Abstract
The nucleosome remodeling factor (NURF) is one of several ISWI-containing protein complexes that catalyze ATP-dependent nucleosome sliding and facilitate transcription of chromatin in vitro. To establish the physiological requirements of NURF, and to distinguish NURF genetically from other ISWI-containing complexes, we isolated mutations in the gene encoding the large NURF subunit, nurf301. We confirm that NURF is required for transcription activation in vivo. In animals lacking NURF301, heat-shock transcription factor binding to and transcription of the hsp70 and hsp26 genes are impaired. Additionally, we show that NURF is required for homeotic gene expression. Consistent with this, nurf301 mutants recapitulate the phenotypes of Enhancer of bithorax, a positive regulator of the Bithorax-Complex previously localized to the same genetic interval. Finally, mutants in NURF subunits exhibit neoplastic transformation of larval blood cells that causes melanotic tumors to form.
Keywords: NURF, chromatin remodeling, ISWI, Drosophila, JAK/STAT pathway
The establishment of the body plan of an animal requires the correct initiation and maintenance of expression of a number of developmental regulators. In insects, segmental identity is conferred by region-specific expression of overlapping sets of homeotic selector genes. Systematic genetic screens for changes in segmental fate have been used to identify a wide class of positive-acting transregulatory factors required for homeotic gene function, the trithorax-group (trxG; Kennison 1995). Members of the trxG define novel chromatin-modifying activities. One class comprises the ATP-dependent chromatin remodeling factors. These are multisubunit protein complexes that utilize the energy of ATP hydrolysis to alter gene architecture by perturbing the dynamic properties of nucleosomes, the basic units of chromatin. They can be divided into broad families depending on the catalytic subunit utilized. Brahma, one of the defining members of the trxG, and its yeast homolog SWI2/SNF2 provide the catalytic subunit of the first group of SWI/SNF chromatin remodelers identified (Tamkun et al. 1992).
Other categories of ATP-dependent chromatin remodeling complexes have been discovered using biochemical approaches. In Drosophila, the ISWI ATPase forms the catalytic core of three protein complexes that have been purified from nuclear extracts: ACF (ATP-utilizing chromatin assembly and remodeling factor; Ito et al. 1997), CHRAC (chromatin accessibility complex; Varga-Weisz et al. 1997) and NURF (nucleosome remodeling factor; Tsukiyama et al. 1995). All catalyze energy-dependent nucleosome sliding in vitro (Corona et al. 1999; Hamiche et al. 1999; Langst et al. 1999), but with distinct results by the different complexes. ACF and CHRAC can assemble and slide nucleosomes to establish regular ordered arrays (Ito et al. 1997; Varga-Weisz et al. 1997). In contrast, NURF-induced sliding disrupts nucleosome periodicity (Tsukiyama and Wu 1995; Hamiche et al. 1999). NURF acts in concert with transcription factors to mobilize nucleosomes at promoters, facilitating transcription activation in vitro (Tsukiyama and Wu 1995; Mizuguchi et al. 1997). These properties suggest that NURF should be required for transcription in vivo.
NURF is a complex of four protein subunits. In addition to NURF140—the ISWI ATPase (Tsukiyama et al. 1995)—NURF contains NURF55, a WD-40 repeat protein found in other protein complexes involved in histone metabolism (Martinez-Balbas et al. 1998); NURF38, an inorganic pyrophosphatase (Gdula et al. 1998); and a large subunit NURF301 (Xiao et al. 2001). NURF301 is distantly related to the ACF1/WCRF1 subunits of the ACF and WCRF chromatin remodeling and assembly complexes (Ito et al. 1999). NURF301 contains the PHD fingers, bromodomain, WSTF/Acf1/cbp146 (WAC), and WSTF/Acf1/KIAA0314/ZK783.4 (WAKZ) motifs found in these proteins, but also possesses an N-terminal HMGI/Y-like domain. Reconstitution experiments reveal that both NURF301 and ISWI contribute to the chromatin remodeling activities of NURF. In addition, NURF301 provides an interface through which NURF can interact with a number of transcription factors, in particular the GAGA transcription factor (Xiao et al. 2001).
We sought to characterize the physiological function of NURF. Clues to this have been provided by mutations that affect the catalytic ISWI subunit. Analysis of iswi mutant animals indicates that ISWI is required for homeotic gene activation and chromatin condensation in vivo (Deuring et al. 2000). However, as ISWI is present in NURF and at least two other Drosophila chromatin remodeling complexes—ACF (Ito et al.,1997) and CHRAC (Varga-Weisz et al. 1997)—functions could not be ascribed to a specific remodeling complex. Given the importance of NURF301 for the in vitro activities of NURF, we focused our studies on NURF301 and isolated mutations in this largest NURF subunit. Here we show that nurf301 is required for transcription activation in vivo. In nurf301 mutants homeotic gene expression is impaired. Moreover, we demonstrate that nurf301 corresponds to an uncharacterized member of the trxG, Enhancer of bithorax [E(bx)]. NURF301 is required for induction of the heat-shock genes and has additional functions during larval blood cell development.
Results
Isolation and characterization of nurf301 mutants
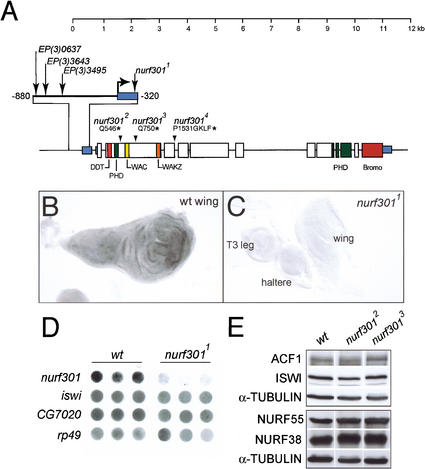
The gene encoding the large NURF subunit, nurf301, was mapped by chromosomal in situ hybridization to the cytological interval 61A. To isolate nurf301 mutants, we first screened P-element lines that had been localized to this region for those that contained inserts within or immediately upstream of nurf301. We identified one homozygous lethal P-element line (l(3)ry122 we call nurf3011), in which a P-element had inserted in the untranslated leader sequence of nurf301 (summarized in Fig. 1A). Subsequently, we recovered a series of EMS-induced mutations that failed to complement nurf3011. The three alleles presented (nurf3012, nurf3013, and nurf3014) encode truncated NURF301 proteins due to the introduction of stop codons at amino acids 546, 750, and 1536, respectively. Homozygous nurf301 mutants (and heteroallelic combinations) were lethal at late-third larval instar or early pupal stages. We also identified three homozygous viable EP insertion lines (Rorth 1996) in which P-elements had inserted upstream of the nurf301 transcription start site. These were used as substrates for imprecise excision to recover lethal excision lines that fail to complement nurf3011. The deficiency Df(3L)3643 was characterized by PCR and shown to remove the ATG of nurf301 and at least two flanking genes downstream of nurf301.
Figure 1.
Characterization of nurf301 mutants. (A) The structure of the nurf301 locus, the position of P-element insertion lines, and the location of EMS-induced lesions are shown. nurf3012 and nurf3013 encode glutamine to stop codon changes at amino acids 546 and 750, respectively. nurf3014 contains a splice-donor site mutation that blocks splicing of the fourth intron. The aberrant transcript introduces four additional amino acids (GKLF) and an in-frame stop codon after proline residue 1531. (B,C) In situ hybridization using a 1.3-kb probe spanning the 3′ end of nurf301 (nucleotides 6800–8000) shows that ubiquitous expression of nurf301 is lost in homozygous mutant nurf3011 larvae (cf. wild-type and mutant third instar discs). (D) Dot-blot analysis of third larval instar total RNA (in triplicate) shows that nurf301 RNA is reduced in nurf3011 mutants relative to wild-type. In contrast, iswi and CG7020, a putative gene located upstream of nurf301 (a fragment from the EST clone LP07661 was used as a probe), are unaffected. rp49 provides a loading control. (E) Protein levels of the NURF subunits. NURF38, NURF55, and ISWI, and the ACF1 subunit of ACF are unchanged in nurf3012 and nurf3013 mutants, relative to α-TUBULIN as shown by Western analysis of extracts from third instar larvae.
We showed by in situ hybridization (Fig. 1B,C) and Northern analysis (Fig. 1D) that the normal expression of nurf301 is reduced in the P-element insertion line nurf3011. In this mutant, transcription of iswi or a predicted gene upstream of nurf301 (CG7020) is unaffected (Fig. 1D). We confirmed that lethality of this line is caused by the P-element insertion as precise excision of the P-element restores viability. In all the nurf301 mutants, the protein levels of the three other NURF subunits (ISWI, NURF55, and NURF38) and the ACF1 subunit of ACF and CHRAC are unchanged (data shown for nurf3012 and nurf3013 in Fig. 1E). This indicates that the nurf301 mutations specifically compromise NURF activity without significantly affecting the other ISWI-containing complexes ACF or CHRAC.
Heat-shock gene activation is impaired in nurf301 mutant animals
NURF had originally been purified from nuclear extracts as a factor required to disrupt chromatin assembled in vitro on the promoters of the Drosophila heat-shock gene hsp70. Using in vitro assays it was shown that NURF cooperates with the GAGA factor to mobilize nucleosomes on the promoter of the heat-shock genes, establishing a nucleosome-free domain over the promoter, thus exposing sites for the heat-shock transcription factor (HSF; Tsukiyama et al. 1994; Tsukiyama and Wu 1995).
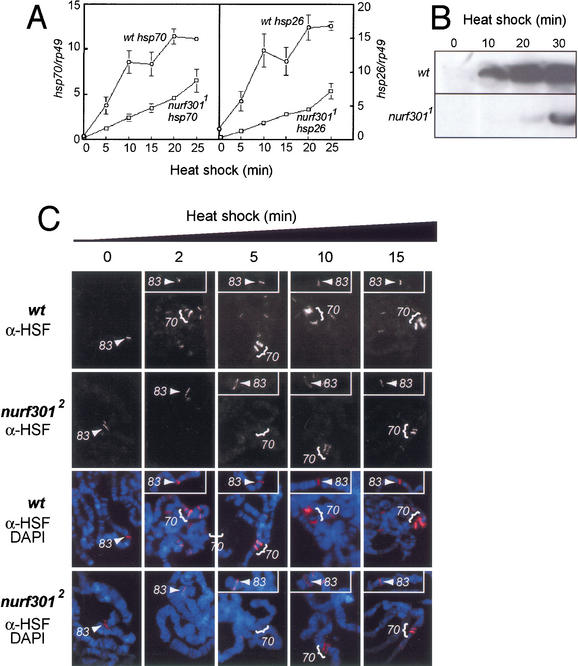
We examined if expression of the heat shock genes is affected in nurf301 mutant animals. RNA dot-blot analysis shows that heat-shock-induced transcription of hsp26 and hsp70 is impaired in nurf3011 larvae (Fig. 2A). Impaired transcription leads to reduced heat-shock protein accumulation. Although HSP70 is detected in wild-type siblings after 10 min heat-shock, no protein is expressed in mutant animals (Fig. 2B). After 20 min of heat-shock, HSP70 is highly expressed in wild-type larvae. In contrast, nurf301 mutant animals only express low levels of HSP70 (Fig. 2B).
Figure 2.
nurf301 is required for heat-shock gene induction. (A) A quantitation of RNA dot-blots shows delay in hsp26 and hsp70 transcription in nurf3011 mutant animals. hsp26 and hsp70 transcript levels are normalized to rp49. Points and bars represent the mean and standard deviation of three independent determinations. (B) Western blotting shows that HSP70 protein synthesis in homozygous mutant animals is reduced relative to the wild-type. (C) HSF was localized to the hsp70 loci (brackets) and hsp83 locus (arrowheads) on polytene chromosomes with antibodies against HSF, following 0-, 2-, 5-, 10- and 15-min heat-shock. HSF-binding to the major heat-shock loci is impaired in nurf3012 mutants.
Consistent with these findings, binding of the heat shock transcription factor HSF is impaired in nurf301 mutants. In wild-type animals, immunofluorescence of salivary gland polytene chromosomes shows that, with the exception of the hsp83 locus at 63B, HSF does not generally bind to polytene chromosomes in the absence of heat stress (Fig. 2C). HSF-binding to hsp83 has been shown to be qualitatively different from the other heat-shock loci (Shopland and Lis 1996). The hsp83 locus does not appear to be GAGA factor-dependent as no (GA.CT)n elements are present in the promoter, nor does GAGA factor accumulate at this locus (Tsukiyama et al. 1994). Instead, HSF-binding is proposed to be facilitated by other transacting factors (Shopland and Lis 1996).
Within several minutes of heat-shock, the bulk of HSF forms trimers competent for specific DNA-binding, and binds to the heat-shock loci and a number of other sites (Westwood et al. 1991). Prominent are the 87A and 87C loci (Fig. 2C, bracketed) which carry two and three copies, respectively, of the hsp70 gene. In nurf301 mutant animals, hsp83 accumulates HSF as in nonshocked conditions. After 2 or 5 min heat-shock, no binding of HSF to hsp70 loci is detectable (Fig. 2C). HSF-binding to the hsp70 loci can only be detected after 10 min of heat-shock, and HSF staining is reduced compared to wild-type animals (Fig. 2C). Impaired HSF-binding is accompanied by reduced accumulation of RNA polymerase II at the hsp70 loci (data not shown).
nurf301 is required for homeotic gene expression
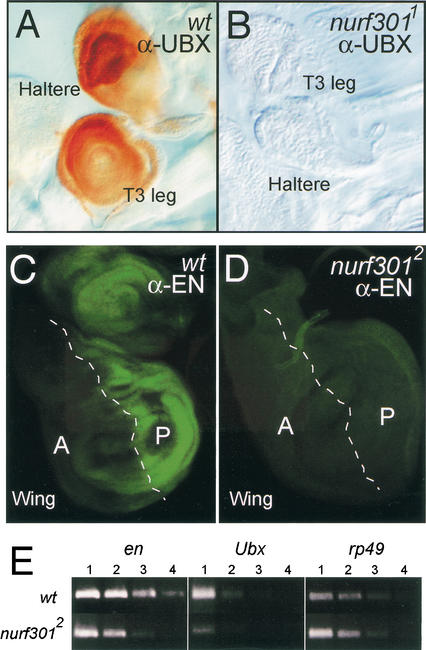
Deuring et al. (2000) had shown that ISWI, the catalytic subunit of NURF, is required for expression of the homeotic gene engrailed (en). However, ISWI is also a component of two other chromatin remodeling complexes, ACF (Ito et al. 1997) and CHRAC (Varga-Weisz 1997). To resolve which ISWI-containing complex is required for homeotic gene expression we examined expression of Ultrabithorax (Ubx) and engrailed (en) in nurf301 mutant animals. We found that when both copies of nurf301 are mutated, in homozygous mutant nurf3011 larvae, expression of the UBX protein becomes undetectable. The normal expression of UBX in the haltere and third leg discs of wild-type third instar larvae is absent in nurf301 mutant animals (Fig. 3A,B). We also showed that expression of the homeotic gene en requires nurf301. The normal expression of EN in the posterior compartment of imaginal discs is abolished in nurf3012 mutants (Fig. 3C,D). Semiquantitative RT–PCR analysis confirms that Ubx and en transcript levels are reduced in nurf301 mutant animals. These results confirms that the defects in homeotic transcription seen in iswi mutants are caused by abrogated NURF function.
Figure 3.
nurf301 is required for homeotic gene expression. (A,B) UBX protein in imaginal discs of the third thoracic segment (brown staining) is undetectable in nurf3011 homozygotes. (C,D) Antibody staining shows that EN protein (revealed in green), which normally can be detected in the posterior compartment of all imaginal discs, is lost in nurf3012 mutant larvae. (E) Semiquantitative RT–PCR analysis confirms that Ubx and en transcript abundance is reduced between 5- and 25-fold in total RNA isolated from nurf301 mutant animals. Lanes represent fivefold serial dilutions of mutant and wild-type total RNA (lane 1, 200 ng; lane 2, 40 ng; lane 3, 8 ng; and lane 4, 1.6 ng).
nurf301 recapitulates E(bx) phenotypes
A positive regulator of the Bithorax-Complex, E(bx), had previously been localized genetically to 61A, the same cytological interval as nurf301 (Bhosekar and Babu 1987; Lewis cited in Lindsley and Zimm 1992). However, unlike numerous regulators of the BX-C, E(bx) had not been cloned. As NURF is required for expression of Ubx, we tested whether nurf301 corresponds to E(bx). Both alleles of E(bx) were no longer extant, so we examined if the mutations we had isolated in nurf301 recapitulated the published morphological properties of E(bx) mutants.
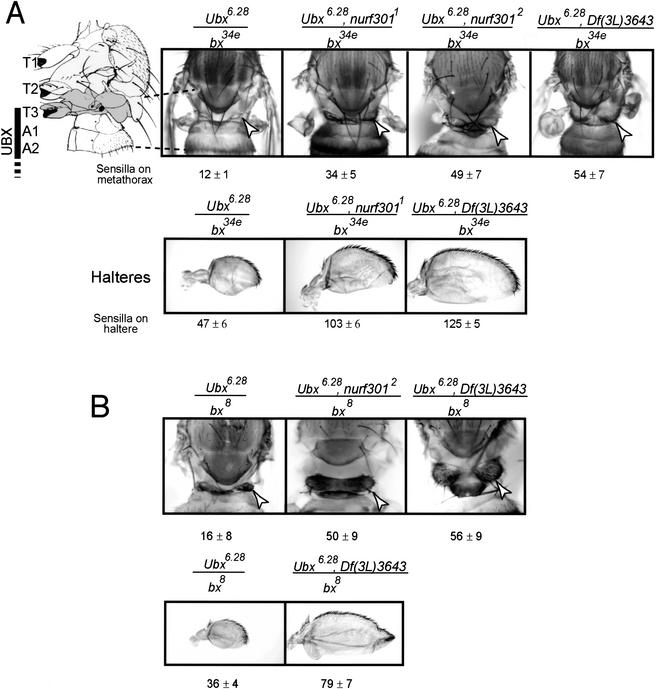
We found that nurf301 mutants, like E(bx), increase the severity of bithorax (bx) mutant phenotypes. bx is a DNA regulatory element required for correct expression of Ubx in regions that give rise to the third (T3), but not second thoracic segment (T2) of the adult fly (Lewis 1978; Bender et al. 1983). This expression distinguishes T3 from T2 identity. Loss or reduction of UBX levels in bx mutant animals (Ubx6.28/bx34e and Ubx6.28/bx8 mutant combinations (shown in Fig. 4A,B) causes a homeotic transformation of the third thoracic segment to the anterior second thoracic segment. Thus, the third thoracic segment, which is normally vestigial and naked, is transformed into the second thoracic segment, increasing its size and causing sensory bristles to develop (Fig. 4A). Moreover, the haltere (T3) is transformed toward wing fate (T2), manifested by increases in size and the development of bristles. The strength of these transformations is increased when one copy of E(bx) also is removed (Bhosekar and Babu 1987). Mutation of one copy of nurf301 similarly enhances bx phenotypes. With one copy of either the nurf3011, nurf3012 or a deficiency that removes nurf301—Df(3L)3643—the strength of the transformation is enhanced (Fig. 4A,B). nurf301 enhances both bx34e and bx8 mutations.
Figure 4.
nurf301 corresponds to E(bx). In the presence of one copy of either nurf3011, nurf3012, or the nurf301 deficiency Df(3L)3643, the strength of the anterior transformation seen in bithorax mutant combinations is increased. Transformation is scored both by increases in the size and bristle number on the metathoracic segment and the severity of the haltere (T3) to wing (T2) transformation. A shows bx34e/Ubx6.28 combinations and B shows bx8/Ubx6.28 combinations. Mean haltere and T3 bristle numbers ± standard deviation for 20 flies of each genotype are indicated. Arrowheads denote the metathorax (T3). Photographs of nota are composites of three images taken at different focal planes of the animals.
Although NURF is required for expression of the homeotic genes in imaginal discs, neither E(bx) nor nurf301 homozygous mutant larvae display obvious homeotic transformations of the larval cuticle. The absence of mutant larval cuticle phenotypes is likely due to the large maternal dowry of nurf301 transcript contributed to embryos (data not shown). Larval cuticular patterning is established before these transcripts have dissipated. We attempted to generate embryos lacking the maternal nurf301 contribution through use of the dominant female sterile technique. Although we could produce germ-line clones using the parental chromosome, we were unable to recover germ-line clones using nurf3011. Like ISWI (Deuring et al. 2000), NURF301 is required for ovary development.
Aberrant male X chromosome morphology in nurf301 mutant animals
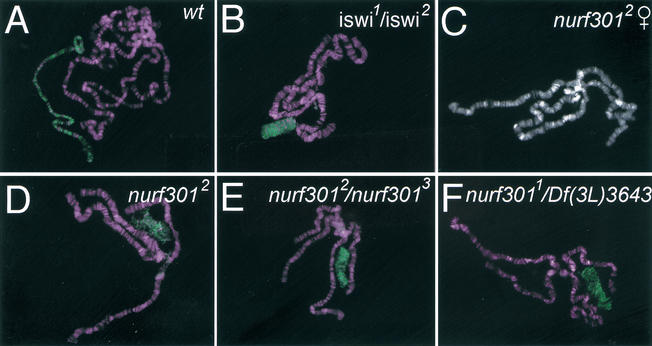
The defects in homeotic transcription seen in nurf301 mutant animals effectively duplicate those reported for animals lacking the catalytic ISWI subunit (Deuring et al. 2000). However, ISWI is also required to maintain higher order chromosome structure. In iswi mutants the male X chromosome is grossly disrupted relative to autosomes as revealed by polytene chromosome preparations (Fig. 5B; Deuring et al. 2000). We found that in nurf301 mutants the male X chromosome is similarly affected. The male X chromosome, identified by anti-MSL2 staining, is reduced in length and breadth, as seen in iswi mutant animals (cf. wild-type and mutant animals in Fig. 5A,D–F). The male X chromosome in homozygous mutant nurf3012 animals is highly aberrant (Fig. 5D). Disorganized male X chromosome morphology also was observed in nurf3012/nurf3013 and hemizygous nurf3011/Df(3L)3643 mutant animals (Fig. 5E,F). Our results demonstrate that the chromosome condensation defect caused by perturbed ISWI function is mediated through the NURF complex. Although the effects on male X chromosome structure suggest that NURF can influence global chromosome structure, we found that NURF function was not required for heterochromatic gene silencing. Reduction in NURF301 levels has no effect on position effect variegation (data not shown). This is consistent with results reported by Cryderman et al. (1999) who showed that iswi is not a modifier of PEV.
Figure 5.
Aberrant male X chromosome in nurf301 mutant animals. Polytene chromosomes prepared from male wt (A), iswi (B), and nurf301 (D–F) mutant animals were stained with antibodies against MSL2 (shown in green) to reveal the male X chromosome. As seen in iswi1/iswi2 mutant animals (Deuring et al. 2000), male X chromosome morphology is perturbed in nurf301 mutants. (C) The X chromosome morphology in female nurf301 mutants is unaffected.
Mutations in NURF subunits affect hematopoietic development
During the course of this analysis we noticed that nurf301 mutant animals display a high incidence of melanotic tumors. Melanotic tumors have previously been reported in a number of mutant backgrounds and are generally caused by neoplastic transformation of the larval blood cells. The circulating cells (hemocytes) of the larval blood or hemolymph provide one tier of the innate immune system of insects by encapsulating or engulfing pathogens (for review, see Immler and Hoffman 2001; Luo and Dearolf 2001; Tzou et al. 2002). A number of mutations have been shown to trigger the overproliferation and premature differentiation of hemocytes. Tumors form when these cells aggregate, or invade and encapsulate normal larval tissues (Gateff 1978; Rizki and Rizki 1979).
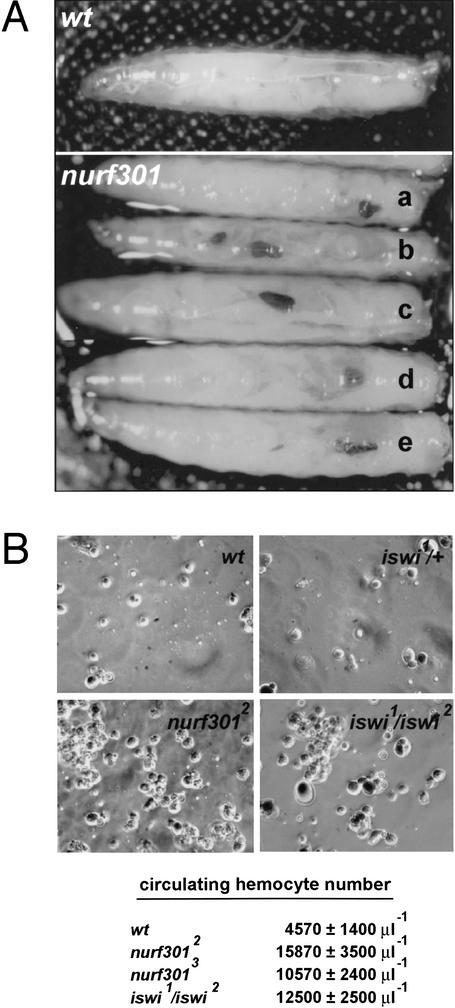
We observed melanotic tumors both in EMS-induced nurf301 mutants that truncate NURF301, the P-element induced mutation that reduces nurf301 transcript levels, and allelic combinations of these mutants (Fig. 6A). Tumor penetrance is extremely high (100% for nurf3012 at 25°C). Consistent with tumor development, circulating hemocyte cell number was increased dramatically in hemolymph isolated from nurf301 mutant animals (Fig. 6B). A large percentage of animals lacking ISWI, the catalytic subunit of NURF, also displayed melanotic tumors confirming that disrupted NURF function induces tumor formation (data not shown). In iswi mutant animals the number of circulating hemocytes is also increased (Fig. 6B). In both nurf301 and iswi mutant hemolymph, small aggregates of hemocytes are often observed. All hemocyte cell types are present, from small round cells (prohemocytes) to crystal cells and lamellocytes.
Figure 6.
Melanotic tumors in NURF mutant animals. Melanotic tumors occur in homozygous mutant nurf301 third instar larvae. (A) Tumors are observed in all combinations of nurf301 mutant alleles. Genotypes displayed are nurf3012/nurf3012 (a), nurf3012/nurf3013 (b), nurf3012/nurf3014 (c), nurf3011/nurf3012 (d), and nurf3011/nurf3013 (e). (B) The circulating hemocyte cell number in hemolymph isolated from nurf301 and iswi mutant third instar larvae is increased considerably with respect to wild-type animals.
In Drosophila, larval blood cell transformation and melanotic tumor formation can be induced by inappropriate activation of either of two distinct signaling cascades: the TOLL or the JAK/STAT pathway. Inappropriate activation and nuclear-localization of the Drosophila NF-κΒ homolog DORSAL, caused either by constitutive activation of the TOLL receptor or removal of the inhibitor, the Drosophila IκΒ CACTUS, leads to melanotic tumors in third instar larvae (Qui et al. 1998). In the second pathway, gain-of-function mutations in HOPSCOTCH (HOP), the Drosophila Janus Kinase (JAK), induce melanotic tumors (Hanratty and Dearolf 1993; Harrison et al. 1995; Hou et al. 1996). HOP gain-of-function mutants cause tumor development by triggering constitutive activation and DNA-binding by the Drosophila STAT transcription factor, STAT92E.
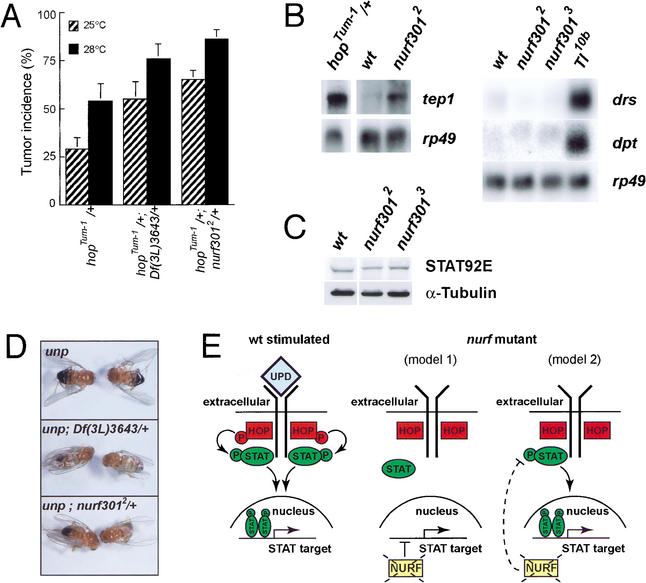
To resolve whether the melanotic tumors seen in the nurf301 mutants were caused by misregulation of either the TOLL or HOP/STAT92E pathways, we determined if nurf301 mutants enhance tumor phenotypes seen in constitutively active TOLL or HOP mutant lines. We found that tumor incidence in animals carrying one copy of a gain-of-function HOP mutation—hopTum-1 —is increased by simultaneous reduction in NURF301 levels (Fig. 7A). In contrast, removal of one copy of NURF301 fails to enhance the TOLL gain-of-function allele Tl10b (data not shown). The results suggest that NURF acts as a negative regulator within the Drosophila JAK/STAT signaling pathway.
Figure 7.
NURF is a negative regulator of the JAK/STAT signaling pathway. (A) Reduction of nurf301 levels enhances gain-of-function hopTum-1 mutant phenotypes. Vials of flies were raised at either 25°C or 28°C. The percentage of adult female flies of each genotype in a vial that display tumors was scored as tumor incidence. Data are the mean and standard deviation of at least six vials of flies. (B) nurf301 mutant animals express elevated levels of tep1 but neither the antimicrobial peptide gene drosomycin (drs) nor diptericin (dpt) is induced. (C) Levels of the STAT92E transcription factor are not elevated in nurf301 mutant animals. (D) Reduction of NURF301 levels suppresses the phenotypes of mutations in a ligand for the JAK receptor UNPAIRED (outstretched). (E) Models for how NURF may down-regulate the HOP/STAT92E signaling pathway. NURF could either repress targets of STAT92E or be required for expression of a negative regulator of STAT92E activation. In either case, in the absence of NURF, STAT92E target genes will be transcribed, mimicking STAT92E activation.
Molecular signatures of both JAK and TOLL activation have been defined. It is known that HOP gain-of-function mutants induce expression of a complement-like protein TEP1 (Lagueux et al. 2000). Overactivation of the TOLL pathway also induces TEP1 synthesis but primarily induces expression of antimicrobial peptides, including DROSOMYCIN (DRS) and DIPTERICIN (DPT). We found that loss of nurf301 induces tep1 but fails to induce drs or dpt (Fig. 7B), demonstrating that NURF301 principally affects the HOP/STAT92E pathway. We tested whether nurf301 interacts genetically with other known components of the HOP/STAT92E pathway. Certain mutations in unpaired (upd, also known as outstretched), which encodes a ligand for the HOP receptor, display a characteristic wings-out phenotype, due to decreased activation of HOP and consequently decreased STAT92E function. When NURF301 levels are simultaneously decreased in these mutant backgrounds, animals are mostly restored to the wild-type (Fig. 7D). These genetic interactions confirm that NURF301 acts as a negative regulator of the HOP/STAT92E pathway, at a point downstream of HOP. Hence, disruption of NURF could affect either STAT92E or the targets of STAT92E. We found that in nurf301 mutants, levels of the STAT92E transcription factor are not elevated (Fig. 7C), suggesting that NURF acts to repress the activity of STAT92E or the expression of some STAT92E target genes.
Discussion
Here we provide a physiological correlate for the demonstrated biochemical properties of the nucleosome remodeling factor (NURF). Our results show that the largest subunit of NURF, NURF301, is required for transcription in vivo. These data confirm an earlier report of Tamkun and co-workers (Deuring et al. 2000) that showed that ISWI, the catalytic subunit of NURF, is required for transcription activation in vivo. However, because ISWI is also a subunit of the ACF (Ito et al. 1999) and CHRAC (Varga-Weisz, et al. 1997) chromatin remodeling complexes, it had not previously been possible to ascribe iswi phenotypes to a specific remodeling factor. As NURF301 is not present in ACF and CHRAC, by analyzing nurf301 mutations we have been able to resolve specific functions of NURF. We show that for the tested phenotypes, mutations in nurf301 and iswi are indistinguishable. We also demonstrate that NURF functions during larval blood cell development. In the absence of either the NURF301 or ISWI subunits, circulating hemocytes undergo a neoplastic transformation that results in the overproliferation of blood cell types. Melanotic tumors are formed when these cells invade and encapsulate normal larval tissues. Our data suggest this is caused by misregulation of the Drosophila JAK/STAT pathway.
NURF is required for transcription in vivo
Our results demonstrate that NURF is also required in vivo for transcription of the heat shock and homeotic genes. This agrees with the demonstrated biochemical properties of NURF. NURF can catalyze nucleosome sliding in vitro, in this manner disrupting chromatin structure (Hamiche et al. 1999). Nucleosome sliding on promoters offers a simple way to clear chromatin from promoter elements and has been shown to potentiate the binding of transcription factors and the recruitment of transcription initiation machinery in vitro (Mizuguchi et al. 1997; Kang et al. 2002). Deployment of NURF at target promoters should be critical for gene activation.
An important question is how NURF is recruited to target sites in vivo. Four genes we show to be dependent on nurf301 for expression—Ubx, en, hsp26, and hsp70—all contain multiple binding sites for the GAGA factor, which is genetically required for their correct expression (Soeller et al. 1993; Farkas et al. 1994; Bhat et al. 1996). On the Drosophila hsp70 and hsp26 promoters (GA.CT)n cognate elements to which the GAGA factor binds are required for HSF-binding. When these sequences are deleted, HSF-binding to transgenes in polytene chromosomes is impaired (Shopland et al. 1995; Shopland and Lis 1996), consistent with the defects we see in nurf301 mutant animals. It is therefore compelling that recent biochemical studies show that NURF and the GAGA factor bind to each other in crude extracts, and that purified NURF301 and GAGA factor interact directly in vitro (Xiao et al. 2001). The principal interacting domains map to an N-terminal region of NURF301 and a stretch flanking the Zn finger DNA-binding motif of GAGA factor (Xiao et al. 2001). These data suggest that NURF is recruited by the GAGA factor through specific, direct interactions with the NURF301 subunit, to catalyze local sliding of nucleosomes at bx, en, hsp26, and hsp70 promoters, increasing accessibility to sequence-specific transcription factors and RNA polymerase II. Curiously, though, reduction of nurf301 levels fails to enhance phenotypes of mutations in Trithorax-like, the gene that encodes the GAGA factor (P. Badenhorst, unpubl.).
Conceptual models of the function of chromatin remodeling machines reinforce the view that these complexes are required exclusively during gene activation to expose or “open-up” chromatin. However, nucleosome sliding could equally be harnessed to repress genes. Phenotypic analysis of the orthologous yeast ISW2 complex suggests that ISWI-containing complexes may function both to activate and repress genes. The yeast ISW complexes appear to be recruited to sites within the genome through direct interaction with DNA-binding proteins to activate and repress genes by repositioning nucleosomes (Goldmark et al. 2000; Kent et al. 2001). Drosophila ISWI has been shown to be associated with transcriptionally silent regions of chromatin in salivary gland nuclei, suggesting that it may be involved in repression in this tissue (Deuring et al. 2000). Our analysis of NURF function during larval blood cell development suggests that NURF can repress targets of the JAK/STAT signaling pathway. We show that in nurf301 mutants the expression of the complement-like protein TEP1 is induced. It remains to be established whether tep1 is a direct target of NURF.
NURF and tumorigenesis
Misregulation of TEP1 is seen in mutants that overactivate the Drosophila JAK/STAT (HOP/STAT92E) signaling cascade and induce the formation of melanotic tumors. Increased signaling through the HOP/STAT92E pathway leads to the overproliferation and aberrant differentiation of larval blood cells that subsequently invade and encapsulate normal host tissue (Hanratty et al. 1993; Harrison et al. 1995). We have found that animals lacking NURF301, or the catalytic ISWI subunit, exhibit an identical neoplastic transformation of larval blood cells. In the absence of NURF, the proliferation and differentiation of hemocytes, and the accumulation of lamellocytes, is triggered.
Our data suggests that NURF normally represses targets of the HOP/STAT92E pathway. Genetic epistasis places nurf301 downstream of HOP. We show that loss of NURF resembles gain-of-function mutations in hop, and that targets of the HOP/STAT92E cascade are up-regulated in nurf301 mutants. Normally, HOP activation leads to the expression and posttranslational modification of STAT92E (Chen et al. 2002). We found that although nurf301 mutants activate the HOP/STAT92E pathway, the levels of the STAT92E transcription factor are unchanged in NURF mutant animals. We suggest that NURF acts downstream of STAT92E. In resting cells, in the absence of HOP/STAT92E signaling, NURF could normally repress STAT92E target genes. When NURF is removed, repression is no longer maintained, and targets are transcribed mimicking the effects of HOP activation. However, STAT activity is also influenced by a number of inhibitory, STAT-binding proteins. Among these are the suppressor of cytokine signaling (SOCS; Callus and Mathey-Prevot 2002) and protein inhibitor of activated STAT (PIAS) family of inhibitors (Betz et al. 2001; Hari et al. 2001). It is possible that NURF is required for expression of one such inhibitor, and that loss of NURF activates the HOP/STAT92E cascade by removing a STAT inhibitor.
The involvement of NURF in larval blood cell development agrees well with recent literature implicating a number of chromatin-modifying or chromatin-associated complexes in hemocyte development and melanotic tumor formation. Mutations in modulo, which encodes an interacting partner of the coactivator CBP, cause melanotic tumors (Bantignies et al. 2001). Of particular significance, screens for mutations that cause hematopoietic defects identified domino (dom), which encodes a member of the SWI2/SNF2 family of DNA-dependent ATPase that is distantly related to ISWI (Ruhf et al. 2001). Mutations in domino cause overproliferation of hemocytes, like NURF mutants. However, unlike NURF mutants, hemocytes fail to enter the hemolymph and remain trapped in enlarged lymph glands that become melanized. It will be interesting to assess the relative contributions of the ISWI and DOM complexes in the regulatory hierarchy of larval blood development.
NURF maintains normal sex chromosome morphology
A striking feature of male animals that lack either NURF301 or the catalytic subunit ISWI is the distorted, bloated morphology of the male X chromosome (Deuring et al. 2000; this study). This implicates NURF in the maintenance of male X chromosome morphology. In flies, X chromosome dosage compensation is achieved by up-regulating transcription from the male X chromosome. One characteristic of the male X chromosome is the specific acetylation of histone H4 at Lys 16 (H4-K16), which is believed to favor a looser chromatin structure that allows increased transcription. These patterns of acetylation are established by the male-specific expression of components of the MSL complex that are tethered on the male X chromosome and subsequently recruit the histone acetyl transferase MOF (for review, see Park and Kuroda 2001).
Genetic studies demonstrate that H4-K16 acetylation antagonizes ISWI function on the X chromosome (Corona et al. 2002). Biochemical characterization of the ISWI-containing ACF and CHRAC complexes has revealed that they can assemble and slide nucleosomes to establish regular ordered arrays (Ito et al. 1997; Varga-Weisz et al. 1997). Regular nucleosome arrays are presumed to provide better substrates for chromatin compaction and, thus, it was speculated that ACF and CHRAC might be the complexes that help compact the male X chromosome (Corona et al. 2002). However, here we show that NURF is the ISWI complex required for normal male X chromosome morphology. Unlike ACF or CHRAC, NURF disrupts regular, ordered arrays of nucleosomes (Tsukiyama et al. 1995). While it is possible that NURF is required for global aspects of higher order chromosome morphology that are needed to maintain normal male X chromosome structure, other local or transcription-based mechanisms could also account for the nurf301 and iswi phenotypes. The dosage compensation machinery is recruited to the male X chromosome at specific, high affinity sites or entry points and subsequently spreads into flanking chromatin (Kageyama et al. 2001). NURF may regulate chromatin accessibility at one or a number of these initiation sites. In the absence of NURF, entry of the dosage compensation machinery at such sites may be changed. Alternatively, NURF may control transcription of components of the sex-determination and dosage compensation pathway. Irrespective, the observed antagonistic relationship between ISWI function and H4-K16 acetylation (Corona et al. 2002) suggests that the action of NURF on the X chromosome is correspondingly influenced by H4 lysine acetylation. This influence on NURF could be direct, as suggested by effects of acetylated H4 tail peptide on ISWI ATPase activity in vitro (Corona et al. 2002).
Physiological activities of the ISWI chromatin remodeling machines
ISWI is the catalytic subunit of at least three protein complexes that have demonstrated in vitro chromatin-remodeling activity—NURF, ACF, and CHRAC. Here we have shown that mutation of a NURF-specific component reproduces the published properties of mutations in iswi. Both iswi and nurf301 are required for homeotic gene expression, proper larval blood cell development, and normal male X chromosome morphology. The specific in vivo functions of the ACF and CHRAC complexes remains to be established.
Clues to the function of ACF and CHRAC may be derived from studies of the human Williams Syndrome Transcription Factor–ISWI complex (WSTF–ISWI). The in vitro activities of the WSTF–ISWI complex are essentially identical to ACF. WSTF–ISWI is targeted to pericentric heterochromatin during replication and is believed to allow heterochromatin reassembly in the wake of the replication fork (Bozhenok et al. 2002). It is tempting to speculate that ACF and CHRAC may have similar functions in Drosophila and could be implicated in the establishment of repressive chromatin structures after replication. As mutations that selectively compromise individual remodeling complexes become available, whole genome expression analysis will allow the relative contributions of specific complexes to gene activation and silencing events in vivo to be dissected.
Materials and methods
Genetics and fly strains
nurf3011 corresponds to l(3)ry122 (Berg and Spradling 1991) and was mapped to the nurf301 untranslated leader sequence, 332 bp upstream of the ATG. The homozygous viable EP insertions EP(3)0637, EP(3)3495, and EP(3)3643 (Rørth 1996) were mapped 880, 720, and 840 bp, respectively, upstream of the ATG of nurf301. The deficiency Df(3L)3643 was generated by imprecise P-element excision from EP(3)3643. EMS-induced mutations in nurf301 were generated by feeding isogenized w1118 males 25mM EMS in 10mM Tris, at pH 7.5 and 1% sucrose for 12 h at room temperature. Mutagenized males were batch-crossed to TM3/TM6B virgins, and F1 male progeny then crossed individually to one of two deficiency lines that uncover nurf301, Df(3L)XZB970 or Df(3L)rH321. F2 progeny were scored for lethal noncomplementation of the deficiency. Alleles of nurf301 were identified by lethal noncomplementation of the P-element allele, nurf3011.
EMS-induced nucleotide changes were determined by amplifying overlapping DNA fragments covering nurf301 using nurf301-specific primers and an Expand Long Range PCR kit (Boehringer, Mannheim) and DNA isolated from homozygous mutant animals as template. DNAs were sequenced and compared with similarly isolated w1118 sequence. Altered nurf301 splicing in nurf3014 mutants was demonstrated by isolating RNA from nurf3014 homozygous mutant larvae using an RNAeasy column (Qiagen) and performing RT–PCR using primers flanking the altered splice-site. First-strand cDNA synthesis was performed using Ready-to-go beads (Amersham). For biochemical experiments, nurf301 mutations were kept over a GFP-marked balancer chromosome [w; TM3, P(w+mCActGFP) JMR2, Ser1]. This allowed large numbers of homozygous nurf301 mutant larvae to be selected by the lack of the GFP marker. Animals for expression analysis were harvested at mid-third larval instar—well prior to the lethal phase. Variegating telomeric transgene inserts are as described in Cryderman et al. (1999). All other fly strains are described in FlyBase (http://flybase.bio.indiana.edu)
Microscopy
Adult cuticles were prepared for microscopy by passage through isopropanol and mounted in Euparol (Asco Laboratories). Mutant and wild-type larval imaginal discs were fixed and immuno-histochemistry performed in parallel as described (Dominguez and Hafen 1997). RNA in situ hybridization was performed as described (Dominquez and Hafen 1997). For polytene chromosome analysis, animals were maintained at 18°C until treatment. For heat-shock experiments, salivary glands were dissected from late-third instar larvae, incubated in prewarmed 36.5°C TB1 buffer (Bonner 1981) for the indicated times and chromosomes prepared and stained as described (Gorman et al. 1995). Rabbit anti-HSF antibody was used at a dilution of 1:1000. For X chromosome staining, polytene chromosomes were prepared as described (Deuring et al. 2000) and stained with rabbit anti-MSL2 antibodies (diluted 1:200). DNA was stained using DAPI and is revealed in purple in the merge.
Expression analysis
For heat-shock gene expression analysis, mutant and wild-type larvae were separated, equilibrated for 12 h, then immersed in 1.5-mL microfuge tubes in a 36.5°C water-bath for the listed times. Samples were snap frozen in liquid nitrogen and stored at −70°C until processing. Total RNA was prepared as described (Andres and Thummel 1994) followed by further purification over an RNAeasy column (Qiagen). Dot blots were performed as described (Sambrook and Russell 2001). Probes against rp49, hsp26, and hsp70 are as described in Marchler and Wu (2001). Probes against diptericin, drosomycin, and tep1 were PCR fragments amplified from genomic DNA using the following primer sets: drs (5′-ATGATGCAGATCAAGTACTTGTTCGCC-3′ and 5′-TTAGCATCCTTCGCACCAGCACTTCAG-3′), dpt (5′-TTCTTCAATTGAGAACAACTGAGATGC-3′ and 5′-GAAG TCTGCCTCAATGTTCCGGGTTAA-3′) tep1 (5′-CTACATTT GATACTGCAGAAAATATAGAAC-3′ and 5′-CCGAGCTCC TGTAGATATTTATATTCGCAT-3′). Hybridization was in SUREhyb hybridization buffer (Ambion) using DNA probes labeled using a STRIPeaze kit (Ambion). For analysis of Ubx and en mRNA levels, total RNA was isolated as described above, subjected to DNase I cleavage (Amplification grade DNase I, Invitrogen), and transcript abundance assayed using an Access RT–PCR kit (Promega). Primer sets and cycle conditions used were: rp49 (5′-TCTTGAGAACGCAGGCGACCGTTGGGGT TG-3′ and 5′-ATCCGCCACCAGTCGGATCGATATGCTA AG-3′, 20 cycles), en (5′-CCTCGAATATTGCACGCCCCC GTCAGAAGG-3′ and 5′-CAACGACGAGAAGCGTCCACG CACCGCGTT-3′, 25 cycles) and Ubx (5′-TCTGATTTCACA CAATACGGCGGCATATCA-3′ and 5′-AATCCCACATACA CCCTACGGATCTAAGGA-3′, 25 cycles).
Acknowledgments
We thank W. Bender, A. Greenleaf, S. Govind, S. Hou, J. Kadonaga, M. Kuroda, E. Lewis, S. Lindquist, J. Tamkun, the Developmental Studies Hybridoma Bank and the Bloomington Stock Center, for kindly providing us with antibodies and fly stocks. We thank J. Kassis, J. Kennison, E. Lewis, M. Mortin, J. Tamkun, and members of our laboratory for helpful suggestions and comments on this manuscript. T. Jones performed chromosome in situ hybridization to localize nurf301. R. Sandaltzopoulos provided cDNAs used in Northern analysis. This work was supported by the NCI intramural research program, and a grant from the Wellcome Trust (U.K.) to P.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL carlwu@helix.nih.gov; FAX (301) 435-3697.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1032202.
References
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 1994;44:565–573. doi: 10.1016/s0091-679x(08)60932-2. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Goodman RH, Smolik SM. The interaction between the coactivator dCBP and Modulo, a chromatin-associated factor, affects segmentation and melanotic tumor formation in Drosophila. Proc Natl Acad Sci. 2001;99:2895–2900. doi: 10.1073/pnas.052509799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W, Akam ME, Karch F, Beachy PA, Peifer M, Spierer P, Lewis EB, Hogness DS. Molecular genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Berg CA, Spradling AC. Studies on the rate and site-specificity of P-element transposition. Genetics. 1991;127:515–524. doi: 10.1093/genetics/127.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A, Lampen N, Martinek S, Young MW, Darnell JE. A Drosophila PIAS homolog negatively regulates stat92E. Proc Natl Acad Sci. 2001;98:9563–9568. doi: 10.1073/pnas.171302098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- Bhosekar S, Babu P. Enhancer of bithorax a mutation of Drosophila melanogaster. J Genet. 1987;66:187–194. [Google Scholar]
- Bonner JJ. Induction of Drosophila heat-shock puffs in isolated polytene nuclei. Dev Biol. 1981;86:409–418. doi: 10.1016/0012-1606(81)90199-8. [DOI] [PubMed] [Google Scholar]
- Bozhenok L, Wade PA, Varga-Weisz P. WSTF–ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 2002;21:2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–4821. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- Chen H-W, Chen X, Oh S-W, Marinissen MJ, Gutkind JS, Hou SX. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to mammalian cytokine receptor family. Genes & Dev. 2002;16:388–398. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002;3:242–247. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DFV, Langst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- Cryderman DE, Morris EJ, Biessmann H, Elgin SC, Wallrath LL. Silencing at Drosophila telomeres: Nuclear organization and chromatin structure play critical roles. EMBO J. 1999;18:3724–3735. doi: 10.1093/emboj/18.13.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Hafen E. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes & Dev. 1997;11:3254–3264. doi: 10.1101/gad.11.23.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- Gateff E. Science 200: 1448–1459. 1978. Malignant neoplasms of genetic origin in ITLDrosophila melanogaster. [DOI] [PubMed] [Google Scholar]
- Gdula DA, Sandaltzopoulos R, Tsukiyama T, Ossipow V, Wu C. Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes & Dev. 1998;12:3206–3216. doi: 10.1101/gad.12.20.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Gorman M, Franke A, Baker BS. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development. 1995;121:463–475. doi: 10.1242/dev.121.2.463. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Hanratty WP, Dearolf CR. The Drosophila Tumorous-lethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Mol Gen Genet. 1993;238:33–37. doi: 10.1007/BF00279527. [DOI] [PubMed] [Google Scholar]
- Hari KL, Cook KR, Karpen GH. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes & Dev. 2001;15:1334–1348. doi: 10.1101/gad.877901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes & Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Mengus G, Gilfillan G, Kennedy HG, Stuckenholz C, Kelley RL, Becker PB, Kuroda MI. Association and spreading of the Drosophila dosage compensation complex from a discrete roX1 chromatin entry site. EMBO J. 2001;20:2236–2245. doi: 10.1093/emboj/20.9.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JG, Hamiche A, Wu C. GAL4 directs nucleosome sliding induced by NURF. EMBO J. 2002;21:1406–1413. doi: 10.1093/emboj/21.6.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA. The Polycomb and trithorax group proteins of Drosophila: Transregulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes & Dev. 2001;15:619–626. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci. 2000;97:11427–11432. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DFV, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or transdisplacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego, CA: Academic Press, Inc.; 1992. [Google Scholar]
- Luo H, Dearolf CR. The JAK/STAT pathway and Drosophila development. BioEssays. 2001;23:1138–1147. doi: 10.1002/bies.10016. [DOI] [PubMed] [Google Scholar]
- Marchler G, Wu C. Modulation of Drosophila heat shock transcription factor activity by the molecular chaperone DROJ1. EMBO J. 2001;20:499–509. doi: 10.1093/emboj/20.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol Cell. 1997;1:141–150. doi: 10.1016/s1097-2765(00)80015-5. [DOI] [PubMed] [Google Scholar]
- Park Y, Kuroda MI. Epigenetic aspects of X-chromosome dosage compensation. Science. 2001;293:1083–1085. doi: 10.1126/science.1063073. [DOI] [PubMed] [Google Scholar]
- Qui P, Pan PC, Govind S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development. 1998;125:1909–1920. doi: 10.1242/dev.125.10.1909. [DOI] [PubMed] [Google Scholar]
- Rizki RM, Rizki TM. Cell interactions in the differentiation of a melanotic tumor in Drosophila. Differentiation. 1979;12:167–178. doi: 10.1111/j.1432-0436.1979.tb01002.x. [DOI] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhf M-L, Braun A, Papoulas O, Tamkun JW, Randsholt N, Meister M. The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development. 2001;128:1429–1441. doi: 10.1242/dev.128.8.1429. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 7.49–7.50. [Google Scholar]
- Soeller WC, Oh CE, Kornberg TB. Isolation of cDNAs encoding the Drosophila GAGA transcription factor. Mol Cell Biol. 1993;13:7961–7970. doi: 10.1128/mcb.13.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to the heat-shock pelements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID and RNA-polymerase II-binding sites. Genes & Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Lis JT. HSF recruitment and loss at most Drosophila heat shock loci is coordinated and depends on proximal promoter sequences. Chromosoma. 1996;105:158–171. doi: 10.1007/BF02509497. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP. A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2 SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2 SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Tzou P, De Gregorio E, Lemaitre B. How Drosophila combats microbial infection: A model to study innate immunity and host–pathogen interactions. Curr Opin Microbiol. 2002;5:102–110. doi: 10.1016/s1369-5274(02)00294-1. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Westwood JT, Clos J, Wu C. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature. 1991;353:822–823. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu DG, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]