biosynthesis (Mikkelsen et al. 2000; Wittstock and Halkier 2002). Furthermore, there has been significant doubt as to whether IAOx-mediated IAA synthesis is generally important because IG synthesis and by inference, IAOx production, are limited mostly to Brassica species.
Herein we present in planta evidence that CYP79B2 and CYP79B3 play critical roles in IAA biosynthesis. Overexpression of CYP79B2 in Arabidopsis leads to phenotypes nearly identical to IAA overproduction mutants such as yucca and sur2/rnt1. Correspondingly, cyp79B2 cyp79B3 double mutants have short hypocotyls and smaller stature, as would be expected for partially IAA-deficient plants. IAA levels are elevated in CYP79B2 overexpressors and reduced in the cyp79B2 cyp79B3 double mutant. This work establishes the role of these cytochrome P450s in IAA biosynthesis and, furthermore, demonstrates that formation of IAOx is a key step in IAA biosynthesis. Together with previous work on YUCCA and the nitrilases, this work provides a framework for further dissection of the IAA biosynthesis machinery.
Results
Overexpression of CYP79B2 in Arabidopsis leads to IAA-overproduction phenotypes
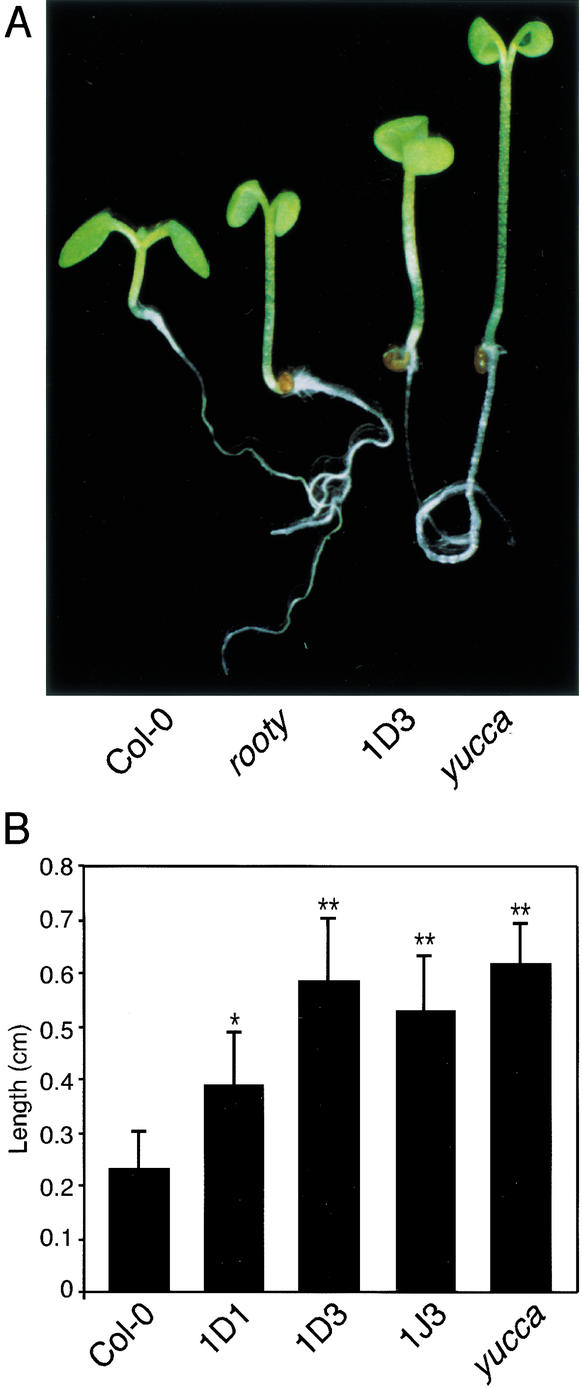
We previously constructed transgenic Arabidopsis that overexpress CYP79B2 under the control of the Cauliflower Mosaic Virus 35S promoter (referred to hereafter as CYP79B2ox; Hull et al. 2000). Light-grown CYP79B2ox lines displayed long hypocotyls and epinastic cotyledons relative to wild-type controls (Fig. 2). Such phenotypes are nearly identical to characterized IAA overproduction mutants such as yucca (Fig. 2A; Zhao et al. 2001), rty/sur1, sur2, and iaaM overexpressors (Klee et al. 1987; Boerjan et al. 1995; King et al. 1995; Barlier et al. 2000), indicating that CYP79B2 may have a role in IAA biosynthesis or regulation.
Figure 2.
CYP79B2 overexpressors display phenotypes consistent with increased IAA synthesis. (A) CYP79B2ox line 1D3 shows elongated hypocotyls and epinastic cotyledons compared with Col-0 and appears more similar to known IAA overproducers, rooty and yucca. (B) CYP79B2ox lines have longer hypocotyls than Col-0. CYP79B2ox lines 1D1, 1D3, and 1J3; yucca; and Col-0 were grown in high light (65–90 μE m−2/sec) on vertically oriented PNS agar medium. Five days after germination, hypocotyl length was measured by using NIH Image software. Each bar is representative of eight samples; error bars indicate S.D. All measurements are in centimeters. The differences between Col-0 and lines 1D3, 1J3, and yucca are significant, with p < 0.01 (*) and p < 0.001 (**) using Neuman-Keuls ANOVA.
IAA-regulated genes are induced by CYP79B2 overexpression
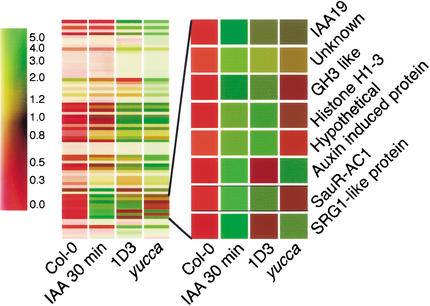
One consequence of IAA overproduction in Arabidopsis is the increased expression of IAA-inducible genes, including IAA/AUX, SAUR, and GH3 (Zhao et al. 2001). By using microarray analysis, we found that expression of these genes in CYP79B2ox line 1D3 is in fact several-fold higher than in wild type (Fig. 3; Table 1). Moreover, the gene expression profiles of yucca and CYP79B2ox are very similar; >70% of the genes that are induced in yucca are also induced in the CYP79B2ox line, 1D3 (Fig. 3; Table 1).
Figure 3.
CYP79B2 overexpression results in the expression of IAA-induced genes similar to the expression pattern of the yucca mutant. Cluster analysis is shown for genes induced in CYP79B2ox line 1D3 and yucca, and by 2-h treatment of Col-0 with 1 μM IAA. Color bar at left indicates the relative expression level. The expanded region shows known IAA-induced genes.
Table 1.
Microarray analysis of gene expression in CYP79B2ox and yucca
| Accession
|
Gene product description
|
CYP79B2ox
|
yucca
|
30 min IAA
|
2 hr IAA
|
4 days IAA
|
|---|---|---|---|---|---|---|
| A. Known auxin inducible genes | ||||||
| U18408 | IAA6 gene | ∼7.6 | ∼3.6 | 6.3 | ∼21.6 | ∼−1.8 |
| AF013294 = A_TM018A10.6 | similar to auxin-induced protein | ∼4.8 | ∼2.7 | ∼4.0 | ∼3.5 | ∼2.5 |
| AC005396 = T26I20.12 | member of auxin-responsive GH3 protein multigene family | ∼4.3 | ∼2.4 | ∼1.9 | ∼22.1 | ∼2.2 |
| U18407 | Arabidopsis thaliana IAA5 gene | ∼13.6 | ∼7.8 | ∼38.9 | ∼33.8 | ∼2.3 |
| AL035656 | small auxin up RNA (SAUR-AC1) | 10 | 6 | 26.6 | 26.1 | 1.6 |
| AC002391 = T20D16.20 | similar to auxin-responsive GH3 protein | 8.8 | 5.8 | 9.1 | 30.1 | 2.6 |
| S70188 | SAUR-AC1 = small auxin up RNA A. thaliana | 6.6 | 4.6 | 19.4 | 16 | 1.5 |
| L15448 | auxin-responsive protein (IAA1) mRNA | 6.1 | 4.8 | 7.1 | 11.3 | 1.6 |
| U49075 | early auxin-induced (IAA19) mRNA, partial cds. | 5.4 | 4.9 | 8.9 | 16.1 | 1.3 |
| AC00621 = T27K22.12 | member of auxin-induced protein multigene family | 3.6 | 2.4 | 3.7 | 11.5 | −1.3 |
| B. Stress related genes | ||||||
| U13949 | heat shock protein AtHSP101 (Athsp101) | ∼5.8 | ∼3.2 | ∼−1.1 | ∼−1.8 | ∼6.2 |
| AC006081 = T2G17.21 | pathogenesis-related protein, PR-1 | ∼5.0 | ∼1.8 | ∼2.1 | ∼1.5 | ∼16.4 |
| AC007212 = F8D23.7 | putative peroxidase | ∼5.5 | ∼3.6 | ∼1.7 | ∼−3.4 | ∼−1.1 |
| AC005314 = T32F12.24 | putative peroxidase | ∼5.1 | −1 | 3 | 5.2 | ∼−1.9 |
| X16076 | hsp17.6 mRNA for 17.6-kD heat-shock protein | ∼4.6 | ∼5.1 | ∼2.8 | ∼−2.0 | ∼2.7 |
| Y14070 | mRNA for heat shock protein 17.6A | ∼4.2 | ∼3.2 | ∼−1.6 | ∼−1.0 | ∼3.6 |
| AL035528 = F18A5.290 | putative disease resistance protein | ∼4.0 | 3.1 | −1 | 2.8 | 3.1 |
| AC004747 = T19L18.4 | putative heat-shock transcription factor | ∼3.8 | ∼1.9 | ∼1.4 | ∼1.6 | ∼1.9 |
| X17293 | HSP17.4 gene for 17.4-kD heat-shock protein | ∼14.9 | ∼6.5 | ∼1.0 | ∼−3.8 | 4.7 |
| AC007661 = T8P21.32 | putative alcohol dehydrogenase | ∼12.6 | ∼2.7 | ∼−1.4 | ∼−1.3 | ∼2.7 |
| AC004138 = T17M13.2 | putative basic blue protein (plantacyanin) | 6.4 | 5.9 | 1.9 | 1.2 | 4.1 |
| AC004561 = F16P2.12 | putative small heat-shock protein | 6.3 | 1.6 | 1.1 | −1 | 1.8 |
| X63443 | mRNA for heat-shock protein hsp 17.6–II | 5.9 | 2.3 | −1.7 | ∼−8.4 | 4.8 |
| AC002521 = T20F6.1 | basic blue protein, 5′ partial | 5.9 | 5.2 | 2.4 | 1.1 | 3.6 |
| AL035528 = | putative disease resistance protein | 5.4 | 3 | 2.5 | 2.8 | 2.4 |
| L04173 | glycine rich protein (RAB18) gene, complete cds. | 4.9 | 5.8 | 2.4 | 1.1 | 3.5 |
| AJ002551 | mRNA for heat-shock protein 70 | 4.8 | 2.8 | 1.4 | −2.4 | 4.3 |
| Y14070 | nRNA for heat-shock protein 17.6A | 4.7 | 2.8 | −1.3 | 1.4 | 3.3 |
| Z99708 = C7A10.720 | similar to hypothetical protein YNL231c, Saccharomyces cerevisiae | 3.6 | 2 | 2 | 1.9 | 4.3 |
| AC004138 = T17M13.2 | putative basic blue protein (plantacyanin) | 3.7 | 3.6 | −1.3 | −1.7 | 2.7 |
| C. Transcription factors | ||||||
| Z95758 | mRNA for AtMYB34 R2R34-MYB transcription factor | ∼4.6 | 2.9 | ∼1.6 | 2.1 | ∼3.3 |
| AF062886 | putative transcription factor (MYB50) mRNA | ∼4.6 | ∼2.9 | ∼5.5 | ∼3.1 | ∼1.6 |
| AC005397 = T3F17.4 | putative AP2 domain transcription factor | ∼3.9 | ∼2.1 | 1.6 | ∼3.1 | ∼1.7 |
| AF062876 | putative transcription factor (MYB34) mRNA | ∼10.7 | ∼6.1 | ∼5.0 | ∼2.6 | ∼5.4 |
| AF062860 | putative transcription factor (MYB4) mRNA | 12.2 | 1.5 | 1.2 | 1.6 | 2.1 |
| U20193 | MADS-box protein AGL12 (AGL12) mRNA | 5.2 | 2.3 | 3.3 | ∼−1.7 | ∼−1.3 |
| AL023094 | Homeodomain-like protein | 4.6 | 2.7 | 4.5 | 3.5 | 4 |
| Z97048 | mRNA for AtMYB13 R2R3-MYB transcription factor | 3.8 | 1.4 | −1.2 | 1.8 | 2.2 |
| AF062876 | putative transcription factor (MYB34) mRNA, partial cds. | 3.8 | 1.4 | 1.2 | −2 | 1.7 |
| D. Others | ||||||
| AC005167 = F12A24.7 | putative glucosyltransferase | ∼4.1 | ∼1.2 | ∼1.8 | ∼1.4 | ∼2.6 |
| AC003033 = T21L14.7 | putative glucanse | 6.8 | 2.4 | 3.5 | ∼−2.8 | 2.8 |
| AL021961 | glucosyltransferase-like protein | 3.7 | 1.7 | 1.1 | 3.6 | 1.2 |
| Y12089 | peroxiredoxin A. thaliana Per1 gene | ∼6.4 | ∼11.0 | ∼1.3 | ∼−1.8 | ∼15.2 |
| M22033 | A. thaliana at2S4 gene encoding albumin 2S subunit 4 | ∼6.2 | ∼3.0 | ∼4.1 | ∼−1.4 | ∼−1.2 |
| Z97339 | indole-3-acetate β-glucosyltransferase like | ∼6.2 | ∼3.3 | ∼1.3 | ∼−1.3 | ∼3.1 |
| AL021889 = T6K21.200 | similarity to predicted protein | ∼6.1 | ∼5.3 | ∼4.0 | 1.3 | ∼3.7 |
| U53221 | GASA5 mRNA | ∼5.2 | ∼2.4 | ∼1.9 | ∼4.5 | ∼6.3 |
| AC005662 = F13H10.19 | late-embryogenesis abundant M17 protein | ∼5.1 | ∼22.7 | ∼−1.1 | ∼−1.3 | ∼15.9 |
| AC004218 = F12L6.3 | unknown protein | ∼5.1 | 2.1 | 4.5 | 10.1 | 1.8 |
| AC007060 = T5I8.22 | putative berberine bridge enzyme from A. thaliana | ∼5.0 | ∼1.9 | ∼1.1 | ∼3.2 | ∼1.3 |
| AL022140 = F1N20.170 | Unknown | ∼4.8 | ∼2.3 | ∼4.9 | ∼3.1 | ∼2.5 |
| X91918 | mRNA for oleosin type4 protein | ∼4.6 | ∼5.8 | ∼2.8 | ∼−4.3 | ∼2.7 |
| AC005310 = F19D11.7 | hypothetical protein | ∼4.8 | ∼3.3 | ∼3.4 | ∼8.3 | ∼4.4 |
| U94998 | class 1 non-symbiotic hemoglobin (AHB1) gene, complete cds. | ∼4.6 | ∼−4.5 | ∼2.4 | ∼4.7 | ∼5.6 |
| AC006587 = T17D12.5 | putative seed storage protein (vicilin-like) | ∼4.5 | ∼6.7 | ∼2.9 | ∼2.4 | ∼2.9 |
| AL035528 = F18A5.290 | putative disease resistance protein | ∼4.0 | 3.1 | −1 | 2.8 | 3.1 |
| Z99708 = C7A10.810 | putative protein | ∼4.0 | ∼1.9 | 2.3 | 2.1 | 1.2 |
| U49919 | lupeol synthase mRNA, complete cds. | ∼3.9 | ∼6.0 | ∼1.7 | ∼7.9 | ∼1.1 |
| X91960 | mRNA for major latex protein type 1 | ∼3.9 | ∼3.7 | ∼1.9 | ∼−1.4 | ∼1.6 |
| Y09006 | Arabidopsis thaliana rpoMt gene | ∼3.9 | ∼2.8 | ∼2.2 | ∼2.3 | ∼3.6 |
| AC003672 = F16B22.9 | unknown protein | ∼3.5 | ∼2.2 | ∼1.9 | ∼1.9 | ∼1.7 |
| AL049746 | ABC transporter-like protein | ∼3.5 | ∼2.6 | ∼1.7 | ∼2.3 | ∼2.4 |
| AC006053 = F3N11.3 | putative selenium-binding protein | ∼3.5 | ∼1.9 | ∼2.9 | ∼4.1 | ∼2.6 |
| AL049658 = T17F15.100 | similarity to predicted proteins | ∼3.5 | ∼3.2 | ∼2.3 | ∼1.9 | ∼2.5 |
| L43080 | A. thaliana pEARLI 1 mRNA, complete cds. | 6.9 | 2.9 | 1.3 | 7 | ∼−2.3 |
| AC006232 = F10A12.10 | putative cysteine proteinase | 6.8 | 5.5 | 4 | −2.7 | 7.5 |
| U73781 | A. thaliana histone H1-3 (His1-3) mRNA | 6.6 | 2.9 | 4.8 | 4.8 | 5.1 |
| AC005170 = T29E15.11 | putative cinnamoyl CoA reductase | 5.7 | 3.1 | 2.1 | ∼−1.3 | ∼3.3 |
| AL096860 = T21L8.170 | similarity to sn-glycerol-3-phosphate permease | 5.6 | 2.9 | 1.8 | 1 | 6.2 |
| AC006439 = T30D6.12 | putative lipid transfer protein | 4.7 | 1.4 | 1.6 | 2.9 | 1.1 |
| AC002343 = T19F6.8 | unknown protein | 4.6 | 3 | 3.4 | 2.4 | 1.4 |
| AC002341 = T14G11.20 | putative cysteine proteinase | 4.3 | 2.2 | 1.9 | 8.3 | 4.9 |
| AC007138 = T7B11.13 | encodes predicted protein of unknown functions | 4.2 | 1.6 | −1.3 | 7.3 | −2.5 |
| AC002391 = T20D16.19 | putative cytochrome P450 | 4.1 | 2.9 | 4.4 | 3.3 | 2.8 |
| D32138 | delta1-pyrroline-5-carboxylate synthase, complete cds. | 3.9 | 2.4 | 2 | −1.2 | 3.4 |
| U96045 | Arabidopsis thaliana APS reductase (PRH) mRNA, complete cds. | 3.8 | 4.5 | 3.2 | 1.3 | 7.8 |
| AL035440 = F10M23.190 | putative protein | 3.8 | 2.2 | 1.8 | −1.1 | 4.7 |
| AC006954 = F25P17.16 | unknown protein | 3.8 | 2.7 | 1.9 | −1.2 | 3 |
| AC007195 = F1P15.3 | unknown protein | 3.7 | 1.2 | 2.1 | ∼−3.6 | 1.1 |
| AC006841 = F3K23.25 | putative dehydrin | 3.7 | 4.3 | −1.7 | 1 | 2.7 |
| AC006282 = F13K3.3 | unknown protein | 3.7 | 3.1 | 1.7 | 3 | 2.9 |
| AC005106 = T25N20.20 | unknown protein | 3.7 | 1 | −1.1 | 2.3 | −1.1 |
| U78721 = T1B8.3 | cadmium-induced protein isolog | 3.7 | ∼2.6 | 2.6 | ∼3.8 | 1.1 |
| AL035601 | cytochrome P450 monooxygenase-like protein | 3.6 | 1.7 | 1.5 | 1.6 | −1.4 |
| AC004667 = T4C15.3 | putative LEA (late embryogenesis abundant) | 3.6 | 2.6 | 1.2 | −2.8 | 3 |
| Z99708 = C7A10.720 | similar to hypothetical protein YNL231c, S. cerevisiae | 3.6 | 2 | 2 | 1.9 | 4.3 |
| X13434 | Arabidopsis thaliana NIA1 mRNA for nitrate reductase NR1 | 3.5 | 2.1 | 2.4 | 4.1 | 1.2 |
| AC006841 = F3K23.8 | putative CONSTANS-like B-box zinc finger protein | 3.5 | 2.3 | 1.9 | −1.9 | 3.7 |
∼, Expression level in wild type is very low, and the signals are near or below background. The fold changes of those genes are only approximate.
Levels of IAA and IAN are elevated by CYP79B2 overexpression
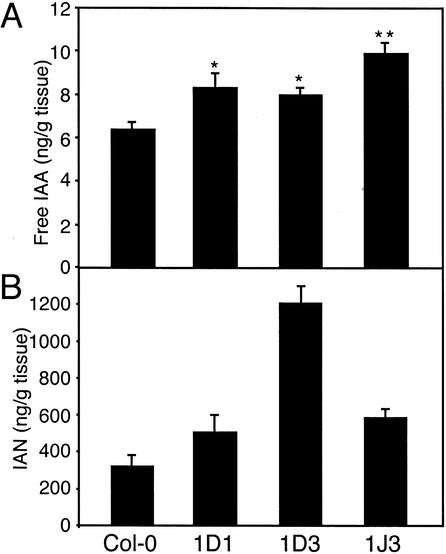
Because the morphological and gene expression phenotypes caused by CYP79B2 overexpression were consistent with IAA overproduction, we directly measured IAA levels in seedlings of CYP79B2ox lines 1D1, 1D3, and 1J3. All three CYP79B2ox lines showed modest but significant increases in free IAA, the biologically active form of IAA (Fig. 4A).
Figure 4.
Quantification of IAA and IAN in CYP79B2ox lines. Free IAA (A) and IAN (B) were measured for CYP79B2ox lines 1D1, 1D3, 1J3, and Col-0 grown in low light (18–30 μE m−2/sec) for 6 d after germination at 21°C. Values are given in nanograms per gram fresh weight of tissue, presented as the mean ± S.E.M. from three or four independent samples. Values for free IAA were subjected to Newman-Keuls ANOVA. Significant differences compared to Col-0 are indicated as p < 0.05 (*) and p < 0.001 (**).
IAN, a proposed intermediate in Trp-dependent IAA biosynthesis (Fig. 1), can be formed from IAOx either directly by the elimination of water or indirectly via the breakdown of IGs (Searle et al. 1982). CYP79B2ox lines 1D1, 1D3, and 1J3 have elevated levels of IAN compared with wild-type plants (Fig. 4B), consistent with IAN being derived from IAOx produced by CYP79B2 and CYP79B3.
IAOx formation may be a rate-limiting step in IAA production
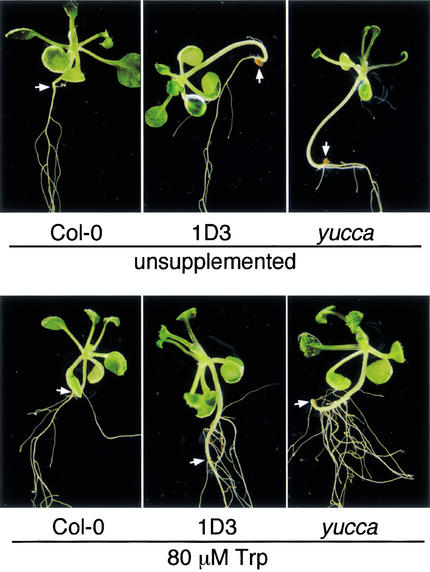
Wild-type Arabidopsis seedlings supplemented with Trp do not show IAA-related phenotypes, suggesting that Trp-dependent IAA biosynthesis is regulated at a step downstream of Trp production. Consistent with this observation, exogenous IAOx induces phenotypes resembling IAA overproduction, suggesting that conversion of Trp to IAOx is a rate-limiting step in the synthesis of IAA (Zhao et al. 2001). We suspected that CYP79B2ox lines, which presumably have increased Trp to IAOx conversion, should show more severe IAA-related phenotypes when supplemented with Trp. Therefore, we examined the effect of exogenous Trp on inducing adventitious root formation in Col-0, yucca, and the CYP79B2ox line 1D3. Adventitious root formation is indicative of increased IAA levels as exemplified by the sur1 and sur2 mutants (Boerjan et al. 1995; Celenza et al. 1995; Delarue et al. 1998). Indeed, 1D3 and yucca both showed a greater propensity for adventitious rooting compared with Col-0 when supplemented with Trp (80 μM); no differences in this regard were observed on unsupplemented medium (Fig. 5). These results are consistent with CYP79B2 overexpression causing increased Trp to IAOx conversion and subsequent IAA formation.
Figure 5.
CYP79B2 overexpression results in increased adventitious root formation when supplemented with Trp. CYP79B2ox line 1D3, yucca, and Col-0 were grown on medium with or without exogenous 80 μM Trp. The root/hypocotyl junction, above which adventious roots can form, is indicated by an arrow; note the adventitious roots on the hypocotyls of 1D3 and yucca when supplemented with Trp. CYP79B2 overexpressor 1J3 appeared similar to 1D3 under both conditions (data not shown). The plants shown are representative of each plant line.
The cyp79B2 cyp79B3 double mutant has decreased levels of IAA
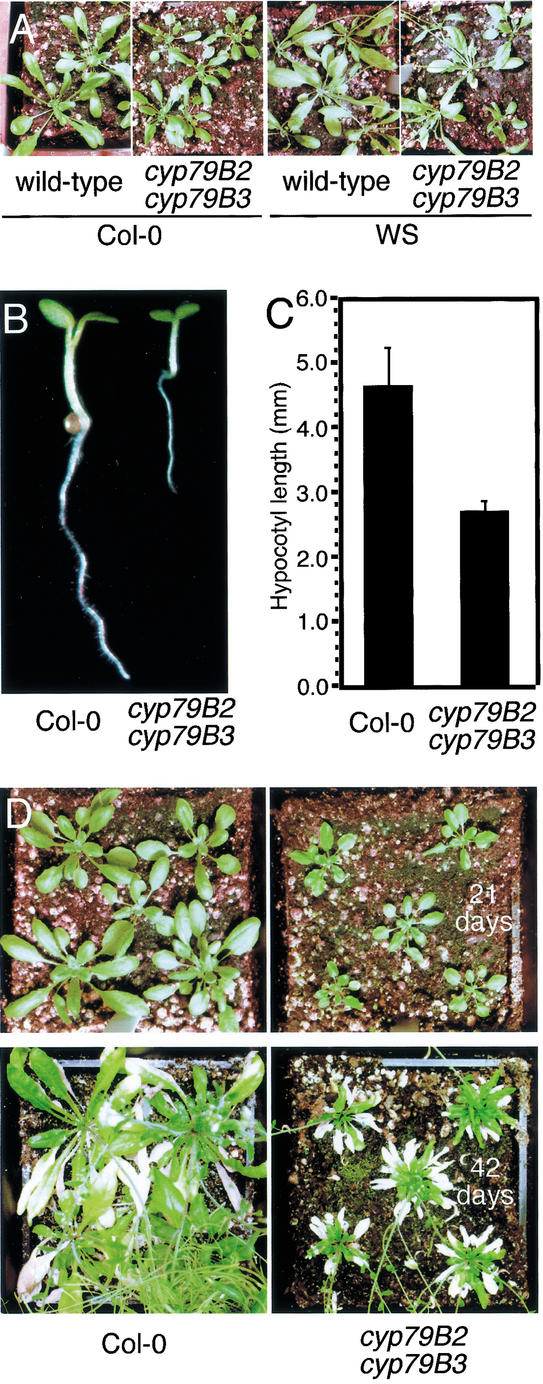
To identify loss-of-function alleles of CYP79B2 and CYP79B3, we screened available T-DNA collections of both the Col-0 and WS accessions and found T-DNA insertions into each gene for each accession. Insertion alleles in the Col-0 background were identified from the SIGnAL T-DNA collection (Salk Institute) and are located at 1512 bp from the ATG for CYP79B2 and at 1425 bp from the ATG for CYP79B3. WS alleles were identified with the assistance of the Arabidopsis Knockout Facility (AKF; http://www.biotech.wisc.edu/Arabidopsis) and are located at 1659 bp from the ATG for CYP79B2 and at 2041 bp from the ATG for CYP79B3. The WS insertions and the Col-0 CYP79B2 insertion are in the second exon, whereas the Col-0 CYP79B3 insertion is in the intron. All four insertions disrupt their respective genes upstream of the region encoding the heme-binding site conserved among cytochrome P450s and thus are expected to be null alleles. Although neither cyp79B2 nor cyp79B3 single mutants have visible phenotypes, the cyp79B2 cyp79B3 double mutants from either accession consistently showed similar subtle growth phenotypes under normal culture conditions (Fig. 6A). cyp79B2 cyp79B3 plants have slightly shorter petioles and smaller leaves, which would be expected if the double mutant produces less IAA.
Figure 6.
Growth phenotypes of the cyp79B2 cyp79B3 double mutant. (A) cyp79B2 cyp79B3 and wild-type plants from the Col-0 and WS accession were grown at 21°C under continuous light (25–35 μE m−2/sec) and were photographed at 25 d after germination. (B,C) The cyp79B2 cyp79B3 double mutant has decreased hypocotyl length compared with wild-type Col-0. Hypocotyls were measured at 4 d after germination and were grown at 26°C. (D) Soil-grown cyp79B2 cyp79B3 double mutant plants have smaller rosette leaves than wild-type Col-0 plants. Plant were grown at 19°C under continuous light (25–35 μE m−2/sec) and were photographed at the days after germination indicated.
The pool size of endogenous free IAA in Arabidopsis responds to temperature. For example, an increase in growth temperature from 20°C to 29°C increases free IAA levels twofold in Arabidopsis hypocotyls (Gray et al. 1998). This increase in IAA presumably leads to the longer hypocotyls and epinastic cotyledons observed in seedlings grown at the higher temperature. We tested the response of the cyp79B2 cyp79B3 double mutant to temperature changes. The hypocotyl length of the cyp79B2 cyp79B3 grown at 26°C was ∼50% of the length of the wild type grown under the same conditions (Fig. 6B,C). This reduction in hypocotyl length likely correlates with decreased levels of endogenous-free IAA (see below). Adult cyp79B2 cyp79B3 plants are also sensitive to temperature changes. cyp79B2 cyp79B3 grown at 19°C has much smaller, slightly curled leaves that are characteristic of known IAA-resistant mutants (Fig. 6D; Hobbie et al. 2000).
We measured IAA in the cyp79B2 cyp79B3 double mutant and in each single mutant to determine if the growth phenotypes seen in the double mutant correlated with reduced IAA levels. At 21°C there was no significant difference between the double mutant and wild type; however, at 26°C there was significantly less free IAA in cyp79B2 cyp79B3 relative to Col-0 (Table 2). In addition, the cyp79B3 mutant showed a modest decrease in free IAA compared with Col-0 at 26°C. These results indicate that the CYP79B2 and CYP79B3 genes play a role in IAA synthesis, but that alternative IAA synthesis pathways remain active. We also measured IAN in each single mutant and in the cyp79B2 cyp79B3 double mutant grown at both 21°C and 26°C. IAN is greatly reduced in the double mutant, suggesting that the CYP79B2/3 pathway plays a more substantial role in IAA biosynthesis than indicated by free-IAA measurements (Table 2).
Table 2.
Quantification of free IAA and IAN from cyp79B2 and cyp79B3 mutants
| Plants
|
Growth temperature, °C
|
Free IAA
|
IAN
|
|---|---|---|---|
| WT Col-0 | 21 | 11.9 ± 0.42 | 855 ± 23 |
| cyp79B2 | 21 | 9.9 ± 0.87 | 283 ± 21 |
| cyp79B3 | 21 | 10.0 ± 1.55 | 347 ± 36 |
| cyp79B2 cyp79B3 | 21 | 11.3 ± 0.55 | 61 ± 14 |
| WT Col-0 | 26 | 7.5 ± 0.5 | 1263 ± 105 |
| cyp79B2 | 26 | 7.0 ± 0.22 (P < 0.05) | 786 ± 39 |
| cyp79B3 | 26 | 6.7 ± 0.13 (P < 0.001) | 714 ± 150 |
| cyp79B2 cyp79B3 | 26 | 4.9 ± 0.15 (P < 0.001) | 53 ± 6 |
Free IAA and IAN levels were determined on seedlings grown aseptically on PNS agar medium under lowpass yellow filters in 18 to 30 μE m−2/sec light at 21°C or 26°C and harvested 6 d after germination. Values are given in nanograms per gram fresh weight of tissue, presented as the mean ± SE from three or four independent samples. Where ANOVA indicated, a significant difference for free IAA between mutant and the respective WT control for a given temperature, the P value is shown in parentheses.
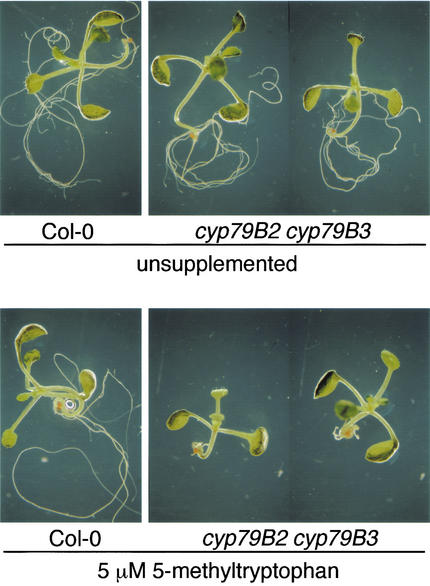
The cyp79B2 cyp79B3 double mutant is hypersensitive to 5MT
We previously reported that overexpression of CYP79B2 leads to 5MT resistance (Hull et al. 2000). Therefore we suspected that the cyp79B2 cyp79B3 double mutant would have increased sensitivity to 5MT. Wild-type plants were minimally sensitive to 5 μM 5MT, whereas the double mutant was extremely sensitive to this concentration of 5MT (Fig. 7) and appears at least threefold more sensitive to 5MT than to wild type (data not shown). The increased 5MT sensitivity of the double mutant suggests that conversion of Trp to IAOx by CYP79B2 and CYP79B3 is an important mechanism in the regulation of both IAA biosynthesis and Trp metabolism.
Figure 7.
The cyp79B2 cyp79B3 double mutant is sensitive to 5-methyltryptophan (5MT). Col-0 and cyp79B2 cyp79B3 plants (Col-0 ecotype) were grown on unsupplemented medium and on medium containing 5 μM 5MT and were photographed 8 d after germination. Shown are representative seedlings from each condition.
IAOx produced by CYP79B2 and CYP79B3 is used for both IAA and IG biosynthesis
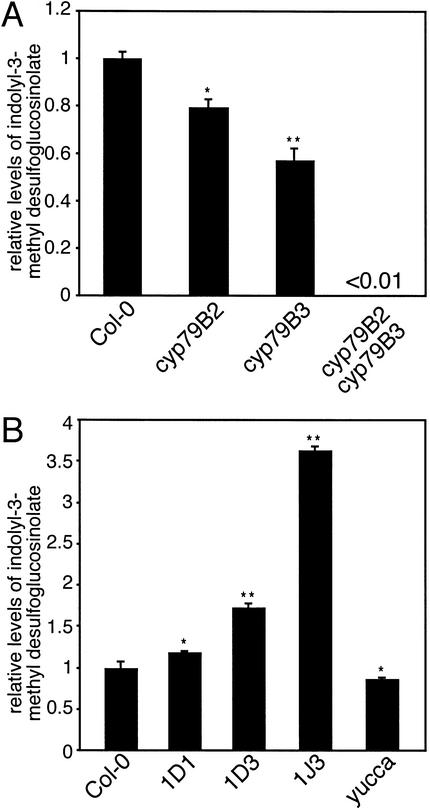
CYP79B2 and CYP79B3 have been implicated in IG biosynthesis (Mikkelsen et al. 2000), but it is unclear whether these enzymes are solely responsible for IG production. IGs are absent in cyp79B2 cyp79B3 plants to the degree detectable by our methodology (Fig. 8A), suggesting that most if not all IGs are derived from IAOx produced by CYP79B2 and CYP79B3. In addition, both the cyp79B2 and cyp79B3 single mutants have significantly reduced IG levels compared with wild type, suggesting that both CYP79B2 and CYP79B3 contribute to IG production (Fig. 8A).
Figure 8.
Analysis of indole glucosinolate production in cyp79B2 and cyp79B3 mutants and in CYP79B2ox lines. Total desulfoglucosinolates from rosette leaves of 3-wk-old plants were analyzed by HPLC as described in Materials and Methods. (A) Single cyp79B2 or cyp79B3 mutants are modestly reduced for indole glucosinolate production, whereas indole glucosinolates are at least 100-fold reduced in the cyp79B2 cyp79B3 double mutant. (B) CYP79B2ox lines accumulate indolyl-3-methyl glucosinolate, whereas yucca has a reduction. Normalized ratios are shown as the mean ± S.E.M. from three independent samples. Significant differences compared to Col-0 are indicated as p < 0.05 (*) and p < < 0.001 (**).
Mikkelsen et al. (2000) showed previously that CYP79B2 overexpression results in increased levels of indolyl-3-methylglucosinolate, the predominant IG in Arabidopsis. In agreement with this previous finding, we found that CYP79B2ox lines 1D1, 1D3, and 1J3 all showed a significant increase in indolyl-3-methyl desulfoglucosinolate (Fig. 8B), but no change in other indole desulfoglucosinolates (data not shown). The yucca mutant is predicted to have higher IAOx levels than those of wild-type plants; however, we did not observe an increase in IGs, and in fact, we found a slight decrease in IGs compared with Col-0 (Fig. 8B). This result, combined with the absence of IGs in cyp79B2 cyp79B3 plants suggests that the role of YUCCA does not extend to IG synthesis.
Discussion
Herein we present direct evidence that CYP79B2 and CYP79B3 are critical enzymes for IAA biosynthesis. From this work we conclude that IAOx is a key intermediate in Trp-dependent IAA biosynthesis, and thus we provide a framework for further investigation of IAA biosynthesis and its regulation.
CYP79B2 and CYP79B3 are key enzymes in Trp-dependent IAA biosynthesis
The first indication that CYP79B2 and CYP79B3 play important roles in IAA biosynthesis is that CYP79B2ox plants have IAA overproduction phenotypes, namely, long hypocotyls and epinastic cotyledons similar to the phenotypes observed for all other known IAA overproduction mutants such as yucca, sur1, and sur2 (Fig. 2). In addition to the phenotypic similarities between CYP79B2ox and other IAA overproduction mutants, CYP79B2ox is also very similar to yucca at the molecular level, as revealed by our microarray studies (Fig. 3; Table 1), further supporting that CYP79B2ox overproduces IAA. Furthermore, direct quantification of IAA indicates that CYP79B2ox, indeed, contains higher levels of free IAA. Together, these studies establish that overexpression of CYP79B2 leads to IAA overproduction and suggest that the conversion of Trp to IAOx, catalyzed by CYP79B2, is a rate-limiting step in IAA biosynthesis. Consistent with our analysis of CYP79B2ox gain-of-function mutants, the cyp79B2 cyp79B3 double loss-of-function mutant has phenotypes indicative of a partial IAA deficiency and, in fact, has decreased levels of free IAA (Fig. 6; Table 2).
Redundancy in IAA biosynthesis
Redundancy between and within putative IAA biosynthetic pathways (Fig. 1) has been proposed as one of the major difficulties in dissecting IAA biosynthesis mechanisms and suggested as a reason why dissection of IAA biosynthesis by forward genetics has not been successful. We previously reported that YUCCA, a flavin monooxygenase that catalyzes a key step in IAA biosynthesis, has several functional homologs in the Arabidopsis genome (Zhao et al. 2001), indicating that redundancy is responsible for the lack of visible phenotypes associated with YUCCA loss-of-function mutants. That CYP79B2 and CYP79B3 are also key enzymes in IAA biosynthesis suggests that there are indeed multiple Trp-dependent IAA biosynthesis pathways. From previous work (Hull et al. 2000) and work presented herein, it is apparent that CYP79B2 and CYP79B3 are also functionally redundant. The cyp79B2 cyp79B3 double null mutant has phenotypes consistent with decreased free-IAA levels, but such phenotypes have not been observed in the single mutants of CYP79B2 and CYP79B3. Although we have shown redundancy within both the YUCCA and CYP79B2/3 pathways, it is not clear whether the YUCCA pathway and the CYP79B2/B3 pathway are redundant with each other at the physiological level; however, we suspect that compensation from other IAA biosynthetic pathways may be one of the reasons that we only observed subtle growth phenotypes in the cyp79B2 cyp79B3 double mutant. Combinations of mutations in various yucca family members with the cyp79B2 cyp79B3 double mutant may be informative in this respect.
It is interesting that plants have so many pathways for IAA biosynthesis. Are all the proposed IAA synthesis pathways simply redundant, or do they have distinct functions in response to environmental and developmental signals? A growing body of evidence suggests that Trp-dependent and Trp-independent pathways are regulated differentially (Quirino et al. 1999; Ljung et al. 2001c). For example, recent studies in Lemna gibba have shown that IAA synthesis pathways are differentially regulated by temperature (Rapparini et al. 2002); Trp-dependent synthesis predominates at low temperatures (15°C), whereas Trp-independent synthesis predominates at high temperatures (30°C). Interestingly, we observed a more severe cyp79B2 cyp79B3 mutant phenotype at 26°C compared with 21°C, suggesting that Arabidopsis might also use temperature to regulate the choice of IAA biosynthetic pathways. Compared with wild type, the cyp79B2 cyp79B3 mutant had shorter hypocotyls and reduced free IAA at 26°C, suggesting that the IAOx Trp-dependent pathway is at least partly responsible for the temperature-induced hypocotyl elongation seen in wild-type plants. Although previous studies in Arabidopsis have shown that IAA synthesis in excised hypocotyls was increased at 29°C grown under high light, and this was responsible for a temperature-dependent increase in hypocotyl length (Gray et al. 1998), we observed a decrease in free IAA isolated from wild-type whole seedlings when grown at 26°C under low light compared with growth at 21°C under low light. We suspect that because our studies examined whole seedlings, the overall levels of free IAA are much lower than those reported for excised hypocotyls. Recent work has shown that IAA concentrations vary greatly between plant tissues, and these levels are also greatly affected by the developmental age of the tissue (Ljung et al. 2001a). Our results suggest that although elevated temperature can induce a hypocotyl-specific increase in IAA production, this local increase in IAA production is masked by our IAA measurements performed on the whole seedling. (We note that in our experiments, the hypocotyl makes up at most 15% of the fresh weight of the seedling.)
Both YUCCA- and CYP79B2/B3-mediated IAA biosynthesis exist in Arabidopsis; however, it is not clear whether all plant species use the same repertoire of IAA biosynthetic pathways to regulate in vivo IAA pools or whether different plant species use distinct sets of IAA biosynthetic pathways in response to similar developmental or environmental cues. Some plant species appear to lack certain sets of IAA biosynthesis pathways and/or have more or less redundancy within a given pathway. For example, we have observed tremendous redundancy in the YUCCA gene family in Arabidopsis, whereas in petunia, YUCCA appears to be a single-copy gene called Floozy (FLZ; Tobeña-Santamaria et al. 2002). A loss-of function flz allele has dramatic defects in vascular tissue and flower development, although IAA levels are not measurably different from wild type. We also noticed that likely orthologs of CYP79B2 and CYP79B3 have been found only in certain plant species (Bak et al. 1998), and thus this pathway for IAA biosynthesis may be limited to those species. For example, we could not find obvious CYP79B2/B3 orthologs in the rice genome, suggesting that CYP79B2/B3 pathway may not be a component of IAA biosynthesis in rice.
IAA overproduction and gene expression
IAA is known to regulate the expression levels of various genes; however, it is difficult to correlate the observed IAA-induced gene expression with particular plant growth and development responses. Comparison of the gene expression patterns of IAA-treated plants with IAA overproduction mutants, such as CYP79B2ox and yucca, may provide clues at the molecular level of why IAA treatment and IAA overproduction often lead to different phenotypes. For example, Arabidopsis plants treated with IAA often have short roots and short hypocotyls, whereas IAA overproduction mutants such as CYP79B2ox and yucca have long hypocotyls and subtle root phenotypes. As shown in Figure 3 and Table 1, many known auxin-inducible genes are induced in yucca and CYP79B2ox. Perhaps more importantly, the gene expression patterns of CYP79B2ox and yucca are different from those of IAA-treated plants. Treatment of Arabidopsis seedlings with IAA for 2 h induces expression of many known IAA-regulated genes, including AUX/IAA and GH3 genes; however, relative to yucca and CYP79B2ox plants, the expression level of these genes is considerably higher (Table 1). Apparently, a transient increase in IAA concentration promotes a transcriptional response that is quite different from continuous in planta IAA overproduction. However, the fact that phenotypes caused by in planta IAA overproduction cannot be phenocopied by continuous exposure to exogenous IAA during germination and seedling growth indicates that continuous exposure to IAA is not the only reason for the observed difference. Furthermore, as shown in Table 1, the gene expression pattern of 4-d-old seedlings grown on 1 μM IAA is very different from that of yucca and CYP79B2ox. It is not clear why exogenous IAA treatment leads to different gene expression patterns from those observed in IAA overproduction mutants such as CYP79B2ox and yucca, although the complications of polar IAA transport and tissue accessibility are likely reasons. Both of these factors could affect local IAA concentrations; subtle changes in IAA concentration can be the difference between inducing cell elongation and cell division or inhibiting these processes. These findings add another level of complexity to studying IAA biosynthesis, as we would expect difficulties in rescuing mutants partially deficient in IAA with exogenous IAA.
Relationship between IAA biosynthesis and Trp homeostasis
An understanding of how IAA biosynthesis is regulated is further complicated by the fact that IAA biosynthetic pathways share intermediates with other metabolic processes, creating the potential for cross-talk between pathways. It is logical that IAA biosynthesis has to be coordinated with Trp biosynthesis and catabolism because IAA biosynthesis uses either Trp or intermediates in Trp biosynthesis as precursors. It is generally believed that the rate-limiting steps for IAA synthesis lie downstream of Trp production because application of Trp normally does not result in IAA-associated phenotypes. That overexpression of CYP79B2 or YUCCA leads to increased levels of free IAA suggests that production of IAOx is a rate-limiting step in IAA synthesis. Although exogenous Trp does not lead to a significant increase in the level of free IAA in wild type or in CYP79B2ox 1D3 (data not shown), exogenous Trp did cause increased adventitious rooting in CYP79B2ox and yucca but not in wild type. These observations indicate that in CYP79B2 or YUCCA overexpressing lines, Trp biosynthesis, not Trp metabolism, has become rate limiting.
We have shown in this and previous work that Trp metabolism and IAA biosynthesis are in fact well coordinated. CYP79B2ox plants are resistant to toxic Trp analogs such as 5MT, whereas cyp79B2 cyp79B3 plants are hypersensitive to 5MT, indicating that the CYP79B2/3 pathway plays a role in regulating the in vivo Trp pool (Hull et al. 2000). Arabidopsis mutants that overproduce Trp biosynthetic genes also induce CYP79B2 expression. For example, the ATR1 gene encodes a Myb transcription factor that, when overexpressed, elevates expression of ASA1 and CYP79B2 (Bender and Fink 1998; Smolen and Bender 2002).
Relationship between IAA biosynthesis and IG biosynthesis
IAA and IG biosynthesis also have common intermediates. CYP79B2 overexpression has been shown previously to result in elevated IG levels (Mikkelsen et al. 2000). Our CYP79B2ox lines that have elevated IAA levels likewise have increased IGs. In addition, the cyp79B2 cyp79B3 double mutant fails to make any detectable IGs, suggesting that IAOx made from CYP79B2 and CYP79B3 is the sole source of IGs in Arabidopsis. These results imply that IAOx derived from YUCCA pathway only contributes to IAA production, and not IG production, suggesting that the tissue or intracellular location of CYP79B2/B3 and YUCCA may play a role in segregating these two pathways. Moreover, the lack of detectable IGs in the cyp79B2 cyp79B3 double mutant indicates that the cyp79B2 and cyp79B3 insertion mutations are indeed null alleles.
Our hypothesis that CYP79B2/3 pathway contributes to both IAA and IG production is further supported by analysis of the sur2/rnt1/atr4 mutants. These are all loss-of-function alleles of the gene encoding CYP83B1, the enzyme immediately downstream of CYP79B2/3 in IG biosynthesis (Barlier et al. 2000; Bak et al. 2001; Hansen et al. 2001; Smolen and Bender 2002). cyp83B1 mutants show dramatic IAA overproduction phenotypes and have elevated IAA levels, while also having a significant decrease in IGs. It has been suggested that CYP83B1 activity serves as a regulator of IAA production, by funneling excess IAOx into IGs (Bak et al. 2001). Our results show that in strong overexpressing CYP79B2ox lines, IGs are increased approximately fourfold, whereas free IAA is only slightly elevated consistent with regulatory role for CYP83B1. However, it is also possible that IAA catabolic pathways mask a more substantial increase in IAA synthesis.
A framework for Trp-dependent IAA biosynthesis
The identification of IAOx as a key intermediate in IAA biosynthesis allows us to significantly strengthen our working model for Trp-dependent IAA biosynthesis in Arabidopsis (Fig. 1). IAOx not only is a sensible link to NHT derived from YUCCA but also ties in to downstream intermediates such as IAN and IAAld (Fig. 1). At present, it is not clear how IAOx is converted to IAA. IAN generally has been considered as the primary intermediate between IAOx and IAA, although IAAld has also been proposed (Rajagopal and Larsen 1972). IAN measurements in CYP79B2ox lines and the cyp79B2 and cyp79B3 single and double mutants are consistent with IAN being a direct IAA intermediate. However, under certain conditions, IGs break down into IAN (Searle et al. 1982), and this less direct route is also consistent with our data because the levels of IAN in CYP79B2ox lines and the cyp79B2 and cyp79B3 single and double mutants mirror IG levels. Although our data do not distinguish between these two models, analysis of cyp83B1 mutants argues against IAN being derived from IGs. The IAN level in the sur2 allele of CYP83B1 is not altered (Barlier et al. 2000), although cyp83B1 mutants have <50% of the wild-type levels of IGs (Bak et al. 2001). Although further analysis will be needed, we favor a model in which IAOx contributes to IGs via CYP83B1, but contributes to IAA in a CYP83B1-independent manner, probably through an IAN or IAAld intermediate.
In summary, we show that gain-of-function and loss-of-function mutations in CYP79B2 and CYP79B3 have a corresponding increase and decrease, respectively, in levels of IAA and its metabolites. These results provide the first in planta evidence that cytochrome P450s 79B2 and 79B3 function in Trp-dependent IAA biosynthesis. These results, taken together with our previous work on the YUCCA family of flavin monooxygenases, strongly argue that Trp-dependent IAA biosynthesis via an IAOx intermediate is a major source of IAA in Arabidopsis. We are now poised to begin assessing redundancy between and within IAA synthetic pathways, to determine the overall effects of IAA synthesis on Trp metabolism, and to begin to examine the developmental and environmental regulation of IAA biosynthesis
Materials and methods
CYP79B2ox lines
Construction of CYP79B2ox lines was described previously (Hull et al. 2000). CYP79B2ox lines 1D3 and 1J3 are considered strong CYP79B2 overexpression lines based on CYP79B2 transcript levels and 5MT resistance, in which line 1D1 is considered a weaker overexpression line based on these same criteria (data not shown).
Growth conditions
Plants used in growth measurement assays were grown on Murashige and Skoog medium or unsupplemented PNS (Haughn and Somerville 1986) on 100 × 15-mm square Petri dishes at 21°C in continuous low light (18–30 μE m−2/sec) with yellow low-pass filter or in high light (65–90 μE m−2/sec). cyp79B2 cyp72B3 double mutant plants were analyzed on PNS containing 5 μM 5MT in 100 × 15-mm square Petri plates sealed with parafilm and grown under low light (18–30 μE m−2/sec). Seeds were sterilized by incubation in 1 mL sterilization solution (30% bleach, 0.01% Triton X-100) for 15 min. The seeds were vernalized 4–6 d at 4°C before being moved to the growth chamber. For hypocotyl measurements, the plates were oriented vertically.
The length of hypocotyls were measured in seedlings 5 d after germination. Pictures were taken of the plants by using a Bio-Rad Gel-Doc with Multi-Analyst software, and National Institutes of Health (NIH) Image 1.62 was used to measure the lengths of hypocotyls using the freehand line in the drawing option. Measurements were calibrated to a 1-cm scale.
Desulfoglucosinolate isolation and analysis
For desulfoglucosinolate isolation, plants were grown in soil (Fafard mix 2) in continuous light until just before bolting. Approximately 200 mg fresh leaf tissue was collected, coarsely cut with scissors, and boiled for 3 min in 1 mL 80% methanol in 15-mL conical tubes. After cooling on ice, another 1 mL of 80% methanol was added, and the samples were boiled for an additional 3 min. The samples were centrifuged at 4700 rpm in a Sorvall SH3000 rotor for 20 min at 20°C, and the supernatant was applied to a DEAE Sephadex A25 column prepared in a pasteur pipette as follows: 0.1 g DEAE Sephadex A25 swelled in 2 mL 0.5 M pyridine acetate was added to the pipette. The column was conditioned with 6 mL 0.5 M pyridine acetate and then washed with 12 mL HPLC grade water. The sample was added to the column and washed with ∼8 mL water until the flow-through was clear. One milliliter aryl sulfatase (4 mg/mL) was added, and the column was sealed and incubated overnight at room temperature. The desulfoglucosinolates were eluted with 8 mL HPLC grade water. To inactivate the sulfatase, the samples were boiled for 3 min; 1.5 mL of each sample was lyophilized to a volume of 200 μL and filtered through a 0.22-μm filter in preparation for HPLC analysis.
HPLC of desulfoglucosinolates was carried out on a Waters Wisp 710 autosampler with Rainin Dynamex model SD-2000 pumps and a Waters 996 phosphodiode array detector set at 229 and 258 nm controlled by Millennium software. Ninety microliters of each sample was separated on a Whatman Partisphere C18 column (110 × 4.7 mm inner diameter, 5-μm particle size; Hogge et al. 1988). The separation conditions were adapted from Hogge et al. (1988). A flow rate of 1 mL/min was used for the following program: 1.2% acetonitrile in water for 5 min, a linear gradient from 1.2% to 22.5% acetonitrile over the next 15 min, 22.5% acetonitrile for 5 min, a linear gradient to 70% acetonitrile for 15 min, and 70% acetonitrile for 5 min. The column was reconditioned for the next sample by a linear gradient to 1.2% acetonitrile over 10 min and washed with this concentration for 15 min until the next injection. Triplicate tissue samples were analyzed for each plant line.
Indole desulfoglucosinolates were distinguished from other glucosinolates by their absorbance at both 229 nm and 258 nm (Kiddle et al. 2001) and by their retention times compared with purified indolyl-3-methyl desulfoglucosinolate and 4-methylsulfinylbutyl desulfoglucosinolate [gifts of J. Gershenzon and M. Reichelt (Max Planck Institute for Chemical Ecology, Jena, Germany)]. Relative abundance of indole desulfoglucosinolates was determined by comparing the ratios of relative peak areas of indolyl-3-methyl desulfoglucosinolate to 4-methylsulfinylbutyl desulfoglucosinolate and normalizing the ratios to Col-0.
IAA and IAN measurements
For IAA and IAN analysis, seeds were plated in 10 mL top agar (PNS without sucrose) on PNS in 15 × 150-mm Petri plates. For each strain, a total of ∼1500 seeds were divided between five plates. Seeds were vernalized for 4–6 d at 4°C before being moved to the growth chamber, where they were grown at 21°C or 26°C in continuous light (18–30 μE m−2/sec) under low-pass yellow filters for 6 d after germination. IAA was purified by the method of Chen et al. (1988) with slight modifications that are described in Tam and Normanly (2002). IAN was purified by the method of Ilić et al. (1996). The conditions for gas chromatography selected ion monitoring mass spectrometry of IAA and IAN are described in Tam and Normanly (2002).
cyp79B2 and cyp79B3 loss-of-function mutations
The AKF at the University of Wisconsin, Madison (http://www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis/IntroductionIndex.html) was used to identify T-DNA insertions into both the CYP79B2 and CYP79B3 genes in the WS background. Primers 79B23′-2 (5′-ACTTGGTTGGGCGATTATA TAAAAGTAGT-3′) and JL-202 (5′-CATTTTATAATAACGCT GCGGACATCTAC-3′) were used to identify a T-DNA insertion at 1659 bp after the start codon of CYP79B2, and primers 79B33′-2 (5′-GAGATACTCGATGTGAATACCTTCTTATT 3′) and XR-21 (5′-TGGGAAAACCTGGCGTTACCCAACTTA AT-3′) were used to identify a T-DNA insertion at 2041 bp after the start codon of CYP79B3. T-DNA insertions were confirmed by Southern blotting and DNA sequencing.
Homozygous cyp79B2 knockouts were identified by using PCR primers 79B23′-2 and 79B25′-2 (5′-TGTATATAAATAG GAAGGTGAAGCTCTCT-3′) to look for the absence of the WT band. In addition, the plants were tested for kanamycin (Km) resistance. Similarly, homozygous cyp79B3 knockouts were identified by using primers 79B33′-2 and 79B35′2 (5′-ATAC TAATTGTTTCTCCTTCTCCTTCTTC-3′) to look for the absence of the wild-type product and by Km resistance. cyp79B2 cyp79B3 double mutants were constructed by crossing the two single mutants to each other.
T-DNA insertion lines of CYP79B2 and CYP79B3 in the Col-0 background were identified from the Salk Institute collection of T-DNA insertion lines by PCR. Primer 79B2-5P (5′-TGGA CAAGTATCATGACCCAATCATCCACG-3′) and LB primer (5′-GGCAATCAGCTGTTGCCCGTCTCACTGGTG-3′) was used to identify a T-DNA insertion at 1512 bp after the ATG of the CYP79B2 gene. The insertion is in the second exon of CYP79B2 gene. Primer 79B3-5P (5′-TGTTCTATGCATGGAC TGGTGGTCAACATG-3′) and LB were used to identify a T-DNA insertion at 1425 bp after the ATG site of CYP79B3 gene. The insertion is in the intron between the two exons of the CYP79B3 gene. We found that the T-DNA insertion in CYP79B3 gene is a tandem T-DNA insertion with LB flanking sequence at both end of the T-DNA insertion. The insertions were confirmed by DNA sequencing of the PCR fragments generated with LB primer and the gene specific primers.
Microarray analysis of gene expression
For microarray studies total RNA was extracted from 5-d-old light-grown CYP79B2ox line 1D3, yucca, and Col-0. RNA extraction, probe preparation, hybridization were carried out according to procedures described by Affymetrix Inc. Affymetrix GeneChip Suite and GeneSpring software were used to analyze the microarray data.
For gene expression patterns induced by exogenous IAA, 5-d-old Col-0 plants grown on a Petri dish were immersed with 1 μM IAA for 30 min or 2 h before they were harvested for total RNA isolation. Gene expression levels of the IAA-treated plants were compared with water-treated Col-0 plants under the same conditions to determine which genes are induced or repressed by exogenous IAA treatment. Gene expression patterns of Col-0 plants germinated and grown on media containing 1 μM IAA were also compared with those of plants germinated and grown on regular 0.5× MS media to determine the effects of continuous exposure of IAA on gene expression.
Acknowledgments
We thank the AKF and ABRC for strains; J. Gershenzon and M. Reichelt for glucosinolate standards; D.J. Waxman for assistance with HPLC and helpful discussions; and J. Perry for helpful suggestions. We also thank J. Nemhauser for help in characterizing cyp79B2 cyp79B3 double null mutants. This work was supported by the National Science Foundation (DBI-0077769; to J.L.C. and J.N.), NIH (GM52413; to J.C.), and the Howard Hughes Medical Institute (HHMI; to J.C.). N.R.G. was supported in part by a Howard Hughes Medical Institute Educational Grant to Boston University (71195-500204). Y.Z. is an HHMI Fellow of the Life Sciences Research Foundation. J.C. is an Associate Investigator of the HHMI.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL celenza@bu.edu; FAX (617) 353-6340.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1035402.
References
- Bak S, Nielsen HL, Halkier BA. The presence of CYP79 homologs in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol Biol. 1998;38:725–734. doi: 10.1023/a:1006064202774. [DOI] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R. CYP83B1, a cytochrome P450 at the metabolic branchpoint in auxin and indole glucosinolate biosynthesis in Arabidopsis thaliana. Plant Cell. 2001;13:101–111. doi: 10.1105/tpc.13.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci. 2000;97:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR. Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci. 1994;91:6649–6653. doi: 10.1073/pnas.91.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, LeClere S, Magidin M, Zolman BK. Inputs to the active indole-3-acetic acid pool: De novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J Plant Growth Regulators. 2001;20:198–216. [Google Scholar]
- Bender J, Fink GR. A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci. 1998;95:5655–5660. doi: 10.1073/pnas.95.10.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Cervera M-T, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D. superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Grisafi PL, Fink GR. A pathway for lateral root development in Arabidopsis. Genes & Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Chen K-H, Miller AN, Patterson GW, Cohen JD. A rapid and simple procedure for purification of indole-3-acetic acid prior to GC-SIM-MS analysis. Plant Physiol. 1988;86:822–825. doi: 10.1104/pp.86.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Van Onckelen H, Caboche M, Bellini C. sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 1998;14:603–611. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediate hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CH, Du L, Naur P, Olsen CE, Axelsen KB, Hick AJ, Pickett JA, Halkier BA. CYP83B1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J Biol Chem. 2001;276:24790–24796. doi: 10.1074/jbc.M102637200. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, Bandyopadhyay A, Estelle M. The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development. 2000;127:23–32. doi: 10.1242/dev.127.1.23. [DOI] [PubMed] [Google Scholar]
- Hogge LR, Reed DW, Underhill EW, Haughn GW. HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thaliana and their identification using thermospray liquid chromatography/mass spectrometry. J Chromatogr Sci. 1988;26:551–556. [Google Scholar]
- Hull AK, Vij R, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilić N, Normanly J, Cohen JD. Quantification of free plus conjugated indoleacetic acid in Arabidopsis requires correction for the nonenzymatic conversion of indolic nitriles. Plant Physiol. 1996;111:781–788. doi: 10.1104/pp.111.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiddle G, Bennett RN, Botting NP, Davidson NE, Robertson AAB, Wallsgrove RM. High-performance liquid chromatographic separation of natural and synthetic desulphoglucosinolates and their chemical validation by UV, NMR and chemical ionisation-MS methods. Phytochem Anal. 2001;12:226–242. doi: 10.1002/pca.589. [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB. A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell. 1995;7:2023–2037. doi: 10.1105/tpc.7.12.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee JH, Horsch RB, Hinchee MA, Hein MB, Hoffmann NL. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes & Dev. 1987;1:86–96. [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001a;28:465–474. doi: 10.1046/j.1365-313x.2001.01173.x. [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol. 2001b;49:249–272. [PubMed] [Google Scholar]
- Ljung K, Östin A, Lioussanne L, Sandberg G. Developmental regulation of indole-3-acetic acid turnover in Scots pine seedlings. Plant Physiol. 2001c;125:464–475. doi: 10.1104/pp.125.1.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem. 2000;275:33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- Normanly J, Bartel B. Redundancy as a way of life: IAA metabolism. Curr Opin Plant Biol. 1999;2:207–213. doi: 10.1016/s1369-5266(99)80037-5. [DOI] [PubMed] [Google Scholar]
- Normanly J, Grisafi P, Fink GR, Bartel B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell. 1997;9:1781–1790. doi: 10.1105/tpc.9.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M, Schönfelder S, Weiler EW. The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode β-cyano-L-alanine hydratase/nitrilase. J Biol Chem. 2001;276:2616–2621. doi: 10.1074/jbc.M007890200. [DOI] [PubMed] [Google Scholar]
- Quirino B, Normanly J, Amasino R. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol. 1999;40:267–278. doi: 10.1023/a:1006199932265. [DOI] [PubMed] [Google Scholar]
- Rajagopal R, Larsen P. Metabolism of indole-3-acetaldoxime in plants. Planta. 1972;103:45–54. doi: 10.1007/BF00394605. [DOI] [PubMed] [Google Scholar]
- Rapparini F, Tam Y, Cohen J, Slovin J. IAA metabolism in Lemna gibba undergoes dynamic changes in response to growth temperature. Plant Physiol. 2002;128:1410–1416. doi: 10.1104/pp.011005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle LM, Chamberlain K, Rausch T, Butcher DN. The conversion of 3-indolylmethylglucosinolate to 3-indolylacetonitrile by myrosinases, and its relavance to the clubroot disease of the Cruciferiae. J Exp Bot. 1982;33:935–942. [Google Scholar]
- Smolen G, Bender J. Arabidopsis cytochrome P450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics. 2002;160:323–332. doi: 10.1093/genetics/160.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YY, Normanly J. Overexpression of a bacterial indole-3-acetyl-L-aspartic acid hydrolase in Arabidopsis thaliana. Physiol Plant. 2002;115:513–522. doi: 10.1034/j.1399-3054.2002.1150405.x. [DOI] [PubMed] [Google Scholar]
- Tobeña-Santamaria R, Bliek M, Ljung K, Sandberg F, Mol JNM, Souer E, Koes R. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes & Dev. 2002;16:753–763. doi: 10.1101/gad.219502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002;7:263–270. doi: 10.1016/s1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]