Abstract
During development of the Drosophila peripheral nervous system, different proneural genes encoding basic helix–loop–helix transcription factors are required for different sensory organs to form. atonal (ato) is the proneural gene required for chordotonal organs and R8 photoreceptors, whereas the achaete-scute complex contains proneural genes for external sensory organs such as the macrochaetae, large sensory bristles. Whereas ectopic ato expression induces chordotonal organ formation, ectopic scute expression produces external sensory organs but not chordotonal organs in the wing. Proneural genes thus appear to specify the sensory organ type. In the ommatidium, or unit eye, R8 is the first photoreceptor to form and appears to recruit other photoreceptors and support cells. In the atonal1 (ato1) mutant, R8 photoreceptors fail to form, thereby resulting in the complete absence of ommatidia. To our surprise, we found that ectopic scute expression in the ato1 mutant induces the formation of ommatidia, which occasionally sprout ectopic macrochaetae. Remarkably, many scute-induced ommatidia lack R8 although they contain outer photoreceptors.
Neural precursors for sensory organs in Drosophila are specified by proneural genes, which are first expressed in clusters of equipotent cells, the proneural clusters, and then restricted to the cells singled out from these proneural clusters to become neural precursors (1–7). Different proneural genes, which code for transcriptional regulators with the basic helix–loop–helix motif, are required for the formation of neural precursors for different sensory organs. For example, the proneural gene atonal (ato) is required for the formation of photoreceptors (4) and chordotonal organs, which are stretch receptors or auditory sensors (3). The proneural genes in the achaete-scute complex, on the other hand, are necessary for the formation of external sensory organs, bristles that are sensitive to physical or chemical stimuli (2).
The proneural genes appear to be capable of specifying the type of neural precursor they induce, even though their expression in the proneural clusters depends on positional information, which in principle may specify the type of neural precursors that emerge from these proneural clusters. For example, ectopic expression of scute results in the formation of external sensory organs exclusively (3, 8, 10) and cannot rescue the chordotonal organ defect in the ato1 mutant (8). In the central nervous system, ectopic expression of achaete or scute, but not ato, can restore MP2 precursors of mutants lacking these precursors (11, 12). Only ato is capable of promoting ectopic chordotonal organ formation (3, 8, 9); its ectopic expression can even convert external sensory organs into chordotonal organs (9). Like chordotonal organs, photoreceptors normally depend on ato for their formation (4). It is thus of interest to determine whether photoreceptors can be induced only by ectopic expression of ato.
Drosophila eye morphogenesis begins at the posterior tip of the eye imaginal disk in early third instar larvae (13, 14). The morphogenetic furrow (MF) moves anteriorly during development. Within and posterior to the MF, regular arrays of ommatidia, the unit eyes of the compound eye, are assembled following a precise spatial and temporal order (14). Of the eight photoreceptors (R1–R8) in each ommatidium, R8 is the “founder photoreceptor” that forms first, and its formation depends on the expression of ato (4). Other photoreceptors are recruited by R8 sequentially, starting with R2 and R5, followed by R3 and R4, then R1 and R6, and finally R7; formation of the eight photoreceptors precedes the appearance of cone cells (14). Thus, the R1–R7 photoreceptors appear to acquire their cell fates by cell–cell communication processes initiated by R8 (13–16). Mosaic analysis demonstrated that the formation of R8 but not R1–R7 requires ato directly. The fact that R1–R7 and R8 all are missing in ato loss-of function mutants supports the notion that R8 is the founder photoreceptor required for the subsequent development of R1–R7 (4).
To form ommatidia (4, 17, 18), as in the formation of other sensory organs (1, 2, 6, 7), early expression of a proneural gene in a proneural cluster endows cells in the cluster with the potential to form neural precursor. Subsequent lateral inhibition mediated by cell–cell interaction singles out one cell in the proneural cluster to become neural precursor and maintain proneural gene expression (1, 2). Early expression of the proneural gene ato in the developing eye is controlled by two separate regulatory regions of the ato gene. Expression of ato is first detected in a continuous stripe of cells in the eye disk, and then is quickly resolved into regularly spaced initial clusters of cells (4, 17, 18). This early expression, immediately anterior to (in front of) the MF, is driven by the 3′F:5.8 enhancer region, located 3′ to the ato coding sequences (18). Within and posterior to (behind) the MF, ato expression is further restricted into intermediate groups and finally into evenly spaced R8 photoreceptors; this later expression pattern requires the 5′ eye enhancer region, located 5′ to the ato coding sequences (18). To test whether ectopic expression of scute can induce photoreceptors, we used either or both of these two regulatory regions of the ato gene to drive scute expression. We also used the GAL4-UAS system (19) for ectopic scute expression in the eye disk. Unexpectedly, we found that ectopic expression of scute induced the formation of photoreceptors in apparently R8-independent manner. This finding suggests the existence of a yet unidentified basic helix–loop–helix gene(s) involved in the formation of R1–R7.
Materials and Methods
DNA Constructs.
The scute coding sequence was obtained by PCR using plasmid pUC19-sc (a gift from Cheng-ting Chien, Academia Sinica, Taipei, Taiwan) as the template and the following primers: 5′-CAG TCA TGA AAA ACA ATA AT-3′ and 5′-GAC GGA TCC TTG GGG ATT AAG TCA, which incorporate a BspHI or a BamHI site, respectively. This fragment then was digested with BspHI and BamHI. A genomic fragment containing the 1.1-kb basic promoter region of ato was obtained by SacII and BglII double digestion of plasmid pBS.Bm4.2 (a gift from Andy Jarman, University of Edinburgh, Scotland; see ref. 18 for description), followed by gel purification and AflIII digestion. The resulting SacII–AflIII fragment was ligated to the above BspHI–BamHI fragment and then inserted into SacII and BamHI sites of the pBluescript to generate pBS.SacII-sc. The nucleotide sequence of the scute coding region derived from PCR was confirmed by DNA sequencing. The cloning strategies for both pCaSpeR.5′eye enhancer-sc (5′eye-sc) and pCaSpeR.3′enhancer-sc (3′F:5.8-sc) rescue constructs are essentially the same as for pCaSpeR.5′eye enhancer-ato (5′eye-ato) and pCaSpeR.3′enhancer-ato (3′F:5.8-ato), respectively (18), except that a 2.1-kb SacII–SmaI fragment excised from pBS.SacII-sc was used in both cases to replace the 2.1-kb SacII–ScaI fragment used to make the ato rescue constructs.
Scanning Electron Micrography.
Fly heads were fixed with 2% glutaraldehyde and 4% formaldehyde in 0.1 M sodium phosphate buffer (pH 7.2) overnight, dehydrated with a graded ethanol series, and critical point-dried in CO2. The samples were sputter-coated with 30 nm of gold palladium and examined with a scanning electron microscope at an accelerating voltage of 5 kV.
Histology.
Fly heads were fixed with 4% formaldehyde in 0.1 M sodium phosphate buffer (pH 7.2) overnight, dehydrated with a graded ethanol series, and embedded in epoxy. Compound eyes were sectioned tangentially at 2 μm and stained with 1% Toluidine blue and 1% sodium borax.
Isolation of the GAL4–7 Line.
The GAL4–7 enhancer trap line was isolated in our lab by Alice Turner and Cheng-ting Chien through mobilizing a previously known enhancer trap line 109C1 in an attempt to establish GAL4 lines that can drive UAS-ato expression and thereby rescue the loss of ommatidia phenotype in the ato1 mutant. The P element is inserted on the third chromosome. This line is viable as homozygotes with no discernible phenotype.
Whole-Mount in Situ Hybridization.
A 0.9-kb Asp-718 fragment from plasmid pBS.84F hs#2 (a gift from Andy Jarman) containing the ato coding region and a 3.4-kb EcoRI–KpnI fragment from the lacZ coding region excised from plasmid pBS.khc:lacZ (20) were used as template for digoxigenin (Boehringer Mannheim) labeling. The labeling was performed according to the manufacturer. Whole-mount in situ hybridization in imaginal discs using digoxigenin-labeled probes was performed essentially as described by Tautz and Pfeifle (21).
Immunohistochemistry.
Imaginal discs from late third instar larvae and early pupae were fixed for 10–20 min with 4% formaldehyde in 0.1 M PBS, washed with PBT (PBS + 0.3% Triton X-100) and blocked with 5% normal goat serum in PBT. Primary antibodies used were: rabbit anti-Ato (17), mouse anti-Boss (22), rabbit anti-Prospero (23), mouse anti-Cut (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City). Dichlorotriazinyl aminofluorescein, lissamine rhodamine B sulfonyl chloride (Jackson Immunoresearch), Alexa 488, or Alexa 568 (Molecular Probes)-conjugated secondary antibodies were used. Discs were mounted in Slow Fade mounting medium (Molecular Probes). All comparative experiments between ato and scute were performed in parallel.
Fly Stocks.
All Drosophila stocks were raised on standard cornmeal-yeast-agar medium at 25°C. ato1 is described by Jarman et al. (4, 17). UAS-ato and UAS-sc were established by Jarman et al. (3) and Chien et al. (8), respectively.
Results
Ectopic Eye Disk Expression of scute and/or ato Driven by the Regulatory Regions of the ato Gene.
The eyes of ato1 mutant are totally devoid of ommatidia. This mutant phenotype can be rescued by expressing ato in ato1 mutant flies via two copies of 5′eye-ato and 3′F:5.8-ato, because of restoration of R8 photoreceptors (Fig. 1 B and F) (18). By contrast, two copies of either or both of the scute transgenes (5′eye-sc and 3′F:5.8-sc) did not restore any Boss-expressing R8 photoreceptors or ommatidia in ato1 mutant (data not shown). To our surprise, ato1 flies carrying two copies of 3′F:5.8-sc and 5′eye-ato developed eyes with significant numbers of ommatidia and R8 photoreceptors (Fig. 1 D and H). Even more remarkably, expression of ato in front of and scute behind the MF leading edge (two copies of 3′F:5.8-ato and 5′eye-sc) in ato1 mutant resulted in a rather robust restoration of the eye without a corresponding restoration of cells expressing the R8 marker Boss, whose expression in wild type is commenced before R7 induction and remains in R8 throughout the period required for sevenless activity (22) (Fig. 1 C and G). Given that either ato transgene alone cannot induce ommatidia (18), these observations indicate that scute can partially substitute for ato in promoting ommatidium development.
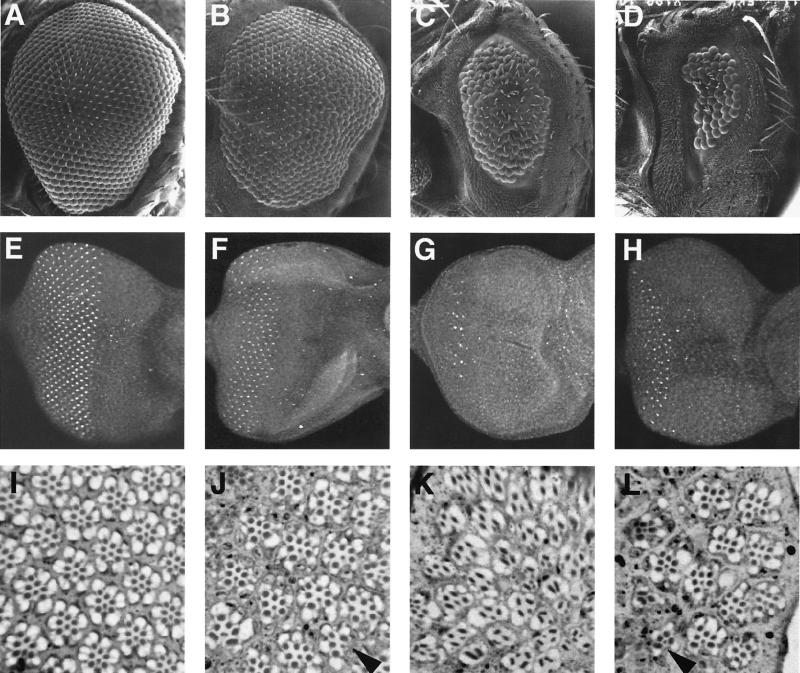
Figure 1.
Ectopic scute expression driven by ato regulatory regions promotes photoreceptor formation. (A–D) Scanning electron microscopy of compound eyes, showing that expression of scute either behind or in front of the MF leading furrow, combined with complementary ato expression, restores some ommatidia in the ato1 mutant (C and D). (E–H) Third instar larval eye discs stained with an antibody against Boss, showing much reduced Boss expression if scute is expressed behind (G) rather than in front of the MF leading edge (H). (I–L) Tangential sections of compound eyes, revealing ommatidia with reduced number of photoreceptors and no R8 in the ato1 mutant expressing ato in front of and scute behind the MF leading edge (K). (A, E, and I) Wild type; (B, F, and J) ato1 mutant expressing ato both in front of and behind the MF leading edge (3′F:5.8-ato; 5′eye-ato,ato1); (C, G, and K) ato1 mutant expressing ato in front of and scute behind the MF leading edge (3′F:5.8-ato; 5′eye-sc,ato1); (D, H, and L) ato1 mutant expressing scute in front of and ato behind the MF leading edge (3′F:5.8-sc; 5′eye-ato,ato1). Arrowheads in J and H indicate the rare ommatidia with fewer than eight photoreceptors. Posterior is to the left. (Magnifications: A–D, ×110; E–H, ×225; I–L, ×900.)
The unexpected ability to induce eye formation but not R8 photoreceptors, because of expression of ato in front of and scute behind the MF leading edge (3′F:5.8-ato; 5′eye-sc) in ato1 mutant, is mirrored by an unusual arrangement of photoreceptors in the ommatidia. Many of these ommatidia contain fewer than eight photoreceptors, and most of the remaining photoreceptors resemble outer photoreceptors by their positions and the large size of their rhabdomeres (rhodopsin-bearing stacks of microvilli) (Fig. 1K). For comparison, the normal trapezoidal arrangement is found in wild-type eyes (Fig. 1I) and eyes of ato1 mutants rescued by both ato transgenes (Fig. 1J) or by expression of scute in front of and ato behind the MF leading edge (3′F:5.8-sc; 5′eye-ato) (Fig. 1L). These results suggest that scute is capable of inducing photoreceptor formation; however, the resulting photoreceptors are not R8.
Ectopic Expression of scute or ato via the GAL4-UAS System and an Enhancer-Trap Line for Early Expression in the Eye Disk.
To further examine the ability of scute to induce photoreceptor formation independent of the founder photoreceptor R8, we introduced UAS-scute (9) into the GAL4–7 line. This enhancer-trap line drives gene expression predominantly in the posterior of the eye disk (Fig. 2B), in many more cells than cells that normally express ato (Fig. 2A). In the hypomorphic ato1 mutant (17, 18), expression of the mutant Atonal protein is detectable only in a narrow stripe of eye disk cells (Fig. 2E) (17) and is incapable of inducing ommatidium development. Expression of UAS-scute in the ato1 mutant, driven by the GAL4–7 line, did not alter the expression pattern of the mutant Atonal (Fig. 2F). Nonetheless, this resulted in compound eyes (Fig. 2D) with surface morphology similar to the compound eyes induced by expression of UAS-ato (3) (Fig. 2C). Thus, scute is capable of inducing ommatidium development.
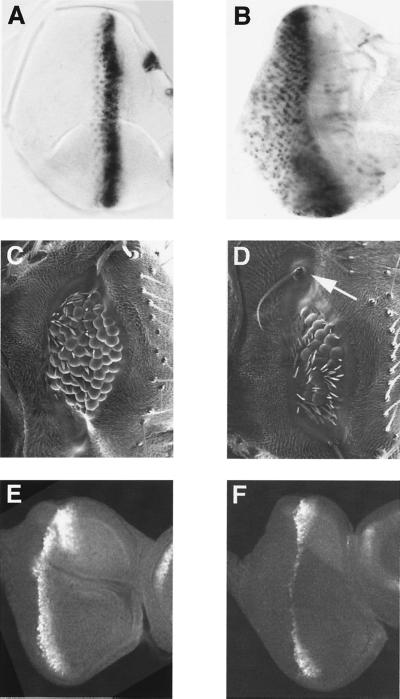
Figure 2.
Ectopic ato or scute expression driven by the GAL4–7 line rescues ommatidia. (A) Third instar larval eye disk hybridized to a digoxigenin-labeled ato probe, showing the endogenous ato expression pattern. (B) Third instar larval eye disk of GAL4–7/UAS-lacZ (nuclear) hybridized to a digoxigenin-labeled lacZ probe. The GAL4–7 line drives reporter gene expression in many more cells as compared with the endogenous ato expression, particularly in the posterior field (A). Scanning electron micrographs of compound eyes reveal that ectopic expression of scute (UAS-sc,ato1/GAL4–7,ato1) in ato1 mutant induces ommatidial formation (D), but to a lesser extent than expression of ato (UAS-ato/+; GAL4–7,ato1/ato1) (C). Occasionally, fully developed macrochaete-like sensory organs (arrow in D and Fig. 5) are seen in different areas of scute-rescued eyes. Third instar eye disk from ato1 mutant (E) and ato1 mutant expressing scute (UAS-sc,ato1/GAL4–7,ato1) (F) show similar patterns of staining with an antibody against Atonal. (Magnifications: A and B, ×225; C and D, ×145; E and F, ×200.)
Interestingly, whereas many ommatidia induced by UAS-ato contain more than the normal number of eight photoreceptors, often including more than one R8 photoreceptor (Fig. 3A), ommatidia induced by UAS-scute contain only 2–5 photoreceptors, usually lacking the centrally located R8 photoreceptors with small rhabdomeres (Fig. 3B).
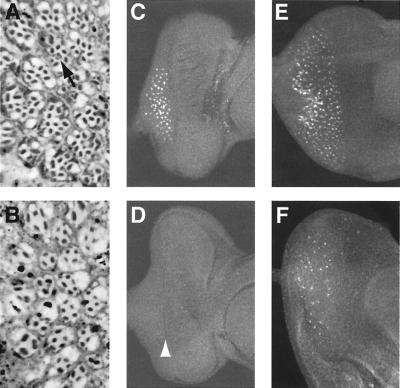
Figure 3.
Ommatidia rescued by ato or scute exhibit different structure and developmental processes. (A, C, and E) ato1 mutant expressing ato (UAS-ato/+; GAL4–7,ato1/ato1 ). (B, D, and F) ato1 mutant expressing scute (UAS-sc,ato1/GAL4–7,ato1). (A and B) Tangential sections of compound eyes. (A) Ommatidia in ato-rescued eye often contain more than eight photoreceptors. Multiple R8-like cells (arrow in A) in one ommatidium frequently are seen. (B) Most ommatidia in scute-rescued eye have only outer photoreceptors, and the number is lower than normal. (C–F) Eye discs from third instar larva (C and D) or early pupae (E and F) stained with an anti-Boss antibody. Third instar larval discs of scute rescued ato1 mutant (D) express no detectable Boss protein, although the MF has clearly advanced (arrowhead in D). Boss is detected only in early pupal discs of ato1 mutant expressing scute (F), in many fewer cells than those in the ato-rescued discs (E). (Magnifications: A and B, ×700; C–E, ×200.)
The R8 Photoreceptors Are Missing in scute Rescued Ommatidia Because Their Precursors Fail to Form.
In ato1 third instar larval eye discs expressing scute via the GAL4–7 line, there are no cells expressing the R8 marker Boss (22) (Fig. 3D). However, these larval eye discs exhibit the advancing MF (arrowhead in Fig. 3D). Moreover, they contain cells expressing Cut (Fig. 4I), the cone cell marker (24), as well as cells expressing Prospero (Fig. 4C), a marker for R7 photoreceptors and cone cells (25). Besides the R7 photoreceptors, other photoreceptors also were present, as indicated by the much greater number of cells stained with the 24B10 mAb (Fig. 4F), a general photoreceptor marker (26). These signs of eye development precede the appearance of just a few Boss-expressing cells in the posterior region of the eye disk after puparium formation (Fig. 3F). Taken together, these observations suggest that eye development is progressing in the absence of R8 photoreceptors.
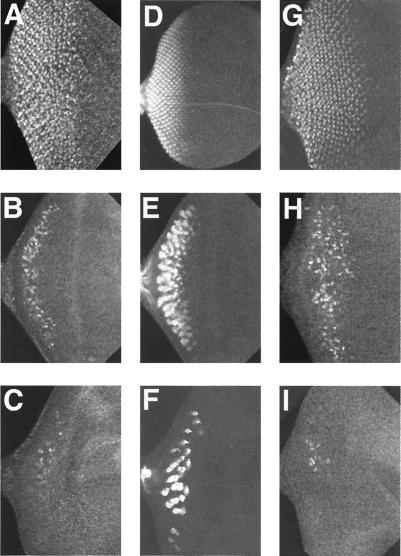
Figure 4.
Expression of prospero, 24B10 and cut in imaginal discs from ato1 mutants ectopically expressing ato or scute. Imaginal discs from third instar larvae of wild type (A, D, and G), ato1 mutant expressing ato (UAS-ato/+; GAL4–7,ato1/ato1) (B, E, and H) or ato1 mutant expressing scute (UAS-sc,ato1/GAL4–7,ato1) (C, F, and I) stained with an anti-Prospero (A–C), 24B10 (D–F), or an anti-Cut (G–I) antibody. All discs are shown with posterior to the left. Both Prospero (C) and 24B10 (F) are apparent in the UAS-sc,ato1/GAL4–7,ato1 discs, albeit in fewer cells as compared with the UAS-ato/+; GAL4–7,ato1/ato1 discs (B and E, respectively). This indicates that some photoreceptor differentiation has occurred in the absence of Boss expression at this stage (Fig. 3D). scute does not seem to activate cut expression directly in this misexpression condition (I). Many more cells express the cone cell marker cut in ato rescued disk (H), consistent with stronger ommatidial rescue by ato. (Magnification: ×200.)
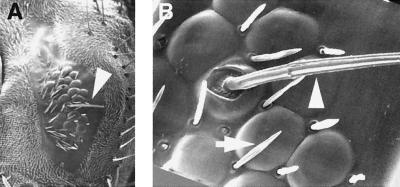
Ectopic scute Expression Also Induces Ectopic Macrochaetae Formation in the Eye.
Normally scute functions as a proneural gene for interommatidial bristles in the eye but is not expressed in cells that form the ommatidia (27). These small bristles develop between ommatidia during the early pupal stage after most photoreceptor cells already have been specified. In ato1 flies expressing scute via the GAL4–7 line, we occasionally observed large bristles in various positions in the eye (arrow in Figs. 2D and 5A). Some are found in the center of well-developed ommatidia (arrowhead in Fig. 5B). These ectopic bristles resemble macrochaetae that are found on the notum of the wild-type fly and are morphologically distinct from the small interommatidial bristles (arrow in Fig. 5B). Thus, ectopic expression of scute in the eye not only induces ommatidia formation in an unusual, R8-independent manner, but also can induce the formation of macrochaetae-like bristles that normally depend on scute although absent from the eye.
Figure 5.
scute induces ectopic external sensory organ formation in the eye. Scanning electron micrographs of a compound eye of ato1 mutant expressing scute (GAL4–7,ato1/UAS-sc,ato1) (A and B). In addition to photoreceptors, scute occasionally induces macrochaete-like external sensory organs (arrowhead in A and B and arrow in Fig. 2D) in various locations in the eye. These ectopic sensory bristles are morphologically distinct from surrounding interommatidial bristles (arrow in B). (Magnifications: A, ×110; B, ×700.)
Discussion
Formation of the Founder Photoreceptor R8 Can Be Induced by ato but Not scute.
The absence of R8 in ato1 mutants expressing scute via the GAL4–7 line, or expressing scute behind the MF leading edge in conjunction with ato in front of the MF, demonstrates that scute cannot substitute for ato in inducing R8 even though it induces ommatidia. Among proneural genes, scute is closely related to other genes of the achaete-scute complex (about 70% amino acid identity) but significantly different from ato (about 45% amino acid identity) (3, 28). The ability of ato, but not scute, to induce chordotonal organs (3, 8, 9) can be accounted for by the differences in their DNA-binding basic domains (8). Indeed, scute induces whereas ato represses expression of cut (9), a gene that functions as a binary switch to allow a neural precursor to choose between producing an external sensory organ (in the presence of cut expression) or a chordotonal organ (in the absence of cut expression) (29, 30).
Ommatidia Development Initiated by Ectopic scute Without Inducing R8 Photoreceptors.
How might ectopic expression of scute induce R8-less ommatidia, given that R8 normally is required for the formation of other photoreceptors and support cells of the ommatidia? It is probably not because of the ability of scute to induce cut expression, because only cone cells in an ommatidium normally express cut (25) and ectopic expression of scute in the eye causes only a small number of cells in the ommatidia to express Cut (Fig. 4I). Presumably, expression of genes that specify the eye primordia, such as the Pax gene eyeless (31), directs scute to act on a different set of downstream genes than those involved in the formation of sensory bristles. Although it is possible that scute induces latent R8 precursors that express none of the known R8 markers but can still recruit the R1–7 photoreceptors, we favor the following possibility. Once R8 is induced by ato during normal eye development, it recruits other photoreceptors by inducing the expression of yet unidentified gene(s) that encode basic helix–loop–helix protein, which shares significant sequence similarity with either Scute or both Scute and Atonal. Ectopic scute expression in ato1 mutants mimics the action of this gene(s) and induces the formation of outer photoreceptors, thereby bypassing the normal requirement of R8 founder photoreceptors. It will be very interesting to identify this hypothetical basic helix–loop–helix gene(s) and study its function in eye development.
Acknowledgments
We thank Alice Turner and Cheng-ting Chien for the gal4–7 enhancer trap line; Cheng-ting Chien for the UAS-sc flies; and Andy Jarman for plasmid pBS.84F hs#2 and the UAS-ato flies. Thanks to Larry Ackerman for the scanning electron micrographs and the adult fly eye sections, and to William Walantus for photographs. We are grateful to members of the Jan laboratory for enlightening discussions and continuous support. Yee-Ming Chan, Nadean Brown, Juergen Knoblich, Monica Vetter, and Liqun Luo are thanked for comments on the manuscript. Y.S. was supported by the Biomedical Sciences Graduate Program at the University of California, San Francisco. L.Y.J. and Y.N.J. are Howard Hughes Medical Institute Investigators.
Abbreviations
- ato
atonal
- MF
morphogenetic furrow
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110154497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110154497
References
- 1.Ghysen A, Dambly-Chaudiere C. Trends Genet. 1989;5:251–255. doi: 10.1016/0168-9525(89)90097-8. [DOI] [PubMed] [Google Scholar]
- 2.Campuzano S, Modolell J. Trends Genet. 1992;8:202–208. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- 3.Jarman A P, Grau Y, Jan L Y, Jan Y N. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- 4.Jarman A P, Grell E H, Ackerman L, Jan L Y, Jan Y N. Nature (London) 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- 5.Gupta B P, Rodrigues V. Genes Cells. 1997;2:225–233. doi: 10.1046/j.1365-2443.1997.d01-312.x. [DOI] [PubMed] [Google Scholar]
- 6.Huang M L, Hsu C H, Chien C T. Neuron. 2000;25:57–67. doi: 10.1016/s0896-6273(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 7.Goulding S E, Lage P, Jarman A P. Neuron. 2000;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 8.Chien C T, Hsiao C D, Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarman A, Ahmed I. Mech Dev. 1998;76:117–125. doi: 10.1016/s0925-4773(98)00116-6. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez I, Hernandez R, Modolell J, Ruiz-Gomez M. EMBO J. 1990;9:3583–3592. doi: 10.1002/j.1460-2075.1990.tb07569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parras C, Garcia-Alonso L A, Rodriguez I, Jimenez F. EMBO J. 1996;15:6394–6399. [PMC free article] [PubMed] [Google Scholar]
- 12.Skeath J B, Doe C Q. Curr Biol. 1996;6:1146–1152. doi: 10.1016/s0960-9822(02)70681-7. [DOI] [PubMed] [Google Scholar]
- 13.Ready D F, Hanson T E, Benzer S. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- 14.Tomlinson A, Ready D F. Dev Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- 15.Zipursky S L, Rubin G M. Annu Rev Neurosci. 1994;17:373–397. doi: 10.1146/annurev.ne.17.030194.002105. [DOI] [PubMed] [Google Scholar]
- 16.Freeman M. Development (Cambridge, UK) 1997;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- 17.Jarman A P, Sun Y, Jan L Y, Jan Y N. Development (Cambridge, UK) 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, Jan L Y, Jan Y N. Development (Cambridge, UK) 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- 19.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 20.Giniger E, Jan L Y, Jan Y N. Development (Cambridge, UK) 1993;117:431–440. doi: 10.1242/dev.117.2.431. [DOI] [PubMed] [Google Scholar]
- 21.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 22.Krämer H, Cagan R L, Zipursky L. Nature (London) 1991;352:207–212. doi: 10.1038/352207a0. [DOI] [PubMed] [Google Scholar]
- 23.Vaessin H, Grell E, Wolff E, Bier E, Jan L Y, Jan Y N. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- 24.Blochlinger K, Jan L Y, Jan Y N. Development (Cambridge, UK) 1993;117:441–450. doi: 10.1242/dev.117.2.441. [DOI] [PubMed] [Google Scholar]
- 25.Kauffman R C, Li S, Gallagher P A, Zhang J, Carthew R W. Genes Dev. 1996;10:2167–2178. doi: 10.1101/gad.10.17.2167. [DOI] [PubMed] [Google Scholar]
- 26.Zipursky S L, Venkatesh T R, Teplow D B, Benzer S. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]
- 27.Cagan R L, Ready D F. Dev Biol. 1989;136:346–364. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- 28.Villares R, Cabrera C V. Cell. 1987;50:415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- 29.Bodmer R, Barbel S, Sheperd S, Jack J W, Jan L Y, Jan Y N. Cell. 1987;51:293–307. doi: 10.1016/0092-8674(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 30.Blochlinger K, Jan L Y, Jan Y N. Genes Dev. 1991;5:1124–1135. doi: 10.1101/gad.5.7.1124. [DOI] [PubMed] [Google Scholar]
- 31.Halder G, Callaerts P, Gehring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]