Abstract
Lymphoid enhancer factor (LEF1), a nuclear mediator of Wnt signaling, is required for the formation of organs that depend on inductive interactions between epithelial and mesenchymal tissues. In previous tissue recombination experiments with normal and Lef1−/− tooth germs, we found that the effect of LEF1 expression in the epithelium is tissue nonautonomous and transferred to the subjacent mesenchyme. Here we examine the molecular basis for LEF1 function and find that the epithelium of the developmentally arrested Lef1−/− tooth rudiments fails to express Fgf4, Shh, and Bmp4, but not Wnt10a. We identify the Fgf4 gene as a direct transcriptional target for LEF1 and show that beads soaked with recombinant FGF4 protein can fully overcome the developmental arrest of Lef1−/− tooth germs. In addition, we find that FGF4 beads induce rapidly the expression of Fgf3 in dental mesenchyme and that both epithelial and mesenchymal FGF proteins induce the delayed expression of Shh in the epithelium. Taken together, these data indicate that a single target of LEF1 can account for the function of LEF1 in tooth development and for a relay of a Wnt signal reception to a cascade of FGF signaling activities, allowing for a sequential and reciprocal communication between epithelium and mesenchyme.
Keywords: LEF1, Wnt, FGF, epithelial–mesenchymal interactions
Development of most vertebrate organs, which are composed of an epithelium and a mesenchyme, requires extensive and precisely regulated communication between these tissues. Inductive tissue interactions play a role in the initiation of organogenesis, in subsequent morphogenesis, and in cytodifferentiation. Classical experiments, involving the separation and recombination of epithelial and mesenchymal tissues, have shown that epithelial–mesenchymal interactions are sequential and reciprocal (Grobstein 1967). Multiple signaling pathways have been implicated in tissue interactions (for review, see Hogan 1999; Jernvall and Thesleff 2000; Fuchs and Raghavan 2002). The complexity of organ development depends further on the precise temporal and spatial coordination of signaling activities, including the integration of different signals and the generation of signaling networks. A sequential and reciprocal exchange of signals between different tissues requires that one tissue respond to signals from the partner tissue by altering its gene expression profile and own signaling activity. Transcription factors may provide this link between signal input and signal generation by mediating the response to inductive signals and regulating the expression of other signaling molecules.
A candidate for a transcription factor that may be involved in the integration of signals and the regulation of signaling networks is lymphoid enhancer factor (LEF1), a member of the LEF-1/TCF subfamily of high mobility group (HMG) domain proteins (Travis et al. 1991; Waterman et al. 1991). LEF1 can act as an architectural transcription factor that activates transcription in association with other proteins. In response to Wnt signals, LEF1 and other members of this family of transcription factors associate with β-catenin and activate Wnt-responsive target genes (Behrens et al. 1996; Huber et al. 1996; Molenaar et al. 1996; for review, see van Noort and Clevers 2002). In addition, LEF1 can interact with the cofactor Aly to activate genes in the absence of Wnt signals (Hsu et al. 1998). Evidence for an integrative function of LEF1 in signaling came from the analysis of the Ultrabithorax enhancer in Drosophila. In this context, the LEF1 ortholog, dTCF, regulates the wingless/WNT response only in the presence of DPP/BMP signals (Riese et al. 1997).
In addition, the analysis of LEF1-deficient mice indicated that LEF1 may have a function in the generation of signaling networks. Targeted inactivation of the Lef1 gene in mice results in a complete block of development of multiple ectodermal appendages, including teeth, vibrissae, hair, and mammary glands (van Genderen et al. 1994). Recombinations of epithelial and mesenchymal tissues from developing teeth of wild-type and Lef1−/− embryos showed that LEF1 is required only transiently in the epithelium in a tissue-nonautonomous manner, which is rather unexpected for a component of a signal reception pathway (Kratochwil et al. 1996). Moreover, recent experiments in chick indicated that Wnt signals in the limb ectoderm activate FGF signals in ectoderm and subjacent mesenchyme (Kawakami et al. 2001). Thus, LEF1 may act not only downstream of Wnt signals, but may also function upstream of other signals, thereby linking distinct signaling pathways.
Tooth development is a well-studied and experimentally accessible model for organogenesis (for reviews, see Thesleff and Sharpe 1997; Jernvall and Thesleff 2000). Organogenesis is initiated with a thickening of the ectodermally derived oral epithelium between embryonic day 10 (E10) and E11. The epithelium invaginates into the mesenchyme that originates from the neural crest of the first branchial arch and forms a “tooth bud” at E12. Beginning morphogenesis at E13 involves both the formation of a signaling center at the base of the epithelial bud, termed “enamel knot,” which expresses multiple signaling molecules, and the formation of a papilla in the mesenchyme. Subsequent differentiation ultimately gives rise to the dentine-secreting odontoblasts of mesenchymal origin and the enamel-secreting ameloblasts derived from the inner dental epithelium.
The role of specific signals in tooth development has been elucidated by targeted gene inactivations and the analysis of the effects of signaling molecules on gene expression. Some epithelial signaling molecules, such as FGF4, FGF8, and BMP4, were found to induce tooth-specific gene expression in dental mesenchyme, suggesting that they may provide inductive signals to the partner tissue (Vainio et al. 1993; Neubüser et al. 1997; Kettunen et al. 2000). In addition, gene targeting of the transcription factors LEF1, MSX1, and PAX9 indicated that these factors regulate distinct steps in the transition from the bud to the cap stage of tooth development. LEF1 was found to function in the epithelium, whereas Msx1 and Pax9 were shown to be expressed and to act in the mesenchyme (Kratochwil et al. 1996; Peters et al. 1998; Bei et al. 2000). MSX1 regulates BMP4 signaling (Bei et al. 2000), but the molecular basis for the relationship between transcription factors and signaling activities remains generally obscure.
Here, we investigate the molecular basis of LEF1 function in inductive signaling during tooth development. We show that the expression of multiple epithelial signal factors is dependent on LEF1, and we identify Fgf4 as a direct transcriptional target of LEF1 and Wnt signaling. Moreover, we find that FGF proteins but not other signaling molecules are capable of fully rescuing development of Lef1−/− tooth germs. Thus, the sole function of LEF1 in odontogenesis may be to activate Fgf4 and to connect the Wnt and FGF signaling pathways at a specific developmental step.
Results
Expression of signaling molecules in Lef1−/− tooth germs
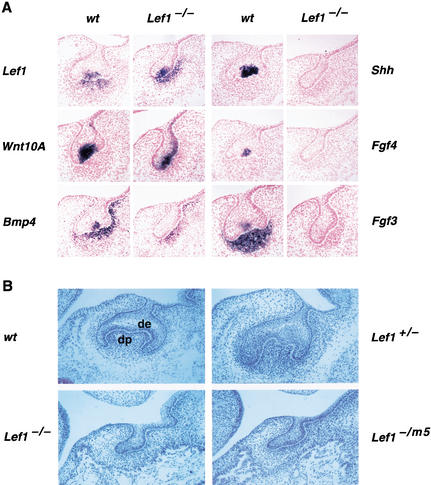
Previous analysis of Lef1−/− embryos indicated that the absence of LEF1 results in a complete block of tooth development after the initiation and formation of an epithelial bud, but prior to the formation of a mesenchymal papilla at embryonic stage E14.5 (van Genderen et al. 1994). Around this stage, the dental epithelium of wild-type embryos forms a bona fide signaling center, termed the enamel knot, which expresses Lef1 and multiple genes for signaling molecules, including Wnt10a and Wnt10b, Bmp2, Bmp4, Bmp7, Fgf3, Fgf4, Fgf9, and Shh (Kratochwil et al. 1996; Vaahtokari et al. 1996; Dassule and McMahon 1998; Kettunen and Thesleff 1998; Sakar and Sharpe 1999; Kettunen et al. 2000). With the aim of examining whether these signaling molecules are expressed in Lef1−/− embryos, we performed in situ hybridizations with probes for representatives of these genes at E14.5. Consistent with the role of LEF1 as a downstream component of the Wnt signaling pathway, both Wnt10a and Wnt10b are expressed in the mutant epithelial tooth bud, albeit at a reduced level (Fig. 1A; data not shown). Likewise, the mutated Lef1 allele, which does not produce protein at a detectable level (van Genderen et al. 1994), is still transcribed in the epithelial bud and in the mesenchyme, suggesting that Lef1 transcription is neither regulated by a positive feedback loop nor by Wnt signaling (Fig. 1A). In addition, transcription of the Lef1 gene and the early enamel knot marker p21 (not shown) in Lef1−/− embryos is localized to the same epithelial region as in wild-type embryos, suggesting that the structure of the enamel knot is formed. In contrast, none of the other signal factors examined is expressed in Lef1-deficient epithelium (Fig. 1A; data for Bmp2, Bmp7, and Fgf9 not shown). The mesenchyme of mutant tooth germs, which does not form a proper papilla despite some occasional condensation, fails to express Fgf3 and Fgf10, whereas the mesenchymal expression of Bmp4 appears to be independent of LEF1 (Fig. 1A; data for Fgf10 not shown). These data suggest that the expression of Fgf3 and Fgf10 requires LEF1-dependent epithelial signals or mesenchymal expression of Lef1. Taken together, this analysis indicates that Lef1−/− tooth germs fail to form a functional enamel knot and fail to express multiple signaling molecules, which are therefore potential candidates for the postulated LEF1-dependent epithelial signal(s).
Figure 1.
(A) Expression of genes encoding signaling molecules in wt and Lef1−/− mandibular molars at E14.5. At this developmental stage, the “enamel knot,” a putative signaling center at the base of the epithelial downgrowth, and the mesenchymal papilla have been formed. Consecutive sections of the same rudiments were used for in situ hybridization with various probes. In the enamel knot of wt tooth germs, expression of Lef1, Wnt10a, Bmp4, Shh, Fgf4, and Fgf3 is detected. The same group of cells also contains Wnt10b, Fgf9, Bmp2, and Bmp7 transcripts (data not shown). With the exception of Lef1, Wnt10a, and Wnt10b (data not shown), epithelial expression of all other genes examined is abrogated in LEF1-deficient molars. In the mesenchyme of both wt and mutant tooth germs, Lef1 and Bmp4 are expressed, whereas the abundant expression of Fgf3 and Fgf10 (data not shown) in the forming papilla is detected in wt but not in mutant molars. (B) Dependence of tooth development on LEF1:β-catenin interaction. Morphology of molar tooth germs from wt and various Lef1 mutant embryos at E16. A normal cap structure of dental epithelium (de) surrounding the dental papilla (dp) is seen in sections of wt and heterozygous Lef1+/− embryos (top). An arrested tooth bud structure is observed in Lef1−/− and Lef1−/m5 embryos. Lef1−/m5 embryos carry one null allele and one allele with point mutations in the region of Lef1 encoding the β-catenin interaction domain of LEF1 (bottom).
Dependence of tooth development on LEF1:β-catenin interaction
Although LEF1/TCF factors serve as nuclear mediators of Wnt signals in association with β-catenin (for reviews, see van Noort and Clevers 2002), both LEF1 and β-catenin have alternative binding partners. Therefore, the question arises as to the dependence of tooth development on the interaction of LEF1 with β-catenin. We have previously shown that mutations of six amino acids in the N terminus of LEF1, termed m5, virtually abrogate both the interaction of LEF1 with β-catenin and the transcriptional activation of LEF1-dependent reporter gene constructs (Hsu et al. 1998). To examine whether tooth formation and other developmental processes are dependent on interaction of LEF1 with β-catenin, we introduced the m5 mutations into the Lef1 locus by gene targeting (W. Roth and R. Grosschedl, unpubl.). E16 embryos carrying one Lef1 null allele and one Lef1m5 allele (Lef1−/m5) showed the same developmental arrest of tooth germs at the bud stage as homozygous Lef1−/− embryos, whereas Lef1+/− embryos formed a normal cap structure of the tooth germs (Fig. 1B). Thus, the role of LEF1 in tooth development is dependent on the interaction with β-catenin and Wnt signaling.
Rescue of Lef1-deficient tooth development by recombinant FGF proteins
In previous experiments in which we performed various combinations of dental epithelium and mesenchyme from wild-type and Lef1-deficient embryos, we have shown that the formation of the mesenchymal tooth papilla requires LEF1 function only in the epithelium (Kratochwil et al. 1996). From these experiments, we concluded that LEF1 acts upstream of an inductive signal from the epithelial enamel knot to the subjacent mesenchyme. If the function of LEF1 in tooth development is to regulate an epithelium-to-mesenchyme signaling activity, it might be possible to rescue tooth development by exogenous application of the missing signal to explanted E13.5 Lef1−/− tooth germs. Toward this end, we placed beads soaked in various signaling molecules between dental epithelium and mesenchyme that have been separated and recombined, or directly onto intact explants. After 2–4 d in culture, the tooth germs were transplanted under the kidney capsule of syngeneic mice to allow their full development.
Beads soaked with recombinant BMP4 or SHH consistently failed to allow development of Lef1−/− tooth germs (Fig. 2; Table 1). However, beads soaked with FGF proteins rescued the development of mutant tooth rudiments with remarkable frequency and quality (Fig. 2; Table 1). The rescued teeth were morphologically and histologically indistinguishable from wild-type explants. Both the ameloblast and the odontoblast layers had developed properly, depositing their characteristic mineralizing extracellular matrix. Exposure of mutant incisor rudiments to FGF4 or FGF8b also generated normal teeth, albeit at a lower frequency (7/33). All types of recombinant FGF proteins tested (FGF4, FGF9, and FGF8b) were effective in rescuing Lef1−/− tooth development (Table 1), with FGF4 inducing a mesenchymal papilla more rapidly than did other types of FGF proteins.
Figure 2.
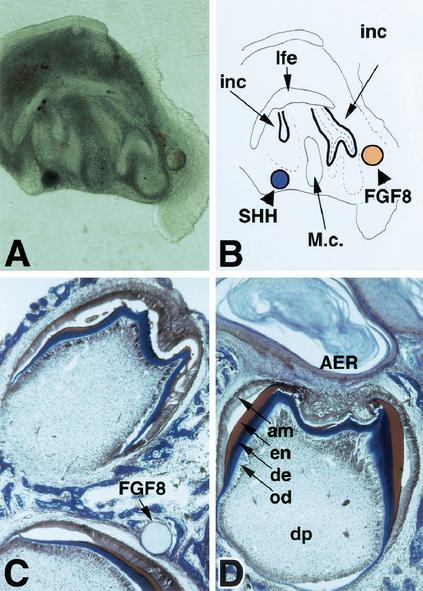
Rescue of arrested Lef1−/− tooth development by recombinant FGF proteins. (A,B) In vitro culture of an E13.5 Lef1−/− mandible that contains a pair of incisor rudiments and has been associated with a bead soaked in SHH (left side, blue Affigel bead) or FGF8 (right side, brown heparin acrylic bead), respectively. A shows a live explant after 3 d of in vitro culture, and B is a graphic presentation of the structures. Note that only the right incisor anlage next to the FGF8 bead has formed a cap (and a fully developed tooth on subsequent transplantation under the kidney capsule). inc, incisor buds; lfe, lip furrow epithelium; M.c., Meckel's cartilage. (C,D) Full development of E14.5 Lef1−/− tooth germs that have been incubated with FGF8-soaked beads (C) or apical ectodermal ridge (AER) of an E10.5 mouse hindlimb (D) in vitro for 3 d and transferred under the kidney capsule for another 9 d. Typical multicusp molars with ameloblast (am) and odontoblast (od) layers, dentin (de, blue), enamel (en, red), and dental pulp (dp) are formed. The limb ectoderm including the AER has persisted in forming a cyst with keratinized layers inside.
Table 1.
Rescue of tooth development in explanted E13/E14 Lef1−/− molar rudiments by recombinant signal factors or heterologous tissues
|
|
Teeth/explants
|
|---|---|
| Signaling molecules | |
| BMP4 | 0/8 |
| SHH | 0/16 |
| FGF4 | 58/59 |
| FGF9 | 10/11 |
| FGF8b | 16/17 |
| FGF7 | 22/25 |
| FGF10 | 11/14 |
| Heterologous tissues | |
| Notochord | 0/27 |
| Limb ectoderm (AER) | 10/12 |
Explants were associated either with beads soaked in recombinant signal factors or with heterologous tissues. After 2 or 3 days in culture, the explants were transplanted under the kidney capsule to allow for full development (see text for details). AER, apical ectodermal ridge.
To control for possible artefacts caused by the addition of nonphysiological concentrations of recombinant signaling molecules in the rescue experiments, we also used heterologous tissues producing a known spectrum of endogenous signaling molecules. A combination of mutant molar rudiments with mouse or chick notochord, which is a physiological source of SHH, failed to support tooth development. In contrast, the apical ectodermal ridge (AER) of E10.5 mouse limb or E3.5 chick wing buds, which is a source of various FGF proteins including FGF2, FGF4, FGF8, and FGF9, yielded fully developed teeth from Lef1−/− molar rudiments (Fig. 2D; Table 1).
Our previous tissue recombinations have shown that Lef1-expressing dental epithelium is required only transiently for the transition from the “bud” to the “cap” stage of tooth development between E13 and E14 (Kratochwil et al. 1996). In examining whether the rescue of tooth development in Lef1−/− organ cultures by FGF4 has similar temporal properties, we found that the application of FGF beads to E13.5 or older mutant molar rudiments yielded normal teeth with high frequency (Tables 1, 2). In contrast, the addition of FGF4 beads to E12.5 mutant explants failed to rescue tooth development, unless the explants had been cultured in vitro for 2 d prior to the application of FGF4 (Table 2). Moreover, removal of the FGF4 beads after 8, 14, or 24 h of incubation still allowed for full tooth development, indicating a similar transient requirement for exogenous FGF4 as for Lef1-expressing epithelium.
Table 2.
Stage specificity of the rescue of Lef1−/− molar tooth development by FGF4
| Stage of FGF4 application
|
Teeth/explants
|
|---|---|
| E12.5 | 0/16 |
| E12.5 + 2 days in vitro | 4/4 |
| E13.5 | 6/6 |
| E14.5 | 52/53 |
| E15.5 | 10/10 |
| E16.5 | 11/24 |
FGF4 beads rescue the development of mutant tooth rudiments at E13.5 and subsequent stages, but not at E12.5 unless the explants are precultured for 2 days prior to the addition of the bead.
Inhibition of FGF signaling abolishes the inductive effect of dental epithelium
If LEF1-dependent epithelial signaling to the mesenchyme was mediated by FGF proteins, an inhibition of FGF signaling would mimic the arrest of tooth development in Lef1−/− mice. Fgf4 and Fgf9 are both expressed in the enamel knot of the dental epithelium and may act in a redundant manner because the targeted inactivation of Fgf9 has no detectable defect in tooth development (Colvin et al. 2001a,b). As the lethality of Fgf4−/− embryos at the preimplantation stage obscures an assessment of a potential redundancy of these genes in tooth organogenesis (Feldmann et al. 1995), we examined whether SU5402, a chemical inhibitor of FGF signaling (Mohammadi et al. 1997), blocks tooth development.
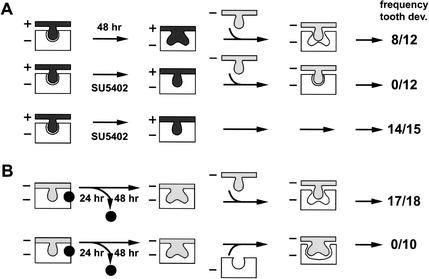
Treatment of wild-type molar rudiments with 25 μM SU5402 resulted in a developmental arrest and eventually in death after a prolonged exposure to the drug. However, the SU5402-treated molar rudiments recovered completely when they were transferred to standard medium after 48 h of exposure to the drug. This observation, together with the transient requirement of LEF1 function in the epithelium, allowed us to adopt the following experimental strategy (Fig. 3A). We incubated tissue recombinants, in which Lef1-deficient dental mesenchyme had been combined with wild-type dental epithelium, with SU5402 for 48 h. Then we combined the mutant mesenchyme with signaling-incompetent Lef1−/− dental epithelium and transferred the tissue recombinant to standard medium. Without treatment of these tissue recombinants with SU5402, normal tooth development was observed (Kratochwil et al. 1996; Fig. 3A, top panel). In contrast, treatment of these tissue recombinants with the drug abrogated tooth development (Fig. 3A, middle panel). SU5402 treatment of control combinations of wild-type epithelium and mutant mesenchyme, in which the wild-type epithelium was not replaced with Lef1−/− epithelium, allowed for normal tooth development (Fig. 3A, bottom panel). This control confirms that the arrest of tooth development in the experimental group was not due to some irreversible tissue damage by the drug. Thus, the inhibition of FGF signaling during the transient association of wild-type epithelium with Lef1−/− mesenchyme prevents the induction of the dental papilla and results in a developmental arrest at a stage similar to that of the Lef1−/− mutation.
Figure 3.
Epithelial-to-mesenchyme FGF signaling. (A) Inhibition of tooth development by the FGF signaling antagonist SU5402. Frequency of tooth development (dev.) in epithelium–mesenchyme double combinations from wt (+) and Lef1−/− (−) tooth germs. (Top panel) Transient (48 h) exposure of Lef1-deficient mesenchyme (white) to wt dental epithelium (dark gray) allows tooth development after its subsequent recombination with mutant epithelium (light gray). (Middle panel) The presence of 25 μM SU5402 during the transient presence of wt epithelium prevents subsequent tooth development under otherwise identical conditions. (Bottom panel) Control experiment indicating that 48 h of exposure to SU5402 per se does not impair subsequent tooth development. The arrest in tooth development is represented by a bud structure of the epithelium. (B) FGF4 acts on the mesenchyme. Lef1−/− tooth germs were incubated with FGF4 beads for 24 h and after a further 48 h incubation without beads, the mutant mesenchyme (top panel) or the mutant epithelium (bottom panel) were exchanged with corresponding fresh mutant tissues (light gray). High frequency (17/18) of tooth development is observed with mutant mesenchyme, but not with mutant epithelium that had been exposed to FGF4.
Epithelial FGF proteins act on the mesenchyme
The rescue of tooth development in explanted Lef1−/− organ cultures by FGF proteins raises the question of whether these signaling molecules act on the epithelial or mesenchymal component of the developing organ. Toward this end, we incubated E14.5 Lef1−/− molar rudiments with FGF4 beads for 24 h, removed the beads, and cultured the explants for an additional 48–72 h to allow for the development of the mesenchymal papilla and epithelial cap (Fig. 3B). Subsequently, we separated the tissues enzymatically and recombined them with fresh mutant partner tissue from E14.5 embryos that had not been exposed to exogenous FGF4. Combination of FGF4-induced mutant mesenchymal papillae with uninduced mutant epithelium allowed for efficient tooth formation (Fig. 3B, top panel). In contrast, the epithelial components of these FGF4-treated mutant molars, which had already formed morphologically identifiable caps, failed to continue their development on combination with uninduced mutant mesenchyme (Fig. 3B, bottom panel). Thus, the exogenously added FGF4 acts predominantly on the mesenchyme and may stabilize a specific developmental program in this tissue, consistent with the requirement of epithelial Lef1 expression for the formation of the mesenchymal papilla (Kratochwil et al. 1996).
Fgf4 is a direct downstream target for LEF1 and Wnt signaling
The observations that Fgf4 expression is abrogated in Lef1−/− tooth germs and that exogenous FGF4 protein overcomes the developmental arrest suggest that FGF4 is acting downstream of LEF1 in tooth development. To examine whether Fgf4 may be directly regulated by LEF1, we searched transcriptional control sequences of the Fgf4 gene for binding sites of LEF1/TCF proteins. A putative LEF1/TCF binding site was identified in a region upstream of two enhancers in the 3′ untranslated region (UTR) of the Fgf4 gene (Fig. 4A). Similar to the 3′ enhancers, which have been shown to direct expression in myotomes and AER of transgenic mice (Fraidenraich et al. 1998), the downstream region, including the putative LEF1/TCF binding site, is conserved between mouse and man. In an electrophoretic mobility shift assay, recombinant LEF1 bound efficiently to a labeled oligonucleotide encompassing the putative binding site in the 3′ region of the Fgf4 gene (Fig. 4B). The specificity of binding was confirmed by the competition of the LEF1–DNA complex with excess of unlabeled oligonucleotide containing the wild-type binding site and by the failure to compete with an oligonucleotide containing a mutated binding site.
Figure 4.
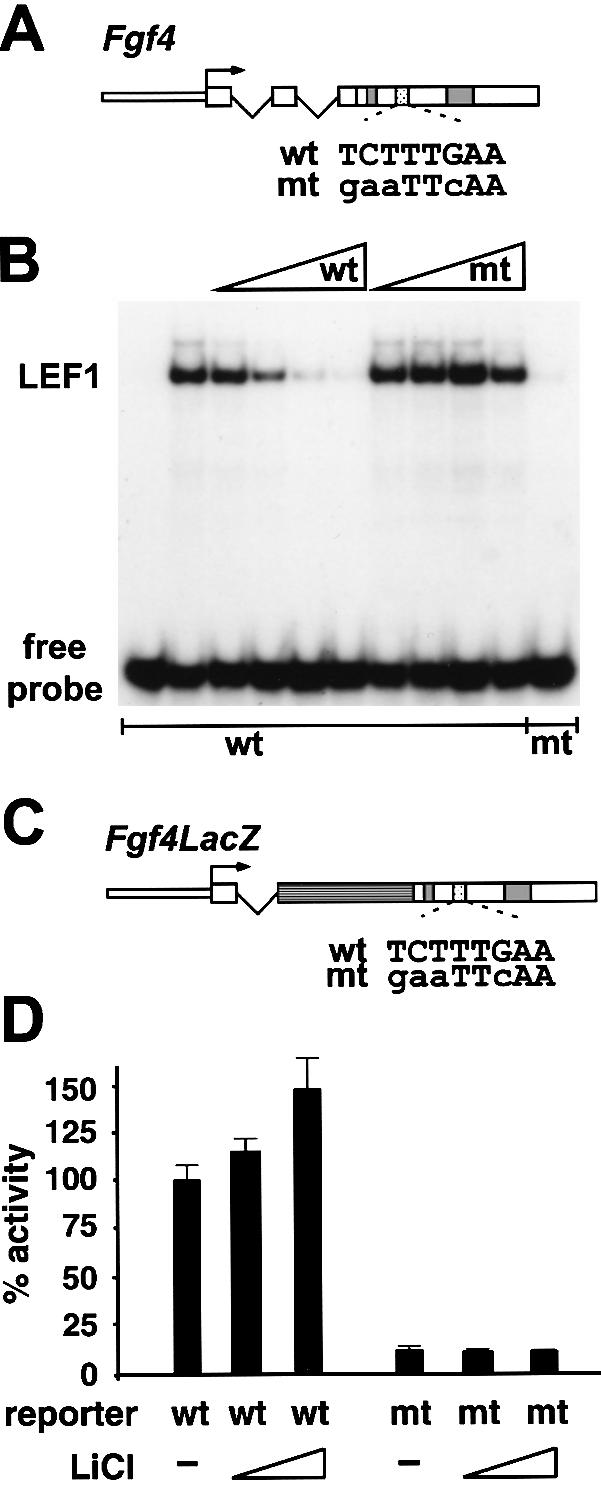
The Fgf4 gene is a direct target for LEF1 and Wnt signaling. (A) Schematic representation of the Fgf4 gene. Sequences in the 3′UTR that are conserved between mouse and man are boxed (gray) and the relative location and sequence of a binding site for LEF1 (stippled) is shown. (B) Electrophoretic mobility shift assay showing binding of 25 ng purified recombinant LEF1 to 32P-labeled double-stranded oligonucleotides encompassing the wt or mt sequence from the Fgf4 3′UTR. The specificity of binding is confirmed by a competition with increasing amounts of unlabeled wt or mt oligonucleotides. (C) Schematic representation of the Fgf4LacZ reporter gene. The bacterial LacZ gene is shown in dark gray. Sequences in the 3′UTR are represented as in panel A. (D) Functional contribution of the LEF1 binding site to the activity of Fgf4 transcriptional control sequences. Reporter gene constructs carrying the wt or mt LEF1 binding sites (stippled box) in the 3′UTR were generated by linking Fgf4 promoter sequences and the Fgf4 3′UTR (open boxes) to bacterial LacZ coding sequences (hatched box). The reporter constructs were stably transfected into F9 cells and lacZ activity was determined in pools of clones. The Wnt-responsiveness of the Fgf4LacZ reporter constructs was examined by treatment of the transfected cell pools with LiCl (10 or 30 mM) as a surrogate Wnt stimulus.
We also examined the functional importance of the LEF1 binding site in the 3′UTR of the Fgf4 gene by stable transfection of gene constructs into F9 embryonic carcinoma cells that express the endogenous Fgf4 gene (Dailey et al. 1994). For the transfection experiments, we used gene constructs in which the Fgf4 coding sequence had been replaced by a β-galactosidase (LacZ) gene and in which the 3′UTR of the Fgf4 gene contains a wild-type (wt) or mutated (mt) LEF1 binding site (Fig. 4C). Analysis of LacZ reporter gene expression in independent pools of stably transfected F9 cells indicated that the mutation of the LEF1 binding site decreased the level of expression by a factor of 4–10 (Fig. 4D shows one representative pool of wt and mt reporters). We also examined whether the LEF1 binding site is required for the responsiveness of the Fgf4 transcriptional control sequences to Wnt signals. Toward this end, we stimulated the pools of transfected cells with LiCl, which inhibits glycogen synthase kinase 3β and can act as a surrogate Wnt signal (Hedgepeth et al. 1997). We observed a modest but reproducible increase in reporter gene expression with the wild type but not with the mutant gene construct (Fig. 4D). Taken together, these data suggest that the Fgf4 gene is a direct transcriptional target for LEF1 and Wnt signaling.
Gene expression in FGF4-rescued Lef1−/− tooth germs
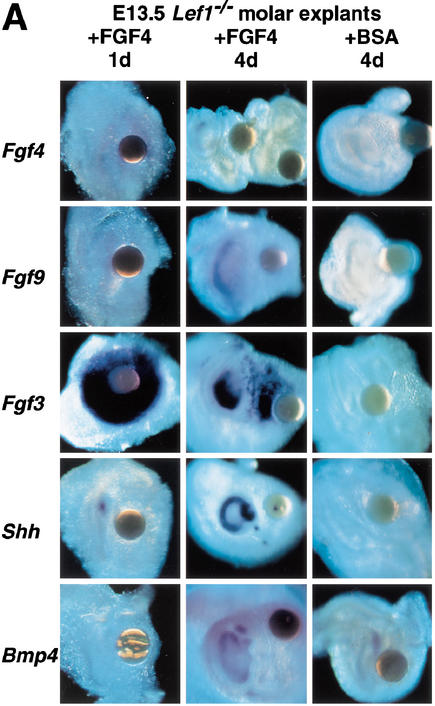
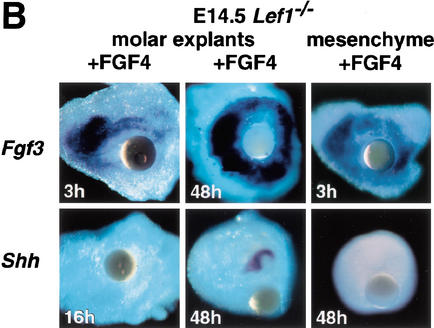
To examine the effects of FGF4 on the expression of signaling molecules implicated in tooth development, we analyzed E13.5 and E14.5 Lef1−/− molar rudiments that had been exposed to FGF4 beads for various times (Fig. 5A). All mutant explants had progressed in development and formed a clearly recognizable epithelial cap over a mesenchymal papilla after 4 d in culture. Strong Fgf3 gene expression was detected already after 1 d. Initially, the hybridization signals were localized around the bead, but after 4 d an additional domain of expression appeared in the newly formed mesenchymal papilla. Histological sections showed that the Fgf3 transcripts were confined to the mesenchyme (data not shown). Induction of Fgf3 expression was even detected after 3 h of bead exposure (Fig. 5B), suggesting that Fgf3 may be an immediate target for FGF4 in the mesenchyme. A similar but weaker induction of mesenchymal Fgf10 expression was observed after 1 d of culture with FGF4 beads (data not shown). Activation of Fgf3 expression in the mesenchyme by FGF4 was also detected in the absence of epithelium, suggesting that FGF4 is sufficient for the induction of Fgf3 (Fig. 5B). Transcripts of the endogenous Fgf4 gene were not detected in 28/35 FGF4-treated explants and detected at very low levels in 7/35 explants, consistent with the regulation of Fgf4 expression by LEF1. Fgf9 transcripts were detected at low levels in 6/6 explants.
Figure 5.
Effects of FGF4 beads on the expression of selected marker genes in explanted molar rudiments of E13.5 Lef1−/− embryos. Gene expression was monitored by whole-mount in situ hybridization after 1–4 d of incubation. (A) Application of FGF4-soaked beads or BSA-soaked control beads to the mutant tooth germs. After 1 d, strong expression of Fgf3 in the mesenchyme and weak expression of Shh in the epithelium was detected. After 4 d, Shh and Fgf9 are expressed in the epithelium, Fgf3 and Bmp4 in the mesenchyme. Note two expression domains of Fgf3: one close to the bead, the other in the mesenchymal papilla surrounded by the epithelial cap. Very rarely, weak Fgf4 expression was seen, as in one of the two explants shown here. None of these genes is activated by control beads soaked in BSA (mesenchymal expression of Bmp4 is not affected in the mutant; see Fig. 1A). (B) Time course of gene activation by FGF4 in explanted molar rudiments or in isolated molar mesenchyme of E14.5 Lef1−/− embryos. Fgf3 transcription is activated within 3 h, even in the absence of dental epithelium. Shh expression is undetectable after 16 h (weak signals are seen after 24 h; see A), and strong signals are observed in the epithelium after 48 h. As expected, Shh transcription is not activated in the mesenchyme.
Shh expression remained undetectable for at least 16 h (Fig. 5). Some Shh transcripts appeared after 1 d, but Shh was expressed consistently and at high levels in the epithelial cap after 4 d. Bmp4 expression was found only in the mesenchyme at low levels and independently of treatment with FGF4 beads, because it was also detected in explants that had been treated with bovine serum albumin (BSA) control beads. Thus, the rapid FGF4-mediated induction of Fgf3 expression in the mesenchyme and the slow induction of Shh expression in the epithelium of Lef1−/− explants suggest that FGF4 acts in two steps.
Induction of epithelial Shh expression by mesenchymal FGF proteins
To gain insight into the question of whether FGF4 regulates Shh expression in the epithelium directly or indirectly via a signaling loop involving mesenchymal FGF3, we examined whether FGF4 beads could induce Shh expression in the absence of mesenchymal FGF3. Because isolated mesenchyme-free dental epithelium cannot be well maintained in vitro for sufficiently long periods, we placed E14 Lef1−/− dental epithelium on heterologous dermal mesenchyme that did not express Fgf3, Fgf7, and Fgf10 (data not shown). Such heterologous tissue recombinants did not form dental structures and their incubation with FGF4 beads failed to induce Fgf3 expression in the mesenchyme and Shh transcription in the epithelium (0/8 tissue recombinants; data not shown). Thus, mesenchymal FGFs seem to play an important role in the signaling cascades of tooth development at E14.5.
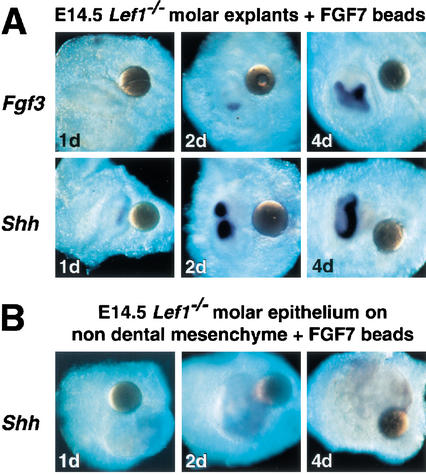
To examine whether mesenchymal FGFs might also overcome the developmental arrest of Lef1-deficient rudiments and induce tooth-specific gene expression, we used FGF7 (KGF) and FGF10 as surrogates for FGF3, which is not available in recombinant form. All three mesenchymal FGF proteins interact with the same epithelial receptor FGFR2-IIIb (or KGFR; Ornitz et al. 1996; Ornitz and Itoh 2001), and Fgf10 is expressed at apparently lower levels but in overlapping domains with Fgf3 in dental mesenchyme (Kettunen et al. 2000). We found that FGF7 and FGF10 beads rescued Lef1−/− tooth development with virtually the same efficiency as the “epithelial” FGF4, FGF8, or FGF9 proteins (Table 1). However, FGF7 induced expression of Shh in the epithelium within 2 d without significant activation of endogenous Fgf3 expression in the mesenchyme (Fig. 6A). This result suggests that the added FGF7 and FGF10 act as surrogates of mesenchymal FGF3 protein, rather than as inducers of the endogenous Fgf3 gene.
Figure 6.
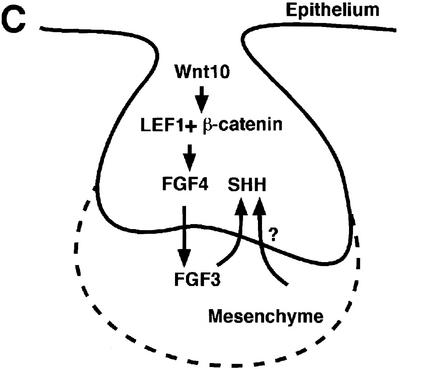
Effects of mesenchymal FGF proteins on gene expression in Lef1−/− molar explants, detected by whole mount in situ hybridization. (A) FGF7 beads fail to activate early expression of Fgf3 in the mesenchyme, but a small expression domain is seen in the epithelium after 2 d. After 4 d, Fgf3 expression is detected in the forming papilla but not around the bead. In contrast, Shh is activated in the epithelium with a similar time course as by FGF4 beads (cf. Fig. 5A). (B) FGF7 cannot induce Shh expression in dental epithelia that have been placed on nondental (dermal) mesenchyme. (C) Model of the signaling cascade governing the transition from the bud (E13) to the cap (E14) stage of tooth development. Intraepithelial Wnt10a and/or Wnt10b signals mediate, via LEF1 and β-catenin, the transcriptional activation of the Fgf4 gene in the epithelium. FGF4 signals to the subjacent mesenchyme. One of the early targets of this epithelial signaling is the Fgf3 gene in mesenchymal cells, and FGF3 together with other signals of dental mesenchyme (presumably including BMP4) in turn induce expression of Shh in the epithelium.
Finally, we examined whether mesenchymal FGF proteins are sufficient to induce epithelial Shh expression. In explants, in which Lef1−/− dental epithelium has been combined with nondental (dermal) mesenchyme, FGF7 did not induce Shh transcription (Fig. 6B), suggesting that additional signals of dental mesenchyme are needed for the induction of Shh. Taken together, these data suggest a model in which LEF1 links the Wnt signal pathway to FGF signaling activity in the epithelial enamel knot, thereby establishing a regulatory signaling loop between epithelial and mesenchymal FGF proteins (Fig. 6C).
Discussion
Our previous tissue recombination experiments with wild-type and Lef1−/− tooth germs revealed that the transcription factor LEF1 is required only at one step in tooth development and acts in a tissue-nonautonomous manner. On the basis of these experiments, we have hypothesized that the sole function of LEF1 in tooth development is to generate an inductive signal from the epithelium to the mesenchyme at E13.5 (Kratochwil et al. 1996). We now provide molecular insight into this process and identify Fgf4 as a transcriptional target of LEF1 and the Wnt signaling pathway. We also show that the arrest of tooth development in organ cultures of Lef1−/− embryos can be fully overcome by exogenous application of recombinant FGF4 protein, suggesting that a single transcriptional target of LEF1 can account for the function of LEF1 in tooth organogenesis.
Fgf4 is a functional and transcriptional target of LEF1
Several lines of evidence suggest that FGF proteins represent the postulated LEF1-dependent epithelial signal that is required for the progression of tooth development at the bud stage. First, various Fgf genes (Fgf3, Fgf4, Fgf9) are coexpressed with Lef1 in the E14 epithelial enamel knot in an LEF1-dependent manner. Second, the inductive effect of wild-type dental epithelium is abolished by SU5402, a small molecule inhibitor of FGF signaling. Third, recombinant FGF proteins can fully rescue the arrest of tooth development in organ cultures of Lef1−/− embryos. This functional rescue is efficient and is also obtained with the AER of mouse and chick limbs, which are physiological sources of FGF proteins (Martin 1998). The effects of FGF proteins on Lef1 tooth germs are specific and not observed with other signal factors that are produced by the wild-type enamel knot. Moreover, FGF beads do not rescue the development of Msx1−/− or Pax9−/− tooth rudiments, which are arrested at a similar stage as Lef1−/− tooth germs (Satokata and Maas 1994; Peters et al. 1998; Bei et al. 2000; H. Peters and K. Kratochwil, unpubl.). Finally, wild-type epithelium and FGF beads have the same temporal specificity in tooth development and they both act to stabilize the developmental potential of the mesenchymal component of tooth germs. This function includes the activation of Fgf3 expression and the completion of the reciprocal epithelial–mesenchymal signaling cascade, which is necessary for full tooth development and typical crown morphogenesis. Taken together, these results indicate that FGF proteins represent the essential epithelial signal missing in the Lef1−/− tooth germs.
The functional rescue of arrested Lef1−/− tooth development by FGF proteins raises the question as to the regulation of other signaling molecules by LEF1. Our experiments suggest that FGF proteins are the only signaling molecules that rescue the arrest of Lef1−/− tooth development. However, other signaling molecules are also regulated by LEF1. Application of exogenous FGF proteins to E13.5 Lef1−/− tooth germs rescues development without inducing epithelial expression of Fgf4 and Bmp4. In addition, Fgf9 is expressed very weakly in FGF4-treated mutant tooth germs, whereas Shh is expressed at high levels. Taken together, these data suggest that Fgf4, Fgf9, and Bmp4 are regulated by LEF1, although we do not know whether Fgf9 and Bmp4 are direct targets of LEF1. The expression of Bmp4 in the dental mesenchyme of Lef1−/− embryos may be accounted for by a redundancy of LEF1 and TCF1, which are coexpressed in the mesenchyme, but not in the epithelium (data not shown).
Another signaling molecule, the TNF-type molecule ectodysplasin, has recently been identified as a transcriptional target of LEF1. Ectodysplasin, encoded in the mouse by Tabby, is down-regulated in Lef1−/− embryos and can be induced by exogenous Wnt6 in an LEF1-dependent manner (Laurikkala et al. 2002). Moreover, the promoter of the human ectodysplasin gene contains functionally important binding sites for LEF1 (Durmowicz et al. 2002). Tabby is coexpressed with Lef1 in nondental oral epithelium of E11 embryos but not in the enamel knot, and Tabby deficiency has only a mild effect on tooth development (Grüneberg 1966; Tucker et al. 2000; Laurikkala et al. 2002). Although the late morphogenesis of the crowns of Tabby mutant molars can be somewhat improved by exogenous FGF10 (Pispa et al. 1999), the impaired expression of Tabby in Lef1−/− embryos is unlikely responsible for the complete arrest of tooth development at the bud stage.
The Fgf4 gene appears to be a direct transcriptional target for LEF1 as it has a functionally important binding site for LEF1/TCF proteins and is stimulated by Wnt signaling. Multiple genes have been shown to be regulated by Wnt signals (for review, see Nusse 1999; van Noort and Clevers 2002). However, only a few genes have also been identified as functional targets, as defined by their ability to rescue defects in components of the Wnt signaling pathway. In addition to FGF4, which can rescue an Lef1 deficiency in tooth development, forced expression of an Engrailed1 transgene has been found to overcome the defects in the formation of the mid/hindbrain boundary that are caused by a targeted mutation of the Wnt1 gene (Danielian and McMahon 1996). Together, these observations raise the interesting possibility that a small number of target genes of Wnt signals and LEF1/TCF transcription factors may be functionally relevant in a specific developmental context. In contrast, relatively large numbers of genes display an altered expression in response to Wnt signals in microarray analysis of tissue culture cells (Ross et al. 2002; Willert et al. 2002).
The target genes of LEF1 and Wnt signaling also appear to differ for individual tissues. The Brachyury gene has been identified as a transcriptional target for LEF1 in the primitive streak of the gastrulating embryo (Yamaguchi et al. 1999; Arnold et al. 2000; Galceran et al. 2001), but this gene is neither expressed in the tooth germ nor at the mid/hindbrain boundary. Moreover, the targeted inactivation of Lef1 does not phenocopy mutations or targeted inactivations of Brachyury, Engrailed1, and Fgf4 (Herrmann et al. 1990; Feldman et al. 1995; Loomis et al. 1996). Although some of the differences in the phenotypes of Lef1−/− and Fgf4−/− mice may be accounted for by redundancies of Lef1 with other Lef/Tcf genes, we have no evidence for a regulation of Fgf4 by LEF1 in all tissues that express Fgf4. In addition, Lef1 but not Fgf4 is expressed at the earliest stages of tooth development, suggesting that LEF1 may collaborate with different proteins in various tissues and developmental stages to regulate target gene expression.
LEF1 acts as a relay of a Wnt signal to an FGF signal
Our experiments suggest that the essential function of LEF1 in tooth development is to connect intraepithelial Wnt signaling with epithelium-to-mesenchyme FGF signaling. A functional link between these two signaling pathways has been implicated in other examples of epithelial–mesenchymal interactions. At the initiation of limb development, Wnt- and β-catenin-mediated signal transduction appears to be involved in the reciprocal FGF signaling between lateral plate mesoderm (LPM) and apical ectoderm (Kawakami et al. 2001). Specifically, Wnt2b and Wnt8c signals activate Fgf10 expression in the LPM, and the subsequent induction of Fgf8 in the overlying AER by FGF10 signals from the LPM is mediated via epithelial Wnt3a (Kawakami et al. 2001). In both directions, Wnt/β-catenin signaling operates within the same tissue and generates downstream FGF signals that are transferred to the neighboring tissue. This situation may be comparable to the enamel knot of the tooth bud, in which Wnt10a and Wnt10b are expressed independently of LEF1 and the function of LEF1 is required to generate FGF signals for the induction of the subjacent mesenchyme.
A similar interaction of the Wnt and FGF signaling pathways was also found to operate in the induction of posterior cell fates during early Xenopus development. Wnt/β-catenin signaling not only induces Fgf expression, but the induction of posterior neural markers by XWnt8 and β-catenin was also shown to depend on downstream FGF signaling (Domingos et al. 2001). Conversely, a study in Fgfr1−/− mice indicated that FGF signaling regulates the canonical Wnt/β-catenin signal transduction pathway during the formation of the primitive streak (Ciruna and Rossant 2001). Thus, FGF signaling and Wnt signaling can be linked in either direction. Although FGF signals are not required for the sustained expression of Wnt10a and Wnt10b in tooth germs of Lef1−/− mice, we cannot rule out the possibility that the initiation of Wnt gene expression is regulated by FGF signals.
Interactions between Wnt signaling and other signaling pathways have also been shown for the development of hair follicles. De novo hair morphogenesis has been found to be controlled by stabilization of β-catenin and Wnt signaling (Gat et al. 1998; Huelsken et al. 2001; Andl et al. 2002; Niemann et al. 2002). In addition, the formation of hair placodes involves a hierarchy of signaling pathways in which Wnt has been placed downstream of Tabby and upstream of Bmp and Shh (Huelsken et al. 2001).
The signaling cascade downstream of Fgf4 in tooth development, which includes the rapid activation of Fgf3 in the dental mesenchyme, is more difficult to resolve because of the functional redundancies of the LEF1/TCF transcription factors and FGF proteins. Lef1 is coexpressed with Tcf1 at multiple sites of the mouse embryo, including the mesenchyme of tooth germs, the primitive streak, and limb buds (Oosterwegel et al. 1993; Galceran et al. 1999). The functional redundancy of these closely related transcription factors is revealed by the phenotype of the Lef1−/−Tcf1−/− double mutation, which is more severe than either single mutation (Okamura et al. 1998; Galceran et al. 1999). In addition, Fgf4 and Fgf9 may act in a redundant manner in tooth development, reminiscent of the redundant function of various Fgf genes in the AER of the limb bud (Moon et al. 2000; Sun et al. 2000). The coexpression of Lef1 with Tcf1 in the mesenchyme may account for the full developmental potential of Lef1−/− tooth mesenchyme and obscure an assessment of a putative role of Wnt signaling in the activation of Fgf3. Therefore, we cannot determine whether the activation of Fgf3 by FGF4 is direct or indirect via Wnt signals.
The localized expression of Fgf3 in wild-type molar mesenchyme subjacent to the epithelial enamel knot and the rapid induction of Fgf3 in Lef1−/− mesenchyme by wild-type dental epithelium or recombinant FGF4 point to an important function of this mesenchymal FGF in tooth development. This assumption is supported by the rescue of Lef1−/− tooth development by recombinant FGF7 and FGF10, which act via the same epithelial receptor, FGFR2b (KGFR), as FGF3 (Ornitz et al. 1996; Celli et al. 1998). In contrast to FGF4, neither FGF7 nor FGF10 were found to induce mesenchymal Fgf3 expression, indicating that they are acting downstream of FGF4 as surrogates for FGF3, rather than as inducers of Fgf3 expression. Fgf3 and Fgf10 are expressed in overlapping patterns in the developing tooth (Kettunen et al. 2000). Tooth development is normal in targeted mutants of Fgf3 and only minimally affected in Fgf10 knockout mice (Mansour et al. 1993; Ohuchi et al. 2000). However, tooth development is arrested at the bud stage in embryos that either express a soluble dominant-negative form of the FGFR2b receptor (Celli et al. 1998), or carry a targeted deletion of exon IIIb of the Fgfr2 gene (DeMoerlooze et al. 2000; Revest et al. 2001). Thus, Fgf3 and Fgf10 appear to act redundantly in tooth development.
One of the targets of mesenchymal FGF signals is epithelial Shh. However, the activation of Shh requires additional factors provided by wild-type or Lef1−/− dental mesenchyme, but not by dermis. One of the factors that cooperate with mesenchymal FGF proteins may be BMP4, which is expressed normally in Lef1−/− dental mesenchyme. Bmp4 is under the transcriptional control of MSX1, and its essential function in tooth development is revealed by the ability of recombinant BMP4 to overcome the arrest of Msx1−/− tooth development at the bud stage (Bei et al. 2000). BMP4 has also been more directly implicated in the induction of epithelial Shh expression in tooth germs (Zhang et al. 2000). In addition, activin has been identified as an early mesenchymal signal in tooth development (Ferguson et al. 1998). Although Shh activation is a good indicator for the development of dental epithelium beyond the bud stage, it cannot be the only gene downstream of the reciprocal FGF signaling activity, because beads soaked in SHH were not sufficient to rescue Lef1−/− tooth development. Moreover, application of exogenous SHH to early tooth germs was found to affect epithelial cell proliferation, and conditional Shh knockout mice show a defect in odontogenesis that occurs after the bud stage and impairs the growth and shape of teeth (Hardcastle et al. 1998; Dassule et al. 2000).
According to our model, Lef1 deficiency interrupts epithelial–mesenchymal interactions in tooth development at a critical stage (E13.5), when the epithelium generates an inductive signal to the mesenchyme that allows for the formation of the dental papilla. The epithelial signal that depends directly on LEF1 and Wnt signals has been identified as FGF4. Targets of this epithelial FGF signaling activity are the Fgf genes that are expressed in the dental mesenchyme (Fgf3 and Fgf10). Together with other signal factors provided by dental mesenchyme, these mesenchymal FGFs in turn induce epithelial Shh expression. Thus, LEF1 serves as a relay of a Wnt signal to an FGF signal in the epithelium, which establishes a network of reciprocal and sequential signaling between epithelium and mesenchyme. This signaling network involves multiple signals that cooperate to allow for the precise and temporally and spatially coordinated differentiation of adjacent tissues during organogenesis.
Materials and methods
Mice
The Lef1 mutation (van Genderen et al. 1994) is maintained on a C57BL/6 background. Timed embryos were obtained by heterozygous matings counting the vaginal plug as E0.5 and using morphological criteria for precise staging. Homozygous mutant embryos can easily be identified from E12.5 because of the absence of whisker hillocks. Both wild-type and heterozygous litter mates were used as donors for normal tissues.
Tissue dissection, recombination, and explantation
Embryos were dissected in Dulbecco's PBS; tooth anlagen were taken from the lower jaw. Limb ectoderm, including the AER, was obtained through trypsin treatment (2.25% crude trypsin, Sigma type II, plus 0.75% pancreatin, Difco, N.F., in PBS for 20 min on ice) either from E10.5 mouse hind limb buds or from E3 chick wing buds. Notochords were dissected from E10.5 to E11.5 mouse or E4 chick embryos. Dental epithelium and mesenchyme were mechanically separated after incubation in 0.1% crude collagenase (Sigma type I) in Dulbecco's minimal essential medium (DMEM, GIBCO-BRL) for 40 min at 37°C. Intact tooth rudiments or tissue recombinations were explanted on gelatinized Nuclepore filters (#110405; pore size 0.1 μm, Whatman) and cultured in DMEM supplemented with 10% fetal calf serum (Hyclone) and 10% chick embryo extract. After in vitro culture for 1–4 d, some explants were transplanted under the kidney capsule of syngeneic adult males to allow for full tooth development (Kratochwil et al. 1996). SU5402 (Calbiochem), an inhibitor of FGF receptor tyrosine kinase activity (Mohammadi et al. 1997), was added to the medium at a final concentration of 25 μM.
Application of recombinant signal proteins
Affigel beads (100–150 μm diameter; Bio-Rad) were soaked in solutions (100 ng/μL) of recombinant BMP4 (gift of E. Wang, Genetics Institute, Cambridge, MA) or SHH (gift of J. Helms, University of California at San Francisco) and placed either between recombined (mutant) dental epithelium and mesenchyme, or on top of intact tooth rudiments close to the dental lamina. Heparin-acrylic beads (Sigma, 200 μm diameter) were soaked in solutions of FGFs (FGF4, FGF7, FGF8b, FGF9, and FGF10, from R&D Systems, all at 100 ng/μL) and were applied in the same way. Control beads were soaked in BSA (100 ng/μL).
Histological procedures and in situ hybridization
For standard histology, explants or transplants were fixed in Bouin's solution, embedded in paraffin, and 7-μm sections were stained with the Azan stain. For hybridizations, embryos or explants were fixed in 4% paraformaldehyde in PBS overnight (o.n.), then washed in PBST (PBS with 0.1% Tween 20). For whole-mount in situ hybridization, tissues were dehydrated in methanol and stored at −20°C until use. Whole-mount in situ hybridizations were done in an automated processor (InsituPro, Abimed) according to standard protocols (Wilkinson 1992). For in situ hybridizations, 7-μm paraffin sections of E14.5 embryos were incubated with Proteinase K (10 mg/mL) in PBS for 20 min after rehydration, then postfixed in 4% paraformaldehyde and acetylated in triethanolamine and acetic anhydride. All probes were DIG-labeled riboprobes (Boehringer, Mannheim) derived from mouse genes, except for rat Shh. The precise nucleotide positions of the probes are available on request.
Electrophoretic mobility shift assay
DNA-binding assays were performed as described (Galceran et al. 2001). The binding reaction contained 100 ng of purified His-6-tagged LEF1 protein and 2 fmole of 5′ end-labeled double-stranded oligonucleotide in 10 mM Hepes (pH 7.9), 50 mM NaCl, 1 mM DTT, 5 mM EDTA, 5% glycerol, 50 mg/mL BSA, and 10 μg/mL poly(dI-dC). The binding reactions were performed in the absence or presence of increasing amounts of unlabeled competitor DNA. The nucleotide sequences of the probes for Fgf4 wt and mt sites, and competitor DNA were as follows. Fgf4 wt, 5′-GACTGATCTTTGAACTCTCTTGCAGC TAGCGATC-3′; Fgf4 mt, 5′-GACTGAgaaTTcAACTCTCTT GCAGCTAGCGATC-3′; competitor wt, 5′-GCACCCTTTG AAGCTCGCTAGCGATC-3′; competitor mt, 5′-GCACCaaT TcAAGCTCGCTAGCGATC-3′. The wild-type LEF1 binding site is shown in upper case and changes in the mutant oligonucleotides are indicated by lower case.
Reporter gene constructs
Fgf reporter constructs were based on construct #1 of Fraidenraich et al. (1998). An SphI–SalI fragment containing 1.2 kb of the mouse Fgf4 promoter was cloned in pBKS. The bacterial LacZ gene was inserted in exon 2 of the Fgf4 gene, and 2.2 kb from the 3′ UTR of the Fgf4 gene (nucleotides 5259–7433) were cloned immediately downstream of the SV40 polyadenylation sequence. The 3′ UTR of the Fgf4 gene contains three homology regions that are conserved between mouse and man. Homology region #2 contains the LEF1 binding site at nucleotide 5766. For additional details, see Fraidenraich et al. (1998). Site-directed mutagenesis was performed using single-stranded DNA from pBS construct #1 and the mutant oligonucleotide 5′-CAA GAGAGTTgAAttcTCAGTCTTG-3′. The mutated nucleotides are indicated in lower case.
Stable transfections and reporter gene assays
Mouse embryonal carcinoma F9 cells were grown on gelatinized plates in DMEM with 10% fetal calf serum. Stable transfections were performed with the linearized reporter gene construct and pCINeo (at a ratio of 5:1) by calcium phosphate coprecipitation. Stable pools were obtained by selection with 800 μg/mL G418 for 16 d. Clones were pooled and 6 × 105 cells were plated in a 6-cm dish. Cells were harvested after 24 h, and beta-galactosidase activity (normalized for protein content) was determined. For the stimulation of cell pools, LiCl was added to the medium at a final concentration of 10 or 30 mM, 18 h prior to harvest. For control cells, NaCl was added at the same concentration.
Acknowledgments
We thank Drs. Andy McMahon, Paul Sharpe, Irma Thesleff, and Annette Neubüser for their generous gifts of DNA probes; Dr. Jill Helms for recombinant SHH; and Dr. Gail Martin for helpful discussions. This work was supported, in part, by funds of the German Research Foundation (DFG).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rgross@lmb.uni-muenchen.de; FAX 49-89-2180-6949.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1035602.
References
- Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Stappert J, Bauer A, Kispert A, Herrmann BG, Kemler R. Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech Dev. 2000;91:249–258. doi: 10.1016/s0925-4773(99)00309-3. [DOI] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Bei M, Kratochwil K, Maas RL. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development. 2000;127:4711–4718. doi: 10.1242/dev.127.21.4711. [DOI] [PubMed] [Google Scholar]
- Celli G, LaRochelle WJ, Mackem S, Sharp R, Merlino G. Soluble dominant-negative receptor uncovers roles for fibroblast growth factors in multi-organ induction and patterning. EMBO J. 1998;17:1642–1655. doi: 10.1093/emboj/17.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001a;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001b;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- Dailey L, Yuan H, Basilico C. Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol Cell Biol. 1994;14:7758–7769. doi: 10.1128/mcb.14.12.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, McMahon AP. Engrailed-1 as a target of the Wnt-1 signaling pathway in vertebrate midbrain development. Nature. 1996;383:332–334. doi: 10.1038/383332a0. [DOI] [PubMed] [Google Scholar]
- Dassule H, McMahon AP. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev Biol. 1998;202:215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- Dassule H, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- DeMoerlooze L, Spencer-Dene B, Revest J-M, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb form of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signaling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Domingos PM, Itasaki N, Jones CM, Mercurio S, Sargent MG, Smith JC, Krumlauf R. The Wnt/β-catenin pathway posteriorizes neural tissue in Xenopus by an indirect mechanism requiring FGF signaling. Dev Biol. 2001;239:148–160. doi: 10.1006/dbio.2001.0431. [DOI] [PubMed] [Google Scholar]
- Durmowicz MC, Cui C, Schlessinger D. The EDA gene is a target of, but does not regulate Wnt signaling. Gene. 2002;285:203–211. doi: 10.1016/s0378-1119(02)00407-9. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Christensen L, Lau AL, Matzuk MM, Sharpe PT. Activin is an essential early mesenchymal signal in tooth development that is required for patterning of the murine dentition. Genes & Dev. 1998;12:2636–2649. doi: 10.1101/gad.12.16.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraidenraich D, Lang R, Basilico C. Distinct regulatory elements govern Fgf4 gene expression in the mouse blastocyst, myotomes and developing limb. Dev Biol. 1998;204:197–209. doi: 10.1006/dbio.1998.9053. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- Galceran J, Fariñas I, Depew MJ, Clevers H, Grosschedl R. Wnt3a−/−-like phenotype and limb deficiency in Lef1−/−Tcf1−/− mice. Genes & Dev. 1999;13:709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galceran J, Hsu SC, Grosschedl R. Rescue of a Wnt mutation by an activated form of LEF-1: Regulation of maintenance but not initiation of Brachyury expression. Proc Natl Acad Sci. 2001;98:8668–8673. doi: 10.1073/pnas.151258098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat U, DasGupta R, Degenstein L, Fuchs E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- Grobstein C. Mechanisms of organogenetic tissue interaction. Natl Cancer Inst Monogr. 1967;26:279–299. [PubMed] [Google Scholar]
- Grüneberg H. The molars of the tabby mouse, and a test of the “single-active X-chromosome” hypothesis. J Embryol Exp Morphol. 1966;15:223–244. [PubMed] [Google Scholar]
- Hardcastle Z, Mo R, Hui C-C, Sharpe PT. The Shh signaling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development. 1998;125:2803–2811. doi: 10.1242/dev.125.15.2803. [DOI] [PubMed] [Google Scholar]
- Hedgepeth CM, Conrad L, Zhang J, Huang HC, Lee VM, Klein PS. Activation of the Wnt signaling pathway: A molecular mechanism for lithium action. Dev Biol. 1997;185:82–91. doi: 10.1006/dbio.1997.8552. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Morphogenesis. Cell. 1999;96:225–233. doi: 10.1016/s0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;8:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Nuclear localization of β-catenin by interaction with transciption factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. β-catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Capdevila J, Büscher D, Itoh T, Esteban CR, Izpisúa Belmonte JC. Wnt signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell. 2001;104:891–900. doi: 10.1016/s0092-8674(01)00285-9. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Thesleff I. Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev Dyn. 1998;211:256–268. doi: 10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Laurikkala J, Itäranta P, Vainio S, Itoh N, Thesleff I. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev Dyn. 2000;219:322–332. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1062>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Kratochwil K, Dull M, Fariñas I, Galceran J, Grosschedl R. Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes & Dev. 1996;10:1382–1394. doi: 10.1101/gad.10.11.1382. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Pispa J, Jung HS, Nieminen P, Mikkola M, Wang X, Saarialho-Kere U, Galceran J, Grosschedl R, Thesleff I. Regulation of hair follicle development by the TNF signal ectodysplasin and its receptor Edar. Development. 2002;129:2541–2553. doi: 10.1242/dev.129.10.2541. [DOI] [PubMed] [Google Scholar]
- Loomis CA, Harris E, Michaud J, Wurst W, Hanks M, Joyner AL. The mouse Engrailed-1 gene and ventral limb patterning. Nature. 1996;382:360–363. doi: 10.1038/382360a0. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Goddard JM, Capecchi MR. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- Martin GR. The roles of FGFs in the early development of vertebrate limbs. Genes & Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Molenaar M, de Wetering M, Oosterwegel M, Peterson J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Moon AM, Boulet AM, Capecchi MR. Normal limb development in conditional mutants of Fgf4. Development. 2000;127:989–996. doi: 10.1242/dev.127.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubüser A, Peters H, Balling R, Martin GR. Antagonistic interactions between FGF and BMP signaling pathways: A mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Niemann C, Owens DM, Hulsken J, Birchmeier W, Watt FM. Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development. 2002;129:95–109. doi: 10.1242/dev.129.1.95. [DOI] [PubMed] [Google Scholar]
- Nusse R. WNT targets. Repression and activation. Trends Genet. 1999;15:1–3. doi: 10.1016/s0168-9525(98)01634-5. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- Okamura R, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:3005.1–3005.12. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubüser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes & Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pispa J, Jung H, Jernvall J, Kettunen P, Mustonen T, Tabata MJ, Kere J, Thesleff I. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev Biol. 1999;216:521–534. doi: 10.1006/dbio.1999.9514. [DOI] [PubMed] [Google Scholar]
- Revest J-M, Spencer-Dene B, Kerr K, DeMoerlooze L, Rosewell I, Dickson C. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- Riese J, Yu X, Munnerlyn A, Eresh S, Hsu SC, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- Ross SE, Erickson R, Gerin I, de Rose P, Bajnok L, Longo K, Misek D, Kuik R, Hanash S, Atkins K, et al. Microarray analysis during adipogenesis: Understanding the effects of Wnt signaling on adipogenesis. Mol Cell Biol. 2002;22:5989–5999. doi: 10.1128/MCB.22.16.5989-5999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakar L, Sharpe PT. Expression of Wnt signaling pathway genes during tooth development. Mech Dev. 1999;85:197–200. doi: 10.1016/s0925-4773(99)00095-7. [DOI] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx-1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Sun X, Lewandoski M, Meyers EN, Liu Y-H, Maxson RE, Jr, Martin GR. Conditional inactivation of Fgf4 reveals complexity of signalling during limb bud development. Nat Genet. 2000;25:83–86. doi: 10.1038/75644. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Sharpe PT. Signaling networks regulating dental development. Mech Dev. 1997;67:111–123. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes & Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Headon DJ, Schneider P, Ferguson BM, Overbeek P, Tschopp J, Sharpe PT. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development. 2000;127:4691–4700. doi: 10.1242/dev.127.21.4691. [DOI] [PubMed] [Google Scholar]
- Vaahtokari A, Åberg T, Jernvall J, Keräinen S, Thesleff I. The enamel knot as a signaling center in the developing mouse tooth. Mech Dev. 1996;54:39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- van Genderen C, Okamura R, Fariñas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-1 deficient mice. Genes & Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- van Noort M, Clevers H. TCF transcription factors, mediators of Wnt-signaling in development and cancer. Dev Biol. 2002;244:1–8. doi: 10.1006/dbio.2001.0566. [DOI] [PubMed] [Google Scholar]
- Waterman ML, Fischer W, Jones KA. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes & Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. Whole-mount in situ hybridization of vertebrate embryos. In: Wilkinson DG, editor. In situ hybridization: A practical approach. Oxford: Oxford University Press; 1992. [Google Scholar]
- Willert J, Epping M, Pollack J, Brown P, Nusse R. A transcriptional reponse to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes & Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang Z, Zhao X, Yu X, Hu Y, Geronimo B, Fromm SH, Chen YP. A new function of BMP4: Dual role for BMP4 in regulation of Sonic hedgehog expression in the mouse tooth germ. Development. 2000;127:1431–1443. doi: 10.1242/dev.127.7.1431. [DOI] [PubMed] [Google Scholar]