Abstract
Yeast possess a plasma membrane sensor of external amino acids that functions as a ligand-activated receptor. This multimeric sensor, dubbed the SPS sensor, initiates signals that regulate the expression of genes required for proper amino acid uptake. Stp1p and Stp2p are transcription factors that bind to specific sequences within the promoters of SPS-sensor-regulated genes. These factors exhibit redundant and overlapping abilities to activate transcription. We have found that Stp1p and Stp2p are synthesized as latent cytoplasmic precursors. In response to extracellular amino acids, the SPS sensor induces the rapid endoproteolytic processing of Stp1p and Stp2p. The processing of Stp1p/Stp2p occurs independently of proteasome function and without the apparent involvement of additional components. The shorter forms of these transcription factors, lacking N-terminal inhibitory domains, are targeted to the nucleus, where they transactivate SPS-sensor target genes. These results define a completely unique and streamline metabolic control pathway that directly routes environmental signals initiated at the plasma membrane to transcriptional activation in the nucleus of yeast.
Keywords: Environmental sensing, regulated proteolysis, subcellular localization, transcription, signaling, Saccharomyces cerevisiae
All cells have the capacity to sense and respond to extracellular environmental cues by changing patterns of gene expression. Metazoan cell proliferation depends on signals derived from receptors in the plasma membrane that respond to specific ligands, for example, growth factors or neighboring-cell membrane proteins. Until recently, it was thought that the nutrient-based decisions affecting growth and development in the yeast Saccharomyces cerevisiae were made in response to assessments of internal metabolite pools. This view has recently been challenged as it has become clear that yeast cells also use plasma-membrane-localized sensing systems to assess concentrations of nutrients in the external environment, including the availability of amino acids, ammonium, and glucose (for review, see Forsberg and Ljungdahl 2001b). Strikingly, the transmembrane components of these sensors—Ssy1p, Mep2p, Snf3p, and Rgt2p—are unique members of nutrient transport protein families (Van Belle and André 2001). With the exception of Mep2p, the ability of these transporter homologs to transduce nutrient (ligand)-induced signals across the plasma membrane appears to be independent of nutrient uptake, and thus these sensor components function analogously to traditional ligand-activated receptors.
A fundamental issue in understanding how cells respond to changes in their environment concerns the mechanistic connections between the proteins that perceive environmental cues and the downstream components that regulate the activity of appropriate transcription factors. In yeast, strikingly few signaling pathways have been characterized that transmit signals originating from plasma-membrane-localized sensing systems, for example, mating pheromone, osmolarity, and pH. With the exception of the two-component osmotic stress phosphorelay system (Li et al. 2002), all of the defined environmental-sensing pathways depend on MAP kinase cascades (for review, see Hohmann 2002). These phosphorylation cascades transduce metabolic signals that increase the expression of target genes by altering the activity of transcription factors, by increasing their promoter-binding affinities, their activation capacities, or their nuclear targeting. Studies regarding the organization of the MAP kinase pathway that regulates mating have provided many of the basic concepts, for example, scaffolding (Choi et al. 1994), that serve as paradigms for understanding similar pathways operating in metazoan cells.
The regulation of amino acid uptake in yeast offers an attractive system to pursue mechanisms of signal transduction. Apart from being metabolites for protein synthesis, amino acids have an important role in nitrogen homeostasis, and yeast possess sophisticated systems to efficiently import amino acids from the external environment. Amino acids are transported into the cell by amino acid permeases (AAPs) that comprise a conserved family of proteins with 18 core members (André 1995; Nelissen et al. 1997). Six of the family members, including BAP2 and BAP3, comprise a subset of more related AAPs that are transcriptionally induced by extracellular amino acids. The amino-acid-induced expression of these permeases requires the SSY1, PTR3, and SSY5 gene products (Barnes et al. 1998; Didion et al. 1998; Jørgensen et al. 1998; Iraqui et al. 1999; Klasson et al. 1999; Forsberg and Ljungdahl 2001a). Available data suggest that Ssy1p, Ptr3p, and Ssy5p function together as a plasma-membrane-localized sensor complex, dubbed the SPS sensor (Forsberg and Ljungdahl 2001a). Ssy1p, the only integral membrane component, is a unique member of the AAP family with an extended cytoplasmically oriented N-terminal domain that is not present in the other family members. Ssy1p does not itself transport amino acids (Didion et al. 1998; Iraqui et al. 1999); instead, it functions as a receptor for external amino acids, initiating signals that are transduced to the peripherally associated plasma-membrane proteins Ptr3p and Ssy5p (Klasson et al. 1999; Bernard and André 2001a; Forsberg and Ljungdahl 2001a).
Two related transcription factors, Stp1p and Stp2p, have been shown to be essential for SPS-sensor-dependent induced expression of the branched-chain amino acid permeases BAP2 and BAP3. In detailed studies focused on the promoters of BAP2 and BAP3, an amino-acid-dependent upstream activating sequence (UASaa) was identified (de Boer et al. 1998). Both Stp1p and Stp2p are able to bind DNA fragments containing the UASaa (de Boer et al. 2000; Nielsen et al. 2001). Similar UASaa motifs are present in the promoters of several other SPS-sensor target genes. The induced transcription of BAP2 and BAP3 is compromised in stp1Δ and stp2Δ null mutants, but is completely absent in stp1Δ stp2Δ double mutants. Together these results suggest that Stp1p and Stp2p are downstream effector components of the SPS sensor that exhibit redundant and partially overlapping activities. The mechanisms, or the physiological signals, that control Stp1p and Stp2p activity have not been identified.
In this paper, we demonstrate that Stp1p and Stp2p are synthesized as latent inactive precursors with negative regulatory domains within their N termini. We show that in response to external amino acids, and in a SPS-sensor-dependent manner, Stp1p and Stp2p are activated via an endoproteolytic-processing event that removes the N-terminal inhibitory domain. The activated shorter forms of Stp1p and Stp2p are targeted to the nucleus, where they transactivate SPS-sensor target genes. The processing occurs without the apparent involvement of additional components. Thus, we propose that the SPS sensor controls gene expression by directly regulating the activity of two effectors by a novel mechanism.
Results
The ASI13-1 mutation defines an inhibitory domain within the N terminus of Stp1p
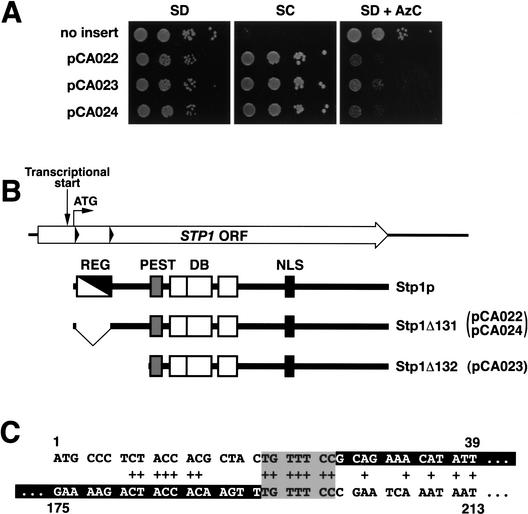
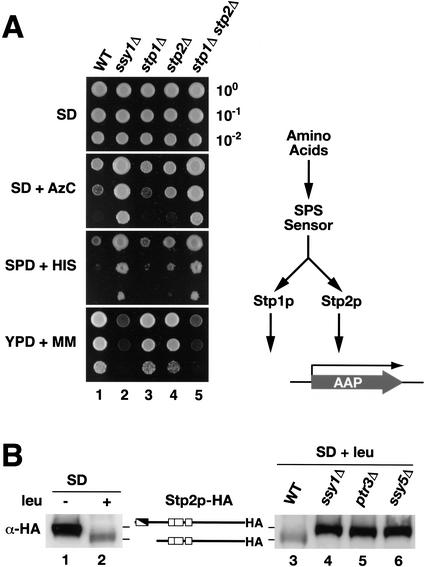
We isolated ASI13-1 (amino acid sensor independent) as a dominant mutation that derepressed the transcription of multiple SPS-sensor-regulated genes in cells lacking a functional SPS sensor (Forsberg et al. 2001). The dominant nature of the mutation suggested that the mutation constitutively activated a downstream component of the SPS-sensor signaling pathway. The gene carrying the ASI13-1 allele was cloned by plasmid-linked complementation (see Materials and Methods). The ssy1 leu2 strain (YMH119) was transformed with a genomic library prepared from the ssy1 leu2 ASI13-1 suppressed strain (YMH233). Strain YMH119 is unable to take up leucine at rates sufficient to support cell growth on SC medium, and exhibits resistance to the toxic amino acid analog azetidine carboxylate (AzC; Fig. 1A, no insert). Three SC+ AzCs transformants were identified. Plasmids pCA022, pCA023, and pCA024 were isolated from these transformants, and when reintroduced into strain YMH119, all transformants grew on SC but were unable to grow in the presence of AzC (Fig. 1A). Plasmids pCA022 and pCA024 were found to contain identical inserts, and pCA023 contained a shorter fragment derived from the same chromosomal region surrounding the STP1 locus (Wang et al. 1992).
Figure 1.
The dominant ASI13-1 (STP1Δ131) allele is an in-frame deletion that removes sequences encoding an inhibitory domain in the N terminus of Stp1p. (A) Phenotypic analysis of strain YMH119 transformed with plasmids pCA022 (STP1Δ131), pCA023 (STP1Δ132), and pCA024 (STP1Δ131). Transformants were spotted onto agar plates containing SD, SC, and SD containing AzC. Plates were incubated at 30°C for 3 d and photographed. (B) Schematic diagram of the STP1 gene. The start site of transcription is located within the ORF (Wang et al. 1992). The positions of the second ATG codon (translational start) and direct sequence repeats (black triangles) within STP1 are indicated. The proteins encoded by wild-type STP1 and the two dominant alleles STP1Δ131 and STP1Δ132 are depicted schematically. The regulatory domain, defined by the sequence between the repeats (REG, white/black diagonal box; amino acids 9–67), is absent in the constitutively active Stp1Δ131p. A putative nuclear localization signal (NLS, black box; amino acids 351–363), DNA-binding domain (DB, open boxes) with three putative zinc fingers (amino acids 160–187, 188–228, and 240–270), and a predicted PEST region (PEST, gray box; amino acids 129–147) are marked. (C) The sequences comprising the direct nucleotide repeats in STP1 are aligned (nucleotides 1–39 and 175–213, respectively). Matching base pairs are marked +, and the sequences removed in the STP1Δ131 deletion allele are indicated as white text within black boxes.
The inserts of pCA022 and pCA024 contain a mutant form of STP1 with an in-frame deletion between codons 8 (Phe) and 67 (Pro) of the STP1 ORF (Fig. 1B). This mutant allele of STP1 encodes a protein lacking 58 amino acids within the N-terminal domain of Stp1p (Fig. 1B). No other mutations could be found within the inserts of pCA022 and pCA024. As this deletion is not present in chromosomal DNA isolated from an isogenic wild-type strain, we considered the possibility that STP1 and ASI13-1 are allelic. To investigate this, a URA3-marked stp1Δ null allele was inserted into the STP1 locus of the suppressed ssy1 leu2 ASI13-1 strain (YMH233); this strain concomitantly lost the ability to grow on SC. This strain was crossed to an ssy1 leu2 strain (YMH119), the resulting diploid was sporulated, and the growth characteristics of the meiotic segregants were analyzed. The Ura+ phenotype segregated 2:2, and no SC+ spores were recovered, indicating that ASI13-1 is allelic to STP1. We therefore refer to ASI13-1 as STP1Δ131.
The sequences flanking the deletion within STP1Δ131 contain imperfect direct repeats (Fig. 1B, black triangles). The presence of these repeats suggest a plausible mechanism to explain the mutagenic event that generated the deletion. A recombination event occurring within these repeats, base pairs 20–26 and 194–200 (Fig. 1C, gray box), could account for the creation of the STP1Δ131 deletion. Additional sequence repeats, immediately adjacent to the site of recombination between base pairs 8–14 and 182–188, respectively, may have contributed to the formation of a DNA structure that facilitated the requisite strand exchange.
The insert of pCA023 was found to contain a 5′-truncated allele of STP1. The junction between the plasmid backbone and the genomic insert occurs at base pair +350 relative to the normal start codon (second ATG) of STP1. We have designated this mutant allele STP1Δ132. The STP1Δ132 allele lacks the endogenous STP1 promoter; therefore, the expression of this mutant allele must be due to the presence of a cryptic promoter present in the plasmid backbone. The ATG codon corresponding to methionine at amino acid residue 130 of wild-type Stp1p is likely to function as the start of translation.
The ability of STP1Δ131 and STP1Δ132 alleles to derepress transcription in a dominant manner suggested that a regulatory domain exists within the N-terminal region that negatively modulates the ability of wild-type Stp1p to activate transcription. The 58 amino acids deleted by the STP1Δ131 mutation (residues 9–66) defines the minimal boundaries of such a regulatory region (Fig. 1B, REG). Accordingly, the deletion of this inhibitory region would account for the observed constitutive activity and dominant behavior of the mutant transcription factor. Furthermore, this finding suggested a simple and testable model to explain SPS-sensor-mediated signaling. We reasoned that if Stp1p acts downstream of the SPS sensor, the events that modulate the activity of the negative regulatory domain of Stp1p should be dependent on the presence of extracellular amino acids and the components that comprise the SPS sensor.
Stp1p is activated by endoproteolytic processing in response to extracellular amino acids
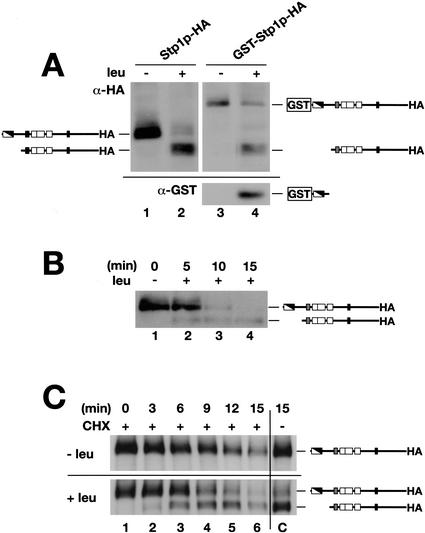
To test whether Stp1p carries an inhibitory domain we examined the electrophoretic behavior of Stp1p in extracts prepared from cells grown in the absence and presence of leucine. Stp1p was tagged at the extreme C terminus with a thrice-reiterated hemagglutinin (HA) epitope. Western analysis of protein extracts prepared from prototrophic cells transformed with this functional HA-tagged construct showed that the electrophoretic gel mobility and behavior of Stp1p was indeed dependent on the presence an inducing amino acid (Fig. 2A). In cells grown in SD, in the absence of an inducing amino acid, a band corresponding to full-length Stp1p was readily detected (Fig. 2A, lane 1). However, when cells were grown in the presence of leucine (SD + leu), the full-length Stp1p band was only weakly detected, and a second, more intense faster-migrating band of ∼10 kD lower molecular weight was observed. Similar results were obtained when an HA epitope was inserted in-frame following the tyrosine residue at position 330, ∼60 amino acids following the third zinc finger domain.
Figure 2.
Characteristics of Stp1p processing. The inhibitory domain of Stp1p is removed in response to amino acids via an endoproteolytic processing event. (A) Immunoblotting of whole-cell extracts from strain CAY59 (stp1Δ) transformed with plasmid pCA047 (Stp1p–HA) or pCA073 (GST–Stp1p–HA) grown in SD or SD supplemented with leucine (SD + leu). Extracts were resolved on a 10% SDS-PAGE gel and serially immunoblotted with anti-HA (upper panel) and anti-GST (lower panel) antibodies. (B) Stp1p activation occurs rapidly. Immunoblotting of whole-cell extracts from strain CAY59 transformed with plasmid pCA047 (Stp1p–HA). Cells were pregrown in liquid SD, and at t = 0 the culture received an aliquot of leucine. At the times indicated, subsamples were removed and whole-cell extracts were prepared. (C) The proteolytic processing of Stp1p does not require de novo protein synthesis. Immunoblotting of whole-cell extracts from strain CAY59 transformed with plasmid pCA047 (Stp1p–HA). Cells were pregrown as in B; at t = 0 the culture was split into four equal volumes. These subcultures received an aliquot of cycloheximide (CHX, final concentration = 100 μg/mL) and leucine as indicated. The immunoreactive forms of Stp1p are schematically represented at their corresponding positions of migration.
In the original characterization of STP1, it was found that transcription initiates within and not upstream of the STP1 ORF (Wang et al. 1992), at a position between the first and second ATG codons (see Fig. 1B). Because these earlier studies were carried out using auxotrophic strains grown in the presence of inducing amino acids, we initially considered the possibility that inducing amino acids may influence the position of transcription initiation. Accordingly, in the absence of inducing amino acids, the start of transcription may shift to a region upstream of the STP1 ORF, thereby enabling the full ORF to be translated. We placed the STP1–HA allele lacking the coding sequence preceding the second ATG codon under the control of the HXT7 promoter. This construct contains the STP1 ORF with only the second ATG codon present. The Stp1p expressed from this construct exhibited an identical pattern of migration as the protein expressed from the endogenous promoter, and also displayed the same amino-acid-dependent shift in mobility (data not shown). These data confirm that STP1 translation normally initiates at the second start codon, a result that indicates that the transcriptional start site, originally mapped by Wang et al. (1992), is not influenced by the presence of inducing amino acids.
We next examined the possibility that the observed leucine-induced shift in Stp1p mobility was the result of a proteolytic processing event that cleaved away the N-terminal negative regulatory domain. GST coding sequences were introduced in-frame to the second codon of the STP1–HA allele. This plasmid expresses a functional fusion protein that complements the slow-growth phenotype of stp1Δ leu2 mutant strains. Immunoblot analysis with anti-HA antibodies showed that the Stp1p band increased with the molecular weight corresponding to the size of the GST tag (Fig. 2A, cf. lanes 1 and 3). When leucine was added to the media, a band corresponding to the faster-migrating, processed form of Stp1p appeared (Fig. 2, cf. lanes 2 and 4). When the same blot was probed with anti-GST antibodies, a small protein band with a size slightly above 35 kD, corresponding to GST fused to ∼10 kD of the N terminus of Stp1p, could be detected in lysates prepared from the strain grown in the presence of leucine. This band was not detected in lysates prepared from the strain grown in media without leucine. Our ability to detect this smaller GST band verified that the amino-acid-induced processing of Stp1p is the result of an endoproteolytic removal of the N terminus. The sizes of both recovered products are consistent with a processing event that cleaves Stp1p at a position ∼10 kD from its N terminus.
Stp1p processing occurs rapidly and in the absence of de novo protein synthesis
When leucine is added to wild-type cells grown in media without supplementary amino acids, the transcription of several SPS-sensor-regulated genes is transiently induced (de Boer et al. 2000; Forsberg and Ljungdahl 2001a). The observed induction is quite rapid: within 10 min there is a 2- to 15-fold increase of target gene mRNA levels. We examined if the time course of leucine-induced Stp1p processing occurred within a similar time frame (Fig. 2B). The results show that the proteolytic processing of Stp1p occurs rapidly; the shorter form of Stp1p was detected within 5 min after leucine addition. The levels of full-length unprocessed Stp1p species decreased concomitantly with the appearance of the processed form, and after 10 min the level of the unprocessed form was barely detectable. Stp1p processing was not detected in parallel uninduced cultures, and in these control cultures the levels of full-length Stp1p did not change.
To directly test the product–precursor relationship of Stp1p processing, we repeated the time course of amino acid induction in the presence of cycloheximide (Fig. 2C). Our results indicate that the full-length unprocessed form of Stp1p is rapidly degraded, exhibiting a half-life of <10 min (Fig. 2C, upper panel). The efficacy of cycloheximide is clearly evident, as the steady-state levels of Stp1p did not decrease in the control culture grown in the absence of cycloheximide (Fig. 2C, upper panel, lane C). The addition of cycloheximide did not affect the rate of leucine-induced proteolysis of Stp1p (Fig. 2C, lower panel). Upon leucine induction, the levels of the full-length unprocessed form of Stp1p rapidly decreased. The shorter form of Stp1p became visible after 3 min of leucine induction. The levels of this processed form increased up to the 12-min time point (Fig. 2C, lower panel, lanes 2–5), and thereafter the levels decreased (Fig. 2C, lower panel, lane 6). These findings establish that the active short form of Stp1p is derived from the full-length form of Stp1p, and that the processing of Stp1p is the consequence of posttranslational events that are independent of de novo protein synthesis.
Amino-acid-induced processing of Stp1p requires a functional SPS sensor
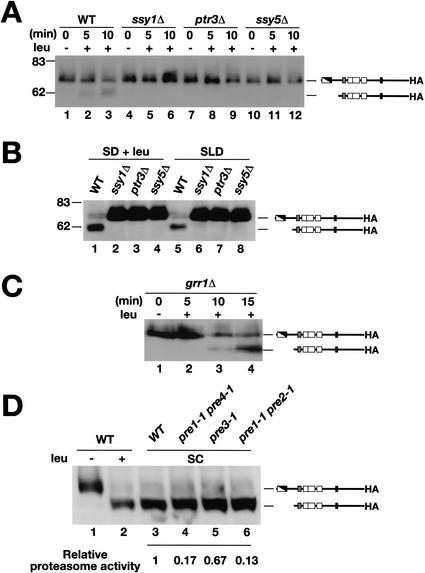
We examined whether the proteolytic processing of Stp1p was dependent on the presence of a functional SPS sensor. The time course of leucine-induced Stp1p processing was monitored in wild-type cells and in cells lacking one of the three SPS sensor components (Fig. 3A). As previously observed (Fig. 2B), the shorter activated form of Stp1p rapidly appeared in wild-type (WT) extracts upon leucine addition (Fig. 3A, lanes 1–3). In contrast, no Stp1p processing was observed in protein extracts prepared from strains carrying null alleles of any of the three SPS sensor components, that is, ssy1Δ, ptr3Δ, and ssy5Δ, respectively (Fig. 3A, lanes 4–12).
Figure 3.
The amino-acid-induced cleavage of Stp1p requires an operational SPS sensor and occurs independently of Grr1p and proteasome function. (A) Immunoblotting of whole-cell extracts from strains wild-type (WT, PLY126), ssy1Δ (HKY20), ptr3Δ (HKY31), and ssy5Δ (HKY77) cotransformed with plasmids pRS317 (LYS2) and pCA047 (Stp1p–HA). Cells were grown and induced with leucine as in Figure 2B. (B) Immunoblotting of whole-cell extracts from the strains grown in SD supplemented with leucine (SD + leu), or in SLD with leucine as the sole nitrogen source. After the preparation of extracts, proteins were resolved on 10% SDS-PAGE gels and immunoblotted with anti-HA antibody. (C) Immunoblotting of whole-cell extracts of CAY86 (grr1Δ) transformed with pCA047. Cells were grown and induced with leucine as in Figure 2B. (D) The migration of Stp1p–HA was analyzed in wild-type (WT, strain WCG4a) transformed with pCA047 and grown in SD before (lane 1) and after (lane 2) induction with leucine. Whole-cell extracts from WT (WCG4a, lane 3) and isogenic strains carrying mutations in the indicated proteasome components (pre1-1 pre4-1, pre3-1, and pre1-1 pre2-1, lanes 4–6, respectively) grown in SC (−ura) were prepared, and the migration of Stp1p–HA (pCA047) was examined by immunoblotting. The relative residual proteasome activity within these strains is indicated; proteasome activity was assessed by comparing the stability of model N-end rule substrates (Bachmair et al. 1986).
To investigate the possibility that the absence of proteolysis was the consequence of diminished rates of leucine uptake, cells were grown in SD supplemented with leucine (SD + leu). In contrast to extracts prepared from wild-type cells (Fig. 3B, WT, lane 1), the lower band corresponding to the short active form of Stp1p was not observed in extracts prepared from SPS-sensor-deficient strains (Fig. 3B, lanes 2–4). To more rigorously exclude the possibility that the lack of Stp1p processing was due to the inability of the SPS mutant strains to import leucine, cells were grown in SLD medium containing leucine as the sole nitrogen source. Under these conditions cells are forced to take up leucine for growth. All of the strains grew in SLD at rates similar to that observed in SD; thus, leucine uptake is not a limiting factor for growth in SLD. As before, the Stp1p present in protein extracts from SPS-sensor-deficient strains was not proteolytically processed (Fig. 3B, lanes 6–8). Based on these findings, we conclude that proteolysis of Stp1p is induced directly by a signal initiated by the SPS sensor, rather than induced indirectly by amino acids imported via SPS-sensor-regulated amino acid permeases.
The endoproteolytic processing of Stp1p is independent of GRR1 and proteasome function
Ubiquitin as well as components of the SCF–Grr1p E3 ubiquitin ligase complex have been reported to be required for induction of pAGP1 and pPTR2 promoted lacZ constructs in response to amino acids (Iraqui et al. 1999; Bernard and André 2001b). SCF (Skp1–Cdc53p/Cullin–F-box) complexes act as E3 ligases that link ubiquitin to various substrates. These latter findings suggest that ubiquitin-mediated degradation, or modification of protein activity, modulates the efficacy of SPS-sensor-derived signals. The obvious link that these proteins have to proteasomal degradation prompted us to examine the possible involvement of Grr1p in the proteolytic activation of Stp1p. Our results indicate that in response to leucine, Stp1p processing proceeds normally in grr1 null mutant cells (Fig. 3C); the short, activated form of Stp1p was readily observable as a faster-migrating band (Fig. 3C, lanes 3,4). The proteolytic processing event occurred within a similar time frame as observed in wild-type cells (cf. Fig. 2B). These results indicate that the proteolytic activation of Stp1p occurs independently of Grr1p.
Next, we investigated the processing of Stp1p in a series of isogenic mutant strains with impaired function in the three major proteolytic activities of the proteasome (Hilt et al. 1993; Heinemeyer et al. 1997; Gueckel et al. 1998). Leucine induced the processing of Stp1p in the isogenic wild-type strain (WCG4a; Fig. 3D, lanes 1,2). Proteasome mutants grow extremely poorly on minimal SD media. We therefore examined the electrophoretic mobility of Stp1p in wild-type and mutant cells grown on complete synthetic media (SC). As expected, because of the high amino acid content of SC, only the short, processed form of Stp1p was observed in the wild-type strain (Fig. 3D, lane 3). Similarly, only the processed form of Stp1p was detected in each of the proteasome mutant strains (Fig. 3D, lanes 4–6). As we did not observe diminished Stp1p processing, even in the pre1-1 pre2-1 double mutant strain with ∼10% of wild-type proteasome activity (Fig. 3D, lane 6), it is unlikely that the amino-acid-induced endoproteolytic cleavage of Stp1p is catalyzed by the proteasome.
Stp1p associates with the plasma membrane in an amino-acid-dependent manner
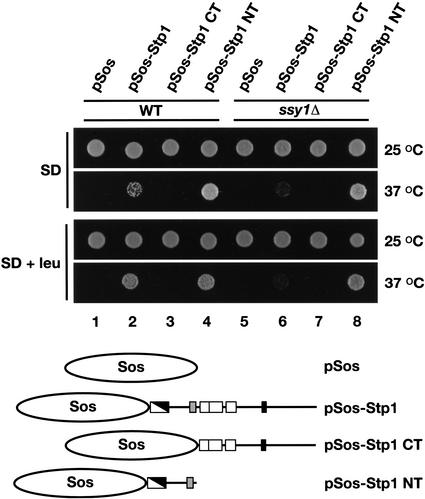
We determined whether Stp1p is able to associate with the plasma membrane using the Sos recruitment system (Aronheim et al. 1997). The Sos recruitment system exploits the ability of the human Cdc25p homolog hSos to suppress the temperature-sensitive cdc25-2 mutation (Petitjean et al. 1990). Fusion proteins that direct hSos to the cytosolic face of the plasma membrane enable cdc25-2 mutants to grow at 37°C. We constructed amino acid prototrophic strains carrying the cdc25-2 mutation in SPS-sensor-competent (WT) and defective (ssy1Δ) strain backgrounds. These strains were transformed with four plasmids; pSos, pSos-Stp1, pSos-Stp1 CT, and pSos-Stp1 NT (see Fig. 4 for a schematic presentation). Ura+ transformants were selected at permissive temperature on SD. Equally dense cell suspensions were prepared in water, and aliquots of each suspension were spotted onto duplicate plates containing SD, or SD supplemented with leucine (SD + leu). One of each duplicate plate was incubated at 25°C (permissive temperature) and the other at 37°C (nonpermissive temperature; Fig. 4).
Figure 4.
Stp1p associates with the plasma membrane. Prototrophic wild-type (WT, CAY187) and ssy1Δ (CAY189) strains carrying a temperature-sensitive cdc25 allele were transformed with plasmids pSos (pCA094), pSos-Stp1 (pCA093), pSos-Stp1 CT (pCA099), and pSos-Stp1 NT (pCA104). The proteins expressed from these plasmids are schematically presented. Transformants were selected and grown on solid SD incubated at 25°C. Cell suspensions were prepared in water (OD600 of 1), and equal aliquots of each suspension were spotted onto SD or SD containing leucine (SD + leu). Culture plates were incubated at 25°C or 37°C as indicated, and after 4 d the plates were photographed.
All wild-type and ssy1Δ transformants grew at similar rates on the SD and SD + leu plates incubated at 25°C. At 37°C, wild-type transformants carrying plasmids expressing the full-length Stp1p (pSos-Stp1) and the N-terminal domain of Stp1p (pSos-Stp1 NT) were able to grow (Fig. 4, dilutions 2 and 4). In contrast, wild-type transformants carrying pSos or pSos-Stp1 CT were unable to form colonies at the nonpermissive temperature (Fig. 4, dilutions 1 and 3). Strikingly, wild-type transformants expressing pSos-Stp1 exhibited more robust growth when cells were grown on SD + leu (Fig. 4, cf. dilution 2 on SD with SD + leu at 37°C). The growth of these transformants carrying the full-length pSos-Stp1 construct was found to depend on the presence of a functional SPS sensor; the ssy1Δ mutant carrying this construct exhibited weak growth (Fig. 4, dilution 6). The growth of transformants expressing the N-terminal fusion construct (pSos-Stp1 NT) was not leucine- or SPS-sensor-dependent, as robust growth was observed on both SD and SD + leu (Fig. 4, dilutions 4 and 8). Together these results suggest that leucine induces the proteolytic processing of Sos-Stp1p in an SPS-sensor-dependent manner, resulting in a protein comprised of Sos fused to the N-terminal domain of Stp1p. Consistent with this interpretation, the full-length pSos-Stp1 construct fully complements stp1Δ mutant phenotypes. These results indicate that Stp1p is able to associate with the plasma membrane via interactions mediated by the N-terminal domain.
Stp1p is targeted to the nucleus upon induction by amino acids
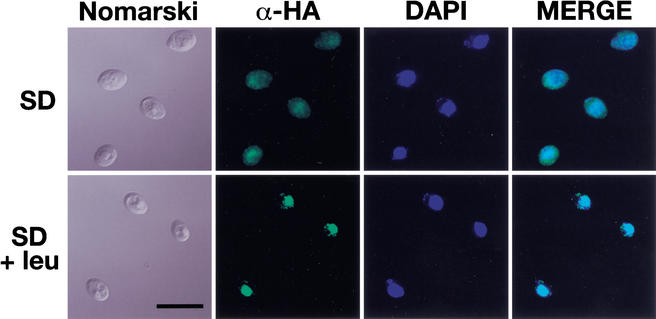
Based on our finding that Stp1p is able to associate with the plasma membrane, we investigated the possibility that the intracellular localization of Stp1p is dependent on the presence of inducing amino acids. A stp1Δ null mutant strain was transformed with a plasmid encoding a functional Stp1p–HA protein. Cells transformed with untagged STP1 were prepared in parallel for microscopic evaluation. When the strains were grown in SD in the absence of an inducing amino acid, the cells transformed with the epitope tagged STP1–HA construct showed weak and diffuse fluorescence that was barely above the background fluorescence observed in cells expressing untagged STP1. The fluorescence was observed throughout the entire cell (Fig. 5, upper panels). However, when the cells were exposed to leucine 30 min prior to fixation, an intense, highly focused fluorescence that colocalized with DAPI-stained DNA was observed (Fig. 5, lower panels). Whole-cell protein extracts were prepared from similarly treated cultures of cells. In uninduced cells only the full-length Stp1p–HA was detected. After the cells had been incubated for 30 min in the presence of leucine, the bulk of immunodetectable Stp1p–HA was found in its proteolytically processed form. In additional control experiments, we localized Stp1p–HA in an ssy1Δ null mutant strain. In this strain lacking a functional SPS sensor, the Stp1p–HA fluorescence was weak and diffuse (identical to that shown in the upper panels of Fig. 5), regardless of whether cells were induced with leucine. In contrast, when cells were transformed with a plasmid encoding a dominant active epitope-tagged Stp1Δ131p–HA construct, the observed fluorescence was highly focused and colocalized with the DAPI staining. The nuclear localization of Stp1Δ131p–HA was independent of the presence of an inducing amino acid. Together, these results indicate that, in response to external amino acids and in an SPS-sensor-dependent manner, Stp1p is processed and subsequently targeted to the nucleus.
Figure 5.
Proteolytically processed Stp1p is targeted to the nucleus. Indirect immunolocalization of full-length (upper panels) and proteolytically processed (lower panels) Stp1p was performed with anti-HA monoclonal antibodies. Strain CAY59 was transformed with pCA078 (pHXT7-STP1-HA) and grown in SD to an OD600 of 0.7. The culture was split into two equal aliquots, and leucine was added to one of the cultures. Both cultures were incubated shaking at 30°C for an additional 30 min, and the cells were fixed. (Panels, left to right) Cells viewed by Nomarski optics; α-HA monoclonal antibody-dependent Alexa Fluor 488 fluorescence; DAPI staining; and the MERGE of α-HA-dependent and DAPI fluorescence. (Lower left panel) Bar, 10 μm.
Stp2p and Stp1p are functionally redundant; Stp2p is proteolytically processed in response to external amino acids
STP2 encodes a protein that is homologous to Stp1p. Both proteins have well-conserved N-terminal domains and three zinc-finger motifs, and both bind promoter sequences containing a UASaa. To ascertain whether these proteins share redundant functions in the broader context of SPS-sensor-dependent signaling, we compared the growth characteristics of isogenic ssy1Δ, stp1Δ, and stp2Δ mutants and stp1Δ stp2Δ double mutants on media that differentially assess the activity of several other SPS-sensor-regulated amino acid permeases, for example, Agp1p and Gnp1p (Fig. 6A). All strains grew equally well on SD supplemented with leucine. As expected, the wild-type strain grew well on YPD + MM but exhibited impaired growth on SD + AzC and SPD + HIS (Fig. 6A, dilution series 1). The stp1Δ and stp2Δ single-mutant strains exhibited a similar pattern of growth as the wild-type strain (dilution series 3 and 4). In contrast, the SPS-sensor-deficient ssy1Δ strain exhibited robust growth on SD + AzC and SPD + HIS, but it grew poorly on YPD + MM (dilution series 2). The stp1Δ stp2Δ double-mutant strain displayed an identical pattern of growth as the ssy1Δ strain (Fig. 6A, cf. dilution series 5 and 2). These results clearly show that Stp1p and Stp2p have redundant function with respect to transducing SPS-sensor-initiated signals (Fig. 6A, pathway diagram).
Figure 6.
Stp1p and Stp2p exhibit overlapping and redundant function. (A) Wild-type (WT, CAY29), ssy1Δ (CAY91), stp1Δ (CAY59), stp2Δ (CAY119), and stp1Δ stp2Δ (CAY123) strains were grown on SD plates. Cells were resuspended in water, 10-fold serial dilutions were prepared, and aliquots of each dilution were applied to SD containing leucine (SD), SD containing leucine and AzC (SD + AzC), SPD containing toxic levels of histidine (SPD + HIS), and YPD containing MM (YPD + MM). In all instances synthetic media were supplemented with uracil. Plates were incubated at 30°C, and after 4 d plates were photographed. (B) SPS-sensor-initiated signals promote the proteolytic processing of Stp2p. Immunoblotting of whole-cell extracts from strain CAY119 transformed with pCA111 (Stp2p–HA) grown in SD or SD supplemented with leucine (SD + leu; left panel). Immunoblotting of whole-cell extracts from strains wild-type (WT, PLY126), ssy1Δ (HKY20), ptr3Δ (HKY31), and ssy5Δ (HKY77) cotransformed with plasmids pRS317 (LYS2) and pCA111 (Stp2p–HA) grown in SD supplemented with leucine (SD + leu; right panel). The immunoreactive forms of Stp2p are schematically represented at their corresponding positions of migration.
To test the possibility that Stp2p carries an inhibitory domain within the structurally conserved N terminus, we examined the electrophoretic behavior of a functional C-terminally epitope-tagged Stp2p in extracts prepared from cells grown in the absence and presence of leucine (Fig. 6B, lanes 1,2). A band corresponding to full-length Stp2p–HA was readily detected in extracts prepared from cells grown in SD. In extracts derived from cells grown in SD supplemented with leucine, a faster-migrating band was observed. These results indicate that in response to amino acids, Stp2p is proteolytically processed similarly as Stp1p. The leucine-induced Stp2p processing was monitored in wild-type cells and in cells lacking one of the three SPS sensor components. As was observed with Stp1p processing (Fig. 3), induction by leucine resulted in the appearance of the shorter form of Stp2p in wild-type extracts (Fig. 6B, lane 3); however, no processing of Stp2p was observed in protein extracts prepared from strains carrying null alleles of any of the three SPS sensor components, that is, ssy1Δ, ptr3Δ, and ssy5Δ, respectively (Fig. 6B, lanes 4–6). These results indicate that the proteolytic processing of Stp2p is dependent on the presence of a functional SPS sensor. Finally, we created an STP2Δ131 allele that encodes a protein lacking 73 amino acids (Pro 2–Phe 74) within the N-terminal domain. This construct exerted dominant suppressing effects when it was introduced into SPS-sensor-deficient strains. These results demonstrate that Stp2p is synthesized as a latent inactive transcription factor that is activated by endoproteolytic processing in response to SPS-sensor-initiated signals.
Genetic analysis of the SPS signaling pathway in an stp2Δ null mutant strain
The redundant function of Stp1p and Stp2p provides an explanation for why mutations in the genes encoding these transcription factors were not found in the genetic screens that identified the SPS sensor components SSY1, PTR3, and SSY5. Other redundant components may exist, perhaps components that specifically affect the function of Stp1p. To directly test this possibility, a genetic screen was carried out using two stp2Δ strains, CAY160 (MATa) and CAY161 (MATα), that carried an AGP1-promoted lacZ construct integrated into the GAP1 locus. In these strains, SPS sensor signaling is solely dependent on STP1, and when grown under conditions that induce SPS sensor signaling, these strains are AzC-sensitive and express high levels of β-galactosidase. We isolated spontaneously arising, and transposon-library-induced (Ross-Macdonald et al. 1997), mutations that enabled cells to become resistant to AzC on SD media supplemented to induce SPS sensor signaling. To exclude mutations uniquely affecting the expression of the AzC-transporting amino acid permease GNP1 (C. Andréasson, E. Neve, and P.O. Ljungdahl, in prep.), the levels of pAGP1-promoted β-galactosidase were monitored. A total of 148 AzCR mutants with no detectable X-gal staining were isolated. All but one of the mutations was found to be recessive. Complementation analysis revealed that recessive mutations defined four complementation groups. A member of each complementation group was mated to ssy1Δ, ptr3Δ, ssy5Δ, and stp1Δ tester strains. The ability of the resulting diploids to grow on SD + AzC was analyzed. In each case, diploids exhibited noncomplementation with one of the mutant tester strains, indicating that we had isolated mutations solely in SSY1, PTR3, SSY5, and STP1 (Table 1). The dominant mutation was subsequently found to be allelic with ssy5 mutations.
Table 1.
Complementation analysis of SPS-sensor signaling pathway
| Group
|
Number of isolates
|
|---|---|
| ssy1 | 55 |
| ptr3 | 23 |
| ssy5 | 49 |
| stp1 | 21 |
| Total | 148 |
Discussion
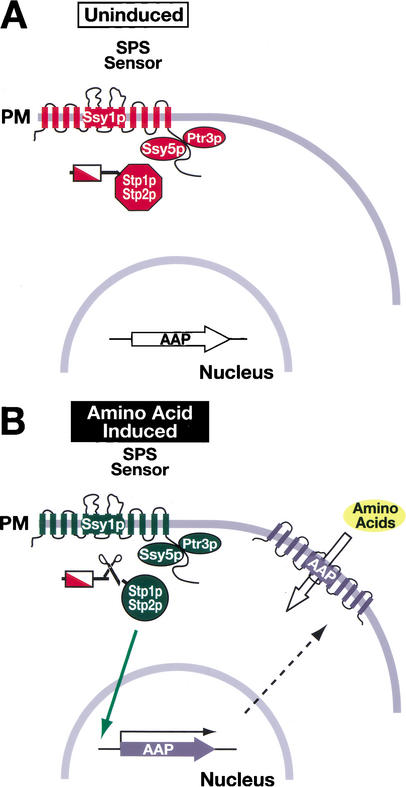
We have uncovered a novel mode of transducing nutrient-derived signals in yeast. The signal transduction pathway that we have characterized is activated in response to extracellular amino acids and transduces signals directly from the plasma membrane to the nucleus without the apparent involvement of previously characterized signal transducing components, that is, kinase cascades, ubiquitin, and the proteasome. Our data are consistent with a model schematically presented in Figure 7. This pathway appears to be strikingly simple. Despite extensive genetic screens carried out in several independent laboratories (Ljungdahl et al. 1992; Grauslund et al. 1995; Didion et al. 1996; de Boer et al. 1998; Jørgensen et al. 1998; Bernard and André 2001a; Forsberg et al. 2001), additional components have not previously been identified. A potential explanation for the lack of additional components being identified is the redundant and overlapping activities of Stp1p and Stp2p. However, in a genetic screen using a strain in which the SPS sensor signaling pathway was solely dependent on Stp1p, we also failed to identify additional components. Thus, the available genetic data support a simple mechanism of signal transduction. Additionally, we directly examined the potential involvement of the proteasome and the SCFGrr1 complex in the proteolytic activation of Stp1p/Stp2p, and our results clearly exclude the participation of these components. The lack of involvement of previously characterized signal transducing components indicates that the SPS sensing pathway represents a new mode of transducing environmental signals in yeast. The simple mechanism of SPS-sensor signaling suggests that yeast, like metazoan cells, possess a far broader repertoire of signaling mechanisms than represented by phosphorylation-dependent processes.
Figure 7.
Model of the SPS-sensor-dependent activation of Stp1p and Stp2p. (A) In cells grown in the absence of inducing amino acids, the SPS sensor of extracellular amino acids is present in the plasma membrane (PM) in its preactivation conformation (Forsberg and Ljungdahl 2001a). The transcription factors Stp1p and Stp2p are synthesized as inactive precursors that localize to the cytosol. The transcription of SPS-sensor-regulated genes, for example, AAPs, occurs at basal levels. Consequently, there are correspondingly low levels of AAPs in the PM, and cells exhibit low rates of amino acid uptake. (B) In the presence of inducing amino acids, the SPS sensor is activated, leading to the endoproteolytic processing of Stp1p and Stp2p. The shorter, activated forms of Stp1p and Stp2p, lacking the inhibitory domains located within their N termini, are targeted to the nucleus (solid arrow), where they function to induce the transcription of SPS-sensor-regulated genes. The increased transcription of AAP genes results in a comcomitant increase in AAPs in the PM (translation and movement to the PM are represented by the dashed arrow), and cells exhibit induced rates of amino uptake.
Our analysis of the activation of Stp1p and Stp2p clearly indicates that these transcription factors belong to the important class of latent cytoplasmic factors (Brivanlou and Darnell 2002). The similarities in the mechanisms that activate Stp1p/Stp2p and other members of this class are interesting. Several of these factors are activated in response to ligands binding receptors at the plasma membrane. For example, the activation of NFκB/Rel-dependent transcription involves two distinct proteolytic events; a proteasomal-dependent degradation of IκB, and the proteolytic activation of NFκB1. The activation of NFκB1 is not well understood. Other well-documented signaling pathways requiring proteasome processing include the Hedgehog and Wnt pathways. Fatty acid metabolism in yeast is regulated by ubiquitin/proteasome-dependent processing (RUP; Hoppe et al. 2001). In this pathway, the endoproteolytic processing of membrane-bound precursor forms of Spt23p and Mga2p releases the soluble transcription-activating domains from their C-terminal membrane anchors. The sterol regulatory element binding protein (SREBP) is released from membranes in two successive rounds of proteolytic processing by site-specific membrane-bound proteases (Brown et al. 2000). This proteasome-independent process, termed RIP (regulated intramembrane proteolysis), is apparently a commonly used mechanism to control diverse cellular processes. Other examples of proteins that undergo RIP include Notch, ErbB-4, and amyloid precursor protein (APP). A common theme emerging from each of these examples is that the proteolytic processing of the latent precursor forms of factors enables the active forms to enter the nucleus, where they transactivate target gene expression. Although we do not fully understand the inhibitory activity present within the N-terminal domain of Stp1p and Stp2p, we assume that the regulatory domains function either directly by interfering with proper transport in or out of the nucleus, or as with NFκB/Rel, by binding to a cytosolic component that sequesters it, thereby preventing nuclear translocation.
The identity of the protease responsible for the activation of Stp1p and Stp2p remains to be elucidated. Because neither Stp1p nor Stp2p has a membrane-spanning domain, the endoproteolytic events that we observe cannot involve RIP. Furthermore, there is no indication of ubiquitylation, and processing occurs in proteasome-defective strains, indicating that proteolysis occurs independently of RUP. Thus, our results define a novel strategy for the mobilization of latent transcription factors. Based on our present understanding of Stp1p and Stp2p activation, we believe that the endoproteolytic processing occurs in close association with the SPS sensor directly at the plasma membrane. This conclusion is based on two independent observations. First, although Stp1p and Stp2p are predicted to be soluble proteins, we have found that the N-terminal domain of Stp1p is able to efficiently direct hSos to the plasma membrane. Furthermore, we have found that the electrophoretic properties of Ssy1p (Forsberg and Ljungdahl 2001a) are altered in mutant cells lacking STP1 (C. Andrésson and P.O. Ljungdahl, unpubl.). This latter finding indicates that in the absence of Stp1p, the SPS-sensor component interactions are affected, suggesting that Stp1p directly associates with one or more of the components of the SPS sensor. We are presently investigating the possibility that the endoproteolytic activity responsible for the processing of Stp1p and Stp2p is inherent in one of the SPS sensor components.
We have found that the phenotypes exhibited by stp1Δ stp2Δ double mutants are identical to those of ssy1Δ null mutants. Recent work using transcription profiling has shown that the altered pattern of gene expression in stp1Δ stp2Δ double mutants mimics the changes observed in an ssy1Δ single mutant (Regenberg et al. 2001). Thus, it is likely that our results regarding the proteolytic activation of Stp1p and Stp2p completely account for the observed specific regulation of SPS-sensor target gene expression. However, a more complex interplay between specific and general factors undoubtedly exists. Consistent with this idea, variations in the quality and quantity of characterized regulatory sequences in the promoters of SPS-sensor-regulated genes are readily obvious when direct comparisons are made. Additionally, depending on growth conditions, cross-talk between intersecting nitrogen-regulated signaling pathways can also be expected to affect the expression of particular SPS-sensor-regulated genes. Not surprisingly, several general factors that affect the expression of SPS-sensor-modulated genes have been described. These include Abf1p, Leu3p, Tup1p, Ssn6p, Uga35p, ubiquitin, and the SCF–Grr1p E3 ubiquitin ligase complex (Iraqui et al. 1999; de Boer et al. 2000; Bernard and André 2001b; Forsberg et al. 2001; Nielsen et al. 2001). The hierarchy in which these factors operate to control gene expression remains to be elucidated.
The fact that yeast plasma membrane nutrient sensors have only recently been discovered reveals how little is understood regarding the molecular signals that enable eukaryotic cells to adapt to changing environments. Although several nutrient-regulated signal transduction pathways are known in yeast, including MAP kinase and cAMP cascades, only in a few instances have the primary plasma membrane sensors been identified. Our identification that the transcription factors Stp1p and Stp2p are the direct downstream components of the SPS-sensor signaling pathway is intriguing, and clearly indicates that these transcription factors belong to the class of latent cytoplasmic factors (Brivanlou and Darnell 2002). Similar modes of activation are known for other latent factors controlling many important cellular processes in multicellular organisms. However, many questions remain as to the precise mechanisms governing the activity of this important and interesting class of proteins. The knowledge gained from analyzing novel sensing systems in yeast may provide the means to better understand similar mechanisms controlling growth in metazoan cells.
Materials and methods
Media and strains
Standard media including YPD and ammonia-based synthetic minimal dextrose (SD) supplemented as required to enable growth of auxotrophic strains were prepared as described (Burke et al. 2000), with the exception that higher concentrations of L-leucine (166 mg/L = 1.3 mM) were used. Ammonia-based synthetic complex dextrose (SC) contains the same components described in Burke et al. (2000); the final concentration of each component was 120 mg/L with the exceptions of adenine (30 mg/L) and L-leucine (245 mg/L). SC also contains para-aminobenzoic acid (12 mg/L). SPD and SLD were prepared and supplemented as SD, but instead of ammonium sulfate, 1 g/L of L-proline or L-leucine was used as the sole nitrogen source, respectively. Where appropriate, 5-fluoroortic acid (5-FOA) was added to 1 g/L to SC, and hygromycin B was used at 300 mg/L in YPD. Media were made solid with 2% (w/v) Bacto agar (Difco), or 2% (w/v) agarose when nitrogen sources other than ammonium were used. Sensitivity to 1 mM L-azetidine-2-carboxylic acid (AzC) was tested on SD supplemented with Lleucine. Sensitivity to high levels of L-histidine was tested on SPD supplemented with 30 mM L-histidine-HCl. Sensitivity to 2-{[({[(4-methoxy-6-methyl)-1,3,5-triazin-2-yl]-amino}carbonyl) amino]-sulfonyl}-benzoic acid (MM) on complex media was tested on YPD prepared as described (Jørgensen et al. 1998).
All yeast strains used in this work (Table 2), except cdc25-2 and proteasome mutant strains, are isogenic descendants of strain PLY115/AA255. In all cases, the correct integration of gene deletion cassettes was confirmed by whole locus PCR analysis. Crosses and subsequent tetrad analysis were performed to verify 2:2 segregation of all deletion markers, and that mutant phenotypes cosegregated with each respective marker gene. To obtain isogenic LYS2 derivatives, PLY127 was transformed with a 6.5-kb PCR product encompassing the LYS2 gene amplified from the prototrophic strain S288C. The Lys+ transformant CAY25 was isolated, and all LYS2 descendants are meiotic segregants from crosses with CAY25. CAY41 was constructed from PLY118 by deleting the entire coding sequence of STP1 with a PCR-amplified CaURA3MX3 cassette (primers prstp1DMX-F and prstp1DMX-R; Goldstein et al. 1999). CAY42 is a Ura− derivative of CAY41 selected as a 5-FOA-resistant loop-out between the two repeated Ashbya gossypii leu2 (Agleu2) sequences flanking the CaURA3MX3 cassette. CAY59 is a Ura− derivative of a meiotic segregant obtained from a cross between CAY41 and CAY25. The hygromycin-B-resistant strain CAY86 was constructed by deleting the entire sequence of GRR1 of CAY29 with a PCR-amplified hphMX4 cassette (primers prgrr1DMX-F and prgrr1DMX-R; Goldstein and McCusker 1999). To obtain CAY90, CAY29 was transformed to Ura+ with an SalI/SpeI linear fragment containing ssy1Δ12∷hisG–URA3–kanr–hisG (pHK031). CAY91 is a 5-FOA-resistant derivative of CAY90. Strain CAY117 carrying an stp2 null allele was obtained by transforming CAY29 with a PCR-amplified hphMX4 cassette (primers prstp2DMX-F and prstp2DMX-R). CAY119 and CAY123 are meiotic segregants from a cross between CAY117 and a strain CAY62. Strains carrying the temperature-sensitive cdc25-2 allele were obtained as follows: strain cdc25H (MATα ade2-101 his3Δ200 leu2-3,112 lys2Δ801 trp1-901 ura3-52 cdc25-2 Gal+) obtained from Stratagene was crossed with CAY91. A temperature-sensitive ura3-52 Gal− meiotic segregant was isolated and saved as CAY186. CAY186 was back-crossed to CAY91, and meiotic segregants CAY187 and CAY189 were isolated.
Table 2.
Yeast strains and plasmids
| Strain
|
Genotype or description
|
Reference
|
|---|---|---|
| CAY29 | MATa ura3-52 | This work |
| CAY42 | MATa ade2 leu2-3,112 lys2Δ201 ura3-52 stp1Δ51::Agleu2 | This work |
| CAY59 | MATa ura3-52 stp1Δ51::Agleu2 | This work |
| CAY86 | MATa ura3-52 grr1Δ50::hphMX4 | This work |
| CAY91 | MATa ura3-52 ssy1Δ13::hisG | This work |
| CAY119 | MATa ura3-52 stp2Δ50::hphMX4 | This work |
| CAY123 | MATa ura3-52 stp1Δ51::Agleu2 stp2Δ50::hphMX4 | This work |
| CAY160 | MATa ura3-52 ade3Δ51 stp2Δ50::hphMX4 gap1Δ::pAGP1-lacZ | This work |
| CAY161 | MATα ura3-52 ade2Δ51 stp2Δ50::hphMX4 gap1Δ::pAGP1-lacZ | This work |
| CAY187 | MATa ura3-52 cdc25-2 gal2 | This work |
| CAY189 | MATa ura3-52 ssy1Δ13::hisG cdc25-2 gal2 | This work |
| HKY20 | MATa lys2Δ201 ura3-52 ssy1Δ13::hisG | Klasson et al. 1999 |
| HKY31 | MATa lys2Δ201 ura3-52 ptr3Δ15::hisG | Klasson et al. 1999 |
| HKY77 | MATa lys2Δ201 ura3-52 ssy5Δ2::hisG | Forsberg and Ljungdahl 2001 |
| PLY126 | MATa lys2Δ201 ura3-52 | Klasson et al. 1999 |
| YMH119 | MATa ade2 leu2-3,112 lys2Δ201 ura3-52 ssy1Δ13::hisG | Forsberg et al. 2001 |
| YMH233 | MATa ade2 leu2-3,112 lys2Δ201 ura3-52 ssy1Δ13::hisG ASI13-1 | Forsberg et al. 2001 |
| Plasmid | ||
| pCA022 | MRPL28, STP1Δ131, and spp41 in pRS316 | This work |
| pCA023 | STP1Δ132, and spp41 in pRS316 | This work |
| pCA024 | MRPL28, STP1Δ131, and spp41 in pRS316 | This work |
| pCA047 | STP1-3xHA in pRS316 | This work |
| pCA073 | GST-STP1-3xHA in pRS316 | This work |
| pCA078 | PHXT7-STP1-3xHA in pRS202 | This work |
| pCA093 | hSOS-STP1 in pCA094 | This work |
| pCA094 | URA3 derivative of pSos | This work |
| pCA099 | hSOS-STP1ΔNT in pCA094 | This work |
| pCA104 | hSOS-STP1ΔCT in pCA094 | This work |
| pCA111 | STP2-3xHA in pRS316 | This work |
Cloning of ASI13-1 (STP1Δ131)
DNA was isolated from strain YMH233 (ssy1Δ leu2 ASI13-1), and a genomic library was constructed in the vector pRS316 as described (Forsberg et al. 2001). Strain YMH119 (ssy1Δ leu2) was transformed with the library ligation mixture, and transformants were selected on SD. From this, 5000 independent transformants were obtained, and transformation plates were replica-plated onto SC (lacking uracil). Four transformants that exhibited robust growth on this media were streaked on SD + AzC. Three of the transformants were unable to grow on SD + AzC, indicating that these strains exhibited both of the ASI13-1 suppression phenotypes. Plasmids pCA022, pCA023, and pCA024 were rescued from these three strains.
Plasmids
The plasmids used in this study are listed in Table 2. The sequences of oligonucleotides used are available at http://www.licr.ki.se/appendix.html. Plasmid pCA027 was constructed by religating SpeI-restricted pCA024; this procedure removed the truncated SPP41 gene present in the original library insert fragment. A 2-kb STP1-containing fragment was PCR-amplified (primers prSTP1-F and prSTP1-R) from genomic DNA prepared from strain PLY118. EcoRI and BamHI sites contained within the primer sequences facilitated the cloning of the amplified fragment into vector pRS316, creating pCA029. The NotI site in the multicloning sites of pCA027 and pCA029 were destroyed by digestion with NotI, Klenow polymerase blunting, and religation. Using single-stranded pCA029 that lacked the NotI sites as template (Kunkel et al. 1987), an NotI site (primer prSTP1-NotI C) was reintroduced immediately before the translational termination codon of STP1. An NotI-flanked cloning cassette, encoding the HA1 epitope reiterated three times, was inserted into the unique NotI site to form plasmid pCA047. Similarly, an NotI site was introduced directly after the STP1 initiator codon using primer prSTP1-NotI N to create pCA042. A Myc epitope was introduced directly after the initiator codon of pCA042: the 5′-end of STP1 was PCR-amplified using primers prSTP1-EagI-Myc-F and prSTP1 SEQ5-R and pCA029 as a template; subsequent restriction of the amplified product with EagI and MluI formed a 0.1-kb fragment that was ligated to an NotI- and MluI-restricted pCA042, creating pCA064. The sequence encoding Glutathion-S-Transferase (GST) from pGEX-2T (Pharmacia) was PCR-amplified (primers prGST-F prGST-R), restricted with EagI (introduced within the primers), and ligated to the NotI site between the initiator codon and the single Myc epitope in pCA064, creating pCA071. pCA073 was created as a fusion of the 5′-GST-tagged STP1, and the 3′-3xHA-tagged STP1, by ligating BstEII- (coding region) and PstI- (vector backbone) restricted fragments. A 390-bp fragment containing the 5′-truncated promoter of HXT7, with an initiator codon followed by six in-frame histidine codons, was PCR-amplified (primers prPHXT7-F and prPHXT7-R) using plasmid p426HXT7-6His as template. Restriction sites introduced within the primer overhangs (EcoRI and EagI) facilitated the insertion of the pHXT7 promoter into pCA064 immediately 5′ of STP1, creating pCA070. Plasmid pCA072 was obtained by subcloning a BstEII/PstI fragment from pCA047 into pCA070; this plasmid contains a pHXT7-promoted STP1-3xHA allele. pCA078 was created by subcloning the whole insert of pCA072 as a BstXI/EcoRI fragment into pRS202 (Connelly and Hieter 1996). The coding region of STP1 was PCR-amplified (primers prSTP1-BamHI-ATG and prSTP1-STOP-BamHI) using pCA029 as template. BamHI sites within the primers facilitated the introduction of STP1 into the vector pSos (Stratagene), forming pCA060, which encodes an in-frame hSos–Stp1p fusion protein. In vivo recombination in yeast between the LEU2 gene of pCA060 and a PCR-amplified URA3 gene (primers prDLEU2toURA3-F and prDLEU2toURA3-R) with ends homologous to the 50-bp outermost parts of LEU2 formed pCA093. pCA094 was constructed by excising the BamHI fragment containing STP1 of pCA093 and religating the vector backbone. pCA099 and pCA104 were constructed by PCR amplification of the 3′ (primers prSTP1-BamHI-DNT and prSTP1-STOP-BamHI) and 5′ (primers prSTP1-BamHI-ATG and prSTP1-DCT-STOP-BamHI) regions of STP1, respectively, and cloning them into BamHI-restricted pCA094. Plasmid pCA103 was constructed as follows: STP2, amplified by PCR using primers prSTP2-KpnI-F and prSTP2-BamHI-R and genomic DNA (PLY118) as a template, was inserted into a KpnI- and BamHI-restricted pRS316 that had previously been modified in the multicloning site by restriction with SpeI and SacI, T4 polymerase blunting, and religation. The epitope-tagged allele of STP2-HA in plasmid pCA111 was created by inserting an EagI-flanked cassette encoding a thrice-reiterated HA1 epitope into an EagI site, introduced by single-stranded mutagenesis (primer prSTP2-EagI C), just prior to the termination codon of STP2.
Complementation tests of plasmid-borne STP1 alleles, encoding epitope-tagged and fusion proteins, were performed in strain CAY42 (stp1Δ leu2). This strain grows slowly on SC media because of reduced L-leucine uptake, primarily a consequence of lower expression of the L-leucine permease genes BAP2 and BAP3. All plasmid-borne alleles of STP1 were also tested to exhibit the proper SC− (i.e., wild-type STP1) or SC+ (i.e., STP1Δ131 and STP1Δ132) phenotype in strain YMH119 (ssy1Δ leu2). Strains were transformed with the plasmids containing STP1 (pCA029) or STP1Δ131 (pCA027) as positive controls and the vector without insert (pRS316) as negative control. Complementation tests of the plasmid-borne STP2 alleles, including the pCA111 encoding epitope-tagged protein, were performed in CAY123 (stp1Δ stp2Δ). Because of decreased uptake of L-isoleucine or L-valine, this double mutant exhibits slow growth on YPD + MM compared with wild-type or stp1Δ single-mutant strains (Jørgensen et al. 1998).
Protein manipulations
Whole-cell protein extracts were prepared according to Silve et al. (1991). For time-course experiments, overnight cultures of cells were diluted in fresh SD to an OD600 of 0.2. Cells were grown to early log phase (OD600 of 0.8–1.0), and the cultures were split into two equal aliquots and placed into fresh prewarmed flasks. An aliquot of a 100× concentrated stock solution of L-leucine (0.13 M) was added to one culture, and an equal aliquot of water was added to the control culture. Then 1-mL aliquots of cell cultures were harvested at the indicated time points, and whole-cell protein extracts were prepared. Extracted proteins were resolved using SDS-PAGE and analyzed by immunoblotting. Immunoblots were incubated with primary antibody in blocking buffer diluted as follows: 12CA5 ascites fluid (anti-HA monoclonal), 1:1500; or anti-GST rabbit polyclonal antibodies diluted to 2 μg/mL. Immunoreactive bands were visualized by chemiluminescence detection of horseradish peroxidase (ECL+ Western Blotting Detection System; Amersham) conjugated to a secondary antibody (anti-rabbit Ig from donkey, or anti-mouse Ig from sheep; Amersham), and quantitated by using the LAS1000 system (Fuji Photo Film Co. Ltd.).
Fluorescence microscopy
Cells were processed for indirect immunofluorescence analysis essentially as described in Burke et al. (2000). In brief, CAY59 transformed with pCA078, expressing a functional HA-epitope-tagged Stp1p, was grown in SD to an OD600 of 0.7. The culture was split into two equal aliquots, and L-leucine was added to one of the cultures from a 100× concentrated stock solution to give a final concentration of 1.3 mM. The cultures were incubated an additional 30 min, after which an aliquot of 37% formaldehyde was added directly to the cultures to a final concentration of 4.5%. Cells were fixed at 30°C for 1 h. The primary antibody used was the 12CA5 anti-HA monoclonal antibody, obtained from ascites fluid, diluted 1:300. The secondary antibody was Alexa Fluor 488 conjugated to goat anti-mouse IgG (H + L; Molecular Probes) diluted 1:500.
Acknowledgments
We thank Mårten Hammar for isolating the ASI13-1 mutation, and the other members of Ljungdahl laboratory, specially Hanna Forsberg and Fredrik Gilstring, for constructive comments throughout the course of this work. The authors thank Bruno André (YCpAGP1-lacZ), Eckhard Boles (p426HXT7–6His), and Stefan Jentsch (N-end rule model substrates, L-β-Gal and M-β-Gal) for their kind gifts of plasmids. Wolfgang Hilt is gratefully acknowledged for providing the proteasome-defective yeast strains. Finally, we thank Adam Antebi for critical comments on the manuscript. This research was supported by the Ludwig Institute for Cancer Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL plju@licr.ki.se; FAX 46-8-33-28-12.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.239202.
References
- André B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein–protein interactions. Mol Cell Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Barnes D, Lai W, Breslav M, Naider F, Becker JM. PTR3, a novel gene mediating amino acid-inducible regulation of peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1998;29:297–310. doi: 10.1046/j.1365-2958.1998.00931.x. [DOI] [PubMed] [Google Scholar]
- Bernard F, André B. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol Microbiol. 2001a;41:489–502. doi: 10.1046/j.1365-2958.2001.02538.x. [DOI] [PubMed] [Google Scholar]
- ————— Ubiquitin and the SCFGrr1 ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 2001b;496:81–85. doi: 10.1016/s0014-5793(01)02412-7. [DOI] [PubMed] [Google Scholar]
- Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science. 2002;295:813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in yeast genetics: A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetichore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer M, Bebelman JP, Goncalves PM, Maat J, Van Heerikhuizen H, Planta RJ. Regulation of expression of the amino acid transporter gene BAP3 in Saccharomyces cerevisiae. Mol Microbiol. 1998;30:603–613. doi: 10.1046/j.1365-2958.1998.01094.x. [DOI] [PubMed] [Google Scholar]
- de Boer M, Nielsen PS, Bebelman JP, Heerikhuizen H, Andersen HA, Planta RJ. Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:974–981. doi: 10.1093/nar/28.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion T, Grausland M, Kielland-Brandt C, Andersen HA. Amino acids induce expression of BAP2, a branched-chain amino acid permease gene in Saccharomyces cerevisiae. J Bacteriol. 1996;178:2025–2029. doi: 10.1128/jb.178.7.2025-2029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion T, Regenberg B, Jørgensen MU, Kielland-Brandt MC, Andersen HA. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:643–650. doi: 10.1046/j.1365-2958.1998.00714.x. [DOI] [PubMed] [Google Scholar]
- Forsberg H, Ljungdahl PO. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p–Ptr3p–Ssy5p sensor of extracellular amino acids. Mol Cell Biol. 2001a;21:814–826. doi: 10.1128/MCB.21.3.814-826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr Genet. 2001b;40:91–109. doi: 10.1007/s002940100244. [DOI] [PubMed] [Google Scholar]
- Forsberg H, Hammar M, Andréasson C, Molinér A, Ljungdahl PO. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor independent (ASI) genes in Saccharomyces cerevisiae. Genetics. 2001;158:973–988. doi: 10.1093/genetics/158.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, Pan X, McCusker JH. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast. 1999;15:507–511. doi: 10.1002/(SICI)1097-0061(199904)15:6<507::AID-YEA369>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Grauslund M, Didion T, Kielland-Brandt MC, Andersen HA. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1269:275–280. doi: 10.1016/0167-4889(95)00138-8. [DOI] [PubMed] [Google Scholar]
- Gueckel R, Enenkel C, Wolf DH, Hilt W. Mutations in the yeast proteasome β-type subunit Pre3 uncover position-dependent effects on proteasomal peptidase activity and in vivo function. J Biol Chem. 1998;273:19443–19452. doi: 10.1074/jbc.273.31.19443. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH. The active sites of the eukaryotic 20S proteasome and their involvement in subunit precursor processing. J Biol Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- Hilt W, Enenkel C, Gruhler A, Singer T, Wolf DH. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. J Biol Chem. 1993;268:3479–3486. [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T, Rape M, Jentsch S. Membrane-bound transcription factors: Regulated release by RIP or RUP. Curr Opion Cell Biol. 2001;13:344–348. doi: 10.1016/s0955-0674(00)00218-0. [DOI] [PubMed] [Google Scholar]
- Iraqui I, Vissers S, Bernard F, de Craene JO, Boles E, Urrestarazu A, André B. Amino acid signaling in Saccharomyces cerevisiae: A permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen MU, Bruun MB, Didion T, Kielland-Brandt MC. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast. 1998;14:103–114. doi: 10.1002/(SICI)1097-0061(19980130)14:2<103::AID-YEA203>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Klasson H, Fink GR, Ljungdahl PO. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol Cell Biol. 1999;19:5405–5416. doi: 10.1128/mcb.19.8.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Li S, Dean S, Li Z, Horecka J, Deschenes RJ, Fassler JS. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via transcription factor, Skn7p. Mol Biol Cell. 2002;13:412–424. doi: 10.1091/mbc.01-09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl PO, Gimeno CJ, Styles CA, Fink GR. SHR3: A novel component of the secretory pathway specifically required for the localization of amino acid permeases in yeast. Cell. 1992;71:463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- Nelissen B, De Wachter R, Goffeau A. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Micro Rev. 1997;21:113–134. doi: 10.1111/j.1574-6976.1997.tb00347.x. [DOI] [PubMed] [Google Scholar]
- Nielsen PS, van den Hazel B, Didion T, de Boer M, Jørgensen M, Planta RJ, Kielland-Brandt MC, Andersen HA. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol Gen Genet. 2001;264:613–622. doi: 10.1007/s004380000347. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Hilger F, Tatchell K. Comparison of thermosensitive alleles of the CDC25 gene involved in the cAMP metabolism of Saccharomyces cerevisiae. Genetics. 1990;124:797–806. doi: 10.1093/genetics/124.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B, Nielsen PS, Kielland-Brandt MC, Nielsen J. Coupling between amino acid sensing and carbon metabolism via Ssy1, Stp1 and Stp2. Yeast. 2001;18:S223. [Google Scholar]
- Ross-Macdonald P, Sheehan A, Roeder GS, Snyder M. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1997;94:190–195. doi: 10.1073/pnas.94.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve S, Volland C, Garnier C, Jund R, Chevallier MR, Haguenauer-Tsapis R. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol Cell Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belle D, André B. A genomic view of yeast membrane transporters. Curr Opin Cell Biol. 2001;13:389–398. doi: 10.1016/s0955-0674(00)00226-x. [DOI] [PubMed] [Google Scholar]
- Wang SS, Stanford DR, Silvers CD, Hopper AK. STP1, a gene involved in pre-tRNA processing, encodes a nuclear protein containing zinc finger motifs. Mol Cell Biol. 1992;12:2633–2643. doi: 10.1128/mcb.12.6.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]