Abstract
The acetylation of histone tails is a primary determinant of gene activity. Histone deacetylase 3 (HDAC3) requires the nuclear receptor corepressor SMRT for HDAC enzyme activity. Here we report that HDAC3 interacts with SMRT only after priming by cellular chaperones including the TCP-1 ring complex (TRiC), which is required for proper folding of HDAC3 in an ATP-dependent process. SMRT displaces TRiC from HDAC3, yielding an active HDAC enzyme. The SMRT–HDAC3 repression complex thus joins the VHL–elongin BC tumor suppression complex and the cyclin E–Cdk2 cell cycle regulation complex as critical cellular machines requiring TRiC for proper assembly and function. The strict control of HDAC3 activity underscores the cellular imperative that histone deacetylation occur only in targeted regions of the genome.
Keywords: Histone deacetylase, TRiC, SMRT, N-CoR, corepressor, HDAC3
Nucleosomes, the basic building block of chromatin, are composed of histone octamers. Posttranslational modification of histone tails by kinases, acetyltransferases, deacetylases, and methylases contributes to remodeling of chromatin structure that regulates gene expression. The “histone code” hypothesis holds that specific modification patterns serve as markers for transcriptional regulatory complexes with different transcriptional consequences (Jenuwein and Allis 2001). Lysine residues are modified by both acetyltransferases and methylases. Acetylation of lysines is associated with transcriptional activation, whereas lysine methylation can be associated with activation or repression (Jenuwein and Allis 2001). Thus the balance among unmodified, acetylated, and methylated lysine is a critical determinant of gene expression. Histone deacetylases (HDACs) reverse the activated state characterized by acetylation, and are associated with transcriptional repression (Hassig and Schreiber 1997). HDACs contribute to the dynamic nature of chromatin remodeling, because they convert acetylated lysine to an unmodified state that is subject to methylation in addition to reacetylation. HDACs also impose direct control on nonchromatin targets such as the p53, E2F, and NF-κB transcription factors (Martinez-Balbas et al. 2000; Chen et al. 2001; Luo et al. 2001; Vaziri et al. 2001) and can influence microtubule-dependent cell motility by deacetylation of α-tubulin (Hubbert et al. 2002).
Consistent with their important role in various cellular processes, HDACs function in multiprotein repression complexes, and their activities are highly regulated. Class II HDACs, such as HDAC4, are retained in the cytoplasm following direct phosphorylation, thus preventing their function in the nucleus (McKinsey et al. 2000). Enzymatic activity of HDAC4 may require complex formation with HDAC3 (Fischle et al. 2002). HDAC3 is inactive on its own but is activated by forming a stable complex with the nuclear hormone receptor corepressor SMRT (or N-CoR; Wen et al. 2000; Guenther et al. 2001), which functions as a subunit in an active HDAC3–SMRT enzyme (Guenther et al. 2001). The deacetylase activating domain (DAD) of SMRT/N-CoR, containing an essential SANT domain, is sufficient for HDAC3 interaction and activation (Guenther et al. 2001; Zhang et al. 2002).
While investigating the role of SMRT in activating the HDAC3 enzyme, we discovered the requirement for a preceding energy-requiring step, involving the TRiC multiprotein chaperone complex. This stringent regulation of HDAC3 activity provides a mechanism to ensure that histone deacetylation occurs specifically in targeted regions of chromatin.
Results and Discussion
An energy-dependent priming step is required for HDAC3 activation by SMRT
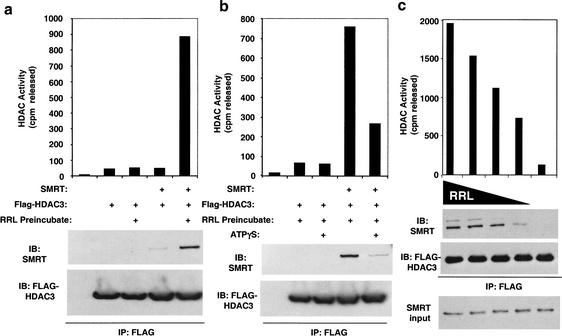
HDAC3 produced in rabbit reticulocyte lysate (RRL) is enzymatically inactive but can be activated by addition of SMRT (Guenther et al. 2001). HDAC3 purified from insect cells was also inactive (Fig. 1a). However, addition of SMRT was insufficient to activate this preparation of HDAC3 unless the purified HDAC3 was first preincubated or “primed” with unprogrammed RRL (Fig. 1a). This priming step was inhibited by nonhydrolyzable ATP (Fig. 1b), indicating that ATP was required. Interaction of HDAC3 with SMRT also required priming by RRL (Fig. 1a,b). The effect of RRL was dose-dependent (Fig. 1c), suggesting that priming might require a stoichiometric rather than a catalytic interaction with one or more components of the RRL.
Figure 1.
HDAC3 purified from insect cells becomes active through a reticulocyte lysate and ATP-dependent interaction with SMRT. (a) Purified recombinant Flag–HDAC3 produced from baculovirus was incubated with Myc–SMRT (amino acids 1–890) after preincubation with or without rabbit reticulocyte lysate (RRL). Flag–HDAC3 complexes were immunoprecipitated with anti-Flag agarose beads and assayed for HDAC activity (top), and subjected to immunoblot analysis (bottom). (b) Baculoviral Flag–HDAC3 was preincubated with RRL in the presence or absence of 2 mM ATP-γ-S, added to Myc–SMRT amino acids 1–890, and immunoprecipitated with anti-Flag agarose beads. Isolated HDAC3 complexes were assayed for HDAC activity (top) and subjected to immunoblot analysis (bottom). (c) Baculoviral Flag–HDAC3 was preincubated with decreasing amounts of rabbit reticulocyte (100 μL to 0 μL), added to Gal4–SMRT amino acids 1–763, and immunoprecipitated with anti-Flag agarose beads. Isolated HDAC3 complexes were assayed for HDAC activity (top) and subjected to immunoblot analysis (bottom).
HDAC3 enzyme activation by SMRT requires prior binding to the ATP-dependent TRiC protein folding machine
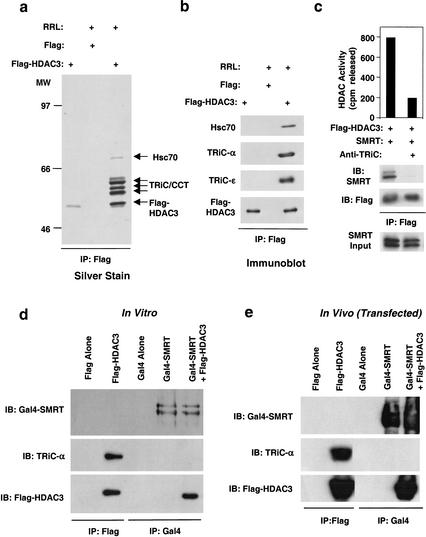
To determine the mechanism of HDAC3 priming, purified Flag–HDAC3 was incubated with RRL and immunopurified using Flag-agarose. Silver staining of the bound material revealed a band corresponding to HDAC3, as well as several other bands whose intensity suggested a stoichiometric complex (Fig. 2a). Direct sequencing revealed one of these proteins to be Hsc70, which has been previously shown to interact with HDAC3 (Johnson et al. 2002). The role of Hsc70 in HDAC3 priming was supported by the effects of deoxyspergualin (DSG), an inhibitor of Hsc70 function (Nadler et al. 1992) that blocked HDAC3 activation and interaction with SMRT (data not shown). Remarkably, all of the other proteins were components of the TCP-1 ring complex (TriC; also called chaperonin-containing TCP-1, or CCT) TRiC is an ∼800-kD, ATP-dependent protein folding machine comprised of eight orthologous subunits that are arranged similarly to bacterial GroEL (Dunn et al. 2001; Hartl and Hayer-Hartl 2002). The association of HDAC3 with TRiC was confirmed by immunoprecipitation of HDAC3 followed by immunoblotting for TRiC constituents (Fig. 2b). Thus, HDAC3 forms a stable and stoichiometric association with the TRiC. In support of this as a general mechanism, a yeast homolog of HDAC3 was recently found to associate with the orthologous TRiC, though the function of this interaction was not addressed (Pijnappel et al. 2001). Addition of antibodies to TRiC constituents prevented RRL from priming purified HDAC3 for SMRT interaction and enzyme activation (Fig. 2c), indicating an essential role of the TRiC in this process.
Figure 2.
HDAC3 interacts with the TRiC complex in reticulocyte lysate. (a) Baculoviral Flag–HDAC3 was immobilized onto anti-Flag agarose beads and incubated in the absence or presence of RRL. After resolution by SDS-PAGE, associated HDAC3 components were visualized by silver stain and identified as Hsc70 and TRiC zeta, epsilon, theta, eta, and delta. (b) Western blot analysis of equal amounts of Flag–HDAC3 and Flag–HDAC3–TriC/Hsc70 complex purified as in a. (c) Purified recombinant Flag–HDAC3 produced from baculovirus was incubated with Gal4–SMRT amino acids 1–763 after preincubation with RRL in the presence of anti-IgG or anti-TRiC antibodies. Flag–HDAC3 complexes were immunoprecipitated with anti-Flag agarose beads, assayed for HDAC activity (top), and subjected to immunoblot analysis (bottom). (d) Western blot analysis of HDAC complexes formed in vitro. Flag–HDAC3 and/or Gal4–SMRT were produced by T7 coupled transcription-translation, immunoprecipitated by anti-Flag or anti-Gal4 agarose beads, and subjected to SDS-PAGE followed by immunoblot. (e) Western blot analysis of HDAC complexes formed in vivo. Plasmids encoding Flag–HDAC3, Gal4–SMRT, or both were transfected into 293T cells, immunoprecipitated by anti-Flag or anti-Gal4 agarose beads, and subjected to SDS-PAGE followed by immunoblot.
TRiC dissociates from the active SMRT–HDAC3 complex
We investigated whether TRiC remains bound to HDAC3 after formation of the active SMRT–HDAC3 complex. Excess Flag–HDAC3 was primed by reticulocyte lysate prior to addition of SMRT. Immunoprecipitated SMRT was associated with HDAC3, but not with TRiC subunits, whereas HDAC3 not associated with SMRT was associated with TRiC (Fig. 2d). Similar results were obtained after overexpression of HDAC3 in 293T cells, where cellular TRiC was found to associate with Flag–HDAC3 alone but not in the context of the SMRT–HDAC3 complex (Fig. 2e).
Geldanamycin inhibits TRiC–HDAC3 complex formation
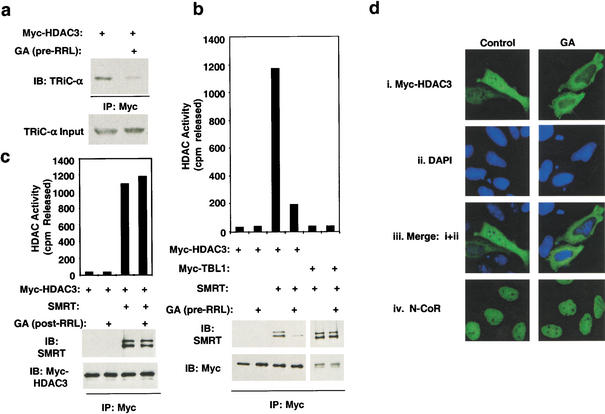
Interestingly, the protein folding inhibitor geldanamycin (GA) prevented the formation of the HDAC3–TRiC complex (Fig. 3a). GA is a relatively specific inhibitor of heat shock protein 90 (Hsp90; Scheibel and Buchner 1998). However, we were unable to demonstrate a stable interaction between HDAC3 and Hsp90 in reticulocyte lysate (data not shown). The Hsp90 homolog GRP94 is also GA-sensitive (Blagosklonny 2002), but GRP94 could not be demonstrated to interact with HDAC3 (data not shown) and is located in the endoplasmic reticulum, making it an unlikely chaperone for HDAC3. Thus, the TRiC–HDAC3 interaction is dependent upon Hsp90 (or another GA-sensitive chaperone). As predicted from its prevention of HDAC3 priming by TRiC, GA also prevented formation of the HDAC3–SMRT complex and activation of HDAC3 by SMRT (Fig. 3b). This effect was specific for HDAC3, as SMRT interaction with TBL1 was unaffected (Fig. 3b). However, when GA was added after formation of the TriC–HDAC3 complex, it was ineffective at preventing formation of the enzymatically active HDAC3–SMRT complex (Fig. 3c). Thus the effect of GA on priming of HDAC3 for SMRT interaction occurs proximal to the formation of the TRiC–HDAC3 complex.
Figure 3.
Geldanamycin blocks HDAC3 interaction with TRiC, activation by SMRT, and nuclear localization. (a) Myc-HDAC3 was translated in RRL with or without geldanamycin (GA; 100 μM), then immunoprecipitated with anti-Myc agarose beads before immunoblotting. (b) Myc–HDAC3 or Myc–TBL1 was produced by T7 coupled transcription-translation in the presence or absence of 100 μM GA. Complexes formed with Gal4–SMRT (1–763) were immunoprecipitated with anti-Myc agarose beads, assayed for HDAC activity (top), and subjected to immunoblot analysis (bottom). (c) GA was added to Myc–HDAC3 produced by T7 coupled transcription-translation. Complexes formed with SMRT amino acids 1–763 were immunoprecipitated with anti-Myc agarose beads, assayed for HDAC activity (top), and subjected to immunoblot analysis (bottom). (d) Immunolocalization of transfected Myc-HDAC3 (i,iii), DAPI (ii,iii), or endogenous N-CoR (iv) in HeLa cells exposed to 10 μm GA or vehicle control.
Prevention of HDAC3 priming blocks nuclear localization of HDAC3
Because GA blocks priming of HDAC3 with TRiC and, consequently, interaction with SMRT/N-CoR, it was used as a tool to determine the effects of priming on the subcellular localization of HDAC3. In agreement with others (Yang et al. 2002), we found that HDAC3 was distributed between the cell nucleus and cytoplasm (Fig. 3d). GA completely prevented nuclear localization of HDAC3 (Fig. 3d). N-CoR was constitutively nuclear in the presence or absence of GA (Fig. 3d, iv), indicating that GA did not generally prevent nuclear import of proteins and, furthermore, that nuclear localization of N-CoR does not require HDAC3. These data suggest that nuclear localization of HDAC3 requires priming by TRiC and, potentially, interaction with N-CoR or SMRT.
Conclusion
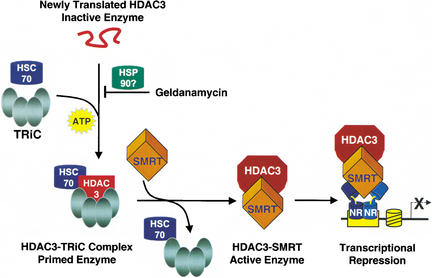
Based on our findings, we propose the following model of HDAC3 activation. After translation, unfolded HDAC3 is likely bound to Hsp90, explaining the GA sensitivity. The HDAC3-Hsp90 association is transient, and Hsp90 is rapidly replaced by the Hsc70-TRiC (Fig. 4). This step is ATP-dependent (Melki et al. 1997), as is the putative Hsp90 interaction. Interestingly, although the HDAC3–TRiC complex is stable in reticulocyte lysate and in cells, Flag–HDAC3 purified from insect cells is not associated with TRiC (Fig. 2a) and also cannot be activated by SMRT (Fig. 1a). It is possible that the amount of HDAC3 produced overwhelms the insect cell TRiC machinery, or that the TRiC complex is lost upon stringent purification of the recombinant HDAC3. Coexpression of HDAC3 and the SMRT DAD in insect cells does lead to formation of a stable, active complex (Guenther et al. 2001). We suggest that this is due either to transient association of HDAC3 with insect TRiC, or to a direct ability of coexpressed SMRT to stabilize newly folded HDAC3. In mammalian systems, HDAC3 is only primed for interaction with SMRT when bound to TRiC. Upon SMRT binding, TRiC dissociates from HDAC3 which together with SMRT now constitutes a stable, enzymatically active deacetylase complex.
Figure 4.
Model depicting the production of an active HDAC3 enzyme. Newly translated, inactive HDAC3 associates with the multiprotein TRiC complex and Hsc70, an energy-dependent step that allows SMRT to interact with and activate HDAC3. TRiC association is prevented by the HSP90 inhibitor geldanamycin, suggesting a role for HSP90, although the interaction of HSP90 with HDAC3 has not been directly shown. The TRiC/Hsc70 complex dissociates from the enzymatically active SMRT–HDAC3 deacetylase complex, which functions as a corepressor for nuclear receptors (NR).
The requirement of TRiC for formation of enzymatically active HDAC3–SMRT complex has important implications. TRiC is also critical for the formation and activity of the VHL–elongin BC elongase complex (Feldman et al. 1999; Hansen et al. 2002) and the cyclin E–cdk2 protein kinase complex (Won et al. 1998). Yet most protein–protein interactions do not require priming by TRiC (Hartl and Hayer-Hartl 2002). Thus, TRiC plays a special, thematic role in the formation of enzymatically active multiprotein complexes that mediate critical cellular functions such as translation (VHL-BC), cell cycle regulation (Cyclin E-cdk2), and transcription (SMRT-HDAC3). Moreover, corepressors have been implicated in the pathophysiology of acute myelogenous leukemias (Minucci et al. 2001). HDAC inhibitors have been used to treat these diseases, but current HDAC inhibitors target multiple HDACs and have numerous side effects (He et al. 2001; Marks et al. 2001). The search for specific HDAC inhibitors requires in vitro reconstitution of active HDACs, and the present work shows that proper folding will be critical. In addition, the chaperonin complexes required for formation of the HDAC3–SMRT repression complex represent novel and potentially HDAC-type-specific therapeutic targets. It is interesting in this context that GA is a chemotherapeutic agent for cancer (Neckers et al. 1999). Finally, the multilevel regulation of HDAC3 deacetylase activity is of critical importance to normal cell function. The control of HDAC3 activity by TRiC as well as a SANT-containing SMRT ortholog is likely to extend to yeast (Pijnappel et al. 2001). Conservation of this strict control of HDAC activity is undoubtedly related to the deleterious consequences of unregulated nuclear HDAC activity. Proper folding of HDAC3 by chaperone machines coupled with the formation of the active repression complex with SMRT/N-CoR thus regulates both HDAC3 localization and enzyme activity. This is in keeping with the recent suggestion that coupling of multicomponent cellular machines coordinates the activity, efficiency, and specificity of each step in gene expression (Maniatis and Reed 2002).
Materials and methods
Plasmids
Flag–HDAC3 was a gift from Drs. Eric Verdin and Wolfgang Fischle. Myc–HDAC3 was produced by cloning HDAC3 cDNA (encoding amino acids 1–428) into pcDNA3.1myc/his (Invitrogen). Myc–TBL1 was produced by cloning TBL1 cDNA (encoding amino acids 1–577) into pcDNA3.1myc/his (Invitrogen). Gal4–mSMRT plasmid containing Gal4–DBD fusion to mSMRT amino acids 1–763 was described previously (Guenther et al. 2001).
Production of active HDAC3 complexes in vitro
Myc-HDAC3, Gal4–SMRT 1–763, and Myc–TBL1 were translated individually with a TNT T7 Quick Coupled Transcription/Translation Kit (Promega) according to the manufacturer's instructions; 100 μL of translated material was used as a source for each protein unless otherwise noted. After translation, the desired proteins were mixed and added to two volumes of buffer D-300 (300 mM KCl, 20 mM HEPES at pH 7.9, 0.25 mM EDTA, 10% glycerol, 0.1% Tween 20) containing protease inhibitors (Boehringer Mannheim Complete cocktail). Complexes were purified by incubation with anti-Myc agarose beads (Santa Cruz Biotechnology) 16–18 h at 4°C. Beads were washed three times in buffer D-300 and once in HD buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 10% glycerol). The beads were then subjected to HDAC assay and immunoblot analysis.
HDAC assays
3H-labeled acetylated histones were from HeLa cells prepared essentially as described (Carmen et al. 1996). Deacetylase activity of immunoprecipitations were assayed by incubating the pelleted beads with 20,000 cpm of 3H-labeled acetylated HeLa histones in a total volume of 200 μL of HD buffer at 37°C for 2 h. The beads were collected and separated from the aqueous reaction mixtures. Reaction mixtures were quenched with 50 μL stop solution (1 M HCl, 0.16 M acetic acid) and extracted with 600 μL ethyl acetate. Released 3H-acetic acid was measured by scintillation counting of the organic phase.
Immunoblot analysis and antibodies
Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes with HMW transfer buffer (50 mM Tris, 380 mM glycine, 0.1% SDS, and 20% methanol). Blots were probed with primary antibodies in TBS containing 0.15% Tween-20 and 3.0% nonfat dry milk, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (Boehringer Mannheim). Alternatively, HRP-Flag (Sigma), HRP-Myc (Invitrogen), or HRP-Gal4 (Santa Cruz)-conjugated primary antibodies were directly used. All blots were visualized by ECL (Amersham Pharmacia Biotech).
Baculoviruses and purification of Flag–HDAC3 and Myc–SMRT
Flag–HDAC3 and Myc–SMRT amino acids 1–890 were cloned into pBlueBac4.5 and recombinant baculoviruses produced with a MaxBac 2.0 Kit (Invitrogen) and purified as described (Guenther et al. 2001). To test the effect of TRiC antibody on HDAC3 priming, Flag–HDAC3 (5 μg) was added to 60 μL RRL containing 10 μg anti-TRiC-α, 10 μg anti-TRiC-γ, and 10 μg anti-TRiC-ɛ (all Stressgen), or 30 μg IgG (Santa Cruz). Priming reactions were allowed to proceed for 2 h at 30°C. SMRT 1–763 was produced in RRL, added to priming reactions, and purified as above.
Identification of HDAC3 components of the HDAC3 complex
Purified Flag–HDAC3 (20 μg) was immobilized onto 50 μL anti-Flag agarose beads by incubation in buffer D-300, followed by extensive washing with buffer D-300. Beads were incubated with 700 μL rabbit reticulocyte lysate (Promega) and 700 μL buffer D-300 in a total volume of 1.4 mL, and then bound material was washed extensively with buffer D-300. Bound material was eluted with 3× 50 μL EB buffer containing 200 μg/mL Flag peptide (Sigma) and subjected to SDS-PAGE, followed by Coomassie or silver stain. Excised bands were subjected to sequence analysis at the Wistar Institute Protein Microchemistry/Mass Spectrometry Facility using microcapillary reverse-phase HPLC nano-spray tandem mass spectrometry on a Finnigan LCQ quadrupole ion trap mass spectrometer.
Cell culture and immunoprecipitation
293T cells were maintained in DMEM (high glucose) supplemented with 10% FBS and L-glutamine (all GIBCO-BRL). Cells were grown at 37°C in 5% CO2. 293T cells were transfected with FuGene6 (Boehringer Mannheim) according to the manufacturer's instructions for 48 h. Cells were washed in PBS and lysed with lysis buffer A (150 mM NaCl, 40 mM Tris-HCl at pH 7.6, 10% glycerol, 0.3% NP-40) and protease inhibitors for 20 min and pelleted. Supernatants were precleared with protein A agarose, then allowed to bind anti-SMRT, anti-Myc (Santa Cruz), or anti-Flag (Sigma) agarose beads for 6–8 h at 4°C. Pellets were washed three times in lysis buffer A and three times in lysis buffer A containing 300 mM NaCl, 0.1% NP-40, and subjected to immunoblot or HDAC assay. Immunoprecipitations of RRL translated proteins were performed in buffer D-300.
Immunofluorescence and confocal microscopy
Hela cells were grown on glass coverslips, and transfected with pDNA3.1–myc-tagged full-length HDAC3. Geldanamycin was added to a final concentration of 10 μM 16 h after transfection, and harvested at the times indicated. At harvest, cells were washed in PBS, and fixed by immersion in ice-cold absolute methanol/acetone (1/1, v/v) for 5 min. The coverslips were washed in PBS containing 0.2% Triton followed by incubation at room temperature for 1–2 h with the respective primary antibodies. Anti-myc antibodies (Roche Biotechnology) and anti-N-CoR antibodies were used at 1:1000, followed by fluorescein-labeled secondary antibodies (Molecular Probes). The secondary antibody was removed and the coverslips again washed in PBS prior to counterstaining nuclei with 4‘,6-Diamidino-2-Phenylindole (DAPI) at 2.5 μg/mL. Coverslips prepared in the same way, in which the primary antibody was omitted, showed no immunofluorescence. Images were captured with a TEC-1 CCD camera (Dage-MTI) mounted on a Nikon TE300 inverted microscope equipped with epifluorescence optics and with IP LabSpectrum v2.0.1 (Scanalytics). Images were acquired, pseudo-colored, and merged with Phase-3 Imaging software.
Acknowledgments
We thank J. Frydman and M. Miller for advice on inhibiting TRiC activity, and X. Hu and Y. Li for helpful discussions. This work was supported by NIH grants DK43806 and DK45586 (M.A.L.) and CA06927 (T.J.Y.), the W.W. Smith Charitable Trust (G.D.K.), and an Appropriation from the Commonwealth of Pennsylvania (T.J.Y.). Protein sequencing and identification was performed by David Speicher and the Wistar protein microsequencing facility.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lazar@mail.med.upenn.edu; FAX (215) 898-5408.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1037502.
References
- Blagosklonny MV. Hsp-90-associated oncoproteins: Multiple targets of geldanamycin and its analogs. Leukemia. 2002;16:455–462. doi: 10.1038/sj.leu.2402415. [DOI] [PubMed] [Google Scholar]
- Carmen AA, Rundlett SE, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- Dunn AY, Melville MW, Frydman J. Review: Cellular substrates of the eukaryotic chaperonin TRiC/CCT. J Struct Biol. 2001;135:176–184. doi: 10.1006/jsbi.2001.4380. [DOI] [PubMed] [Google Scholar]
- Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J. Formation of the VHL–elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen WJ, Ohh M, Moslehi J, Kondo K, Kaelin WG, Welch WJ. Diverse effects of mutations in exon II of the von Hippel-Lindau (VHL) tumor suppressor gene on the interaction of pVHL with the cytosolic chaperonin and pVHL-dependent ubiquitin ligase activity. Mol Cell Biol. 2002;22:1947–1960. doi: 10.1128/MCB.22.6.1947-1960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Schreiber SL. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- He LZ, Tolentino T, Grayson P, Zhong S, Warrell RP, Jr, Rifkind RA, Marks PA, Richon VM, Pandolfi PP. Histone deacetylase inhibitors induce remission in transgenic models of therapy-resistant acute promyelocytic leukemia. J Clin Invest. 2001;108:1321–1330. doi: 10.1172/JCI11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johnson CA, White DA, Lavender JS, O’Neill LP, Turner BM. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J Biol Chem. 2002;277:9590–9597. doi: 10.1074/jbc.M107942200. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: Causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Batelier G, Soulie S, Williams RC., Jr Cytoplasmic chaperonin containing TCP-1: Structural and functional characterization. Biochemistry. 1997;36:5817–5826. doi: 10.1021/bi962830o. [DOI] [PubMed] [Google Scholar]
- Minucci S, Nervi C, Lo Coco F, Pelicci PG. Histone deacetylases: A common molecular target for differentiation treatment of acute myeloid leukemias? Oncogene. 2001;20:3110–3115. doi: 10.1038/sj.onc.1204336. [DOI] [PubMed] [Google Scholar]
- Nadler SG, Tepper MA, Schacter B, Mazzucco CE. Interaction of the immunosuppressant deoxyspergualin with a member of the Hsp70 family of heat shock proteins. Science. 1992;258:484–486. doi: 10.1126/science.1411548. [DOI] [PubMed] [Google Scholar]
- Neckers L, Schulte TW, Mimnaugh E. Geldanamycin as a potential anti-cancer agent: Its molecular target and biochemical activity. Invest New Drugs. 1999;17:361–373. doi: 10.1023/a:1006382320697. [DOI] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart AF. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes & Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel T, Buchner J. The Hsp90 complex—A super-chaperone machine as a novel drug target. Biochem Pharmacol. 1998;56:675–682. doi: 10.1016/s0006-2952(98)00120-8. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won KA, Schumacher RJ, Farr GW, Horwich AL, Reed SI. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol Cell Biol. 1998;18:7584–7589. doi: 10.1128/mcb.18.12.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Tsai SC, Wen YD, Fejer G, Seto E. Functional domains of histone deacetylase-3. J Biol Chem. 2002;277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]