Abstract
Repression of E2F transcription activity by the retinoblastoma (Rb) tumor suppressor through its interaction with the transactivation domain of the E2F transcription factor is one of the central features of G1/S arrest in the mammalian cell cycle. Deregulation of the Rb–E2F interaction results in hyperproliferation, lack of differentiation, and apoptosis, and can lead to cancer. The 2.2-Å crystal structure of the Rb pocket complexed with an 18-residue transactivation-domain peptide of E2F-2 reveals that the boomerang-shaped peptide binds to the highly conserved interface between the A-box and the B-box of the Rb pocket in a bipartite manner. The N-terminal segment of the E2F-2 peptide in an extended β-strand-like structure interacts with helices from the conserved groove at the A–B interface, whereas the C-terminal segment, which contains one 310 helix, binds to a groove mainly formed by A-box helices. The flexibility in the middle of the E2F-2 peptide is essential for the tight association of E2F to the Rb pocket. The binding of Rb to the E2F-2 peptide conceals several conserved residues that are crucial for transcription activation of E2F. We provide the structural basis for the Rb-mediated repression of E2F transcription activity without the requirement of histone-modifying enzymes.
Keywords: Rb, E2F-2, transactivation, crystal structure
Temporal regulation of gene expression in the G1-to-S transition of the cell cycle is a complicated process in which the formation of a network of protein–protein interactions imposes control at multiple steps. A central feature of this network is an interaction between E2F family members and the retinoblastoma tumor suppressor (for review, see Dynlacht 1997; Dyson 1998; Harbour and Dean 2000a). E2F is a heterodimeric transcription factor that activates the genes involved in cell cycle progression, DNA replication, DNA synthesis, and apoptosis. These genes include cell cycle regulators, such as c-myc, cyclin A, cyclin E, and cdc2 (Hiebert et al. 1989; Thalmeier et al. 1989; Dalton 1992; Furukawa et al. 1994; Schulze et al. 1995); products that are required for DNA replication, such as Cdc6, ORC1, MCM6, dihydrofolate reductase, thymidine kinase, and DNA polymerase α (Blake and Azizkhan 1989; Mudryj et al. 1990; Pearson et al. 1991; Dou et al. 1992; DeGregori et al. 1995); and factors involved in apoptosis, such as Arf, p73 and Apaf1 (Harbour and Dean 2000b; Irwin et al. 2000; Guo et al. 2001). Overexpression of E2F induces quiescent cells to enter the S phase. The mice lacking E2F-1 develop various types of tumors (Yamasaki et al. 1996), whereas overexpression of E2F-1 in transgenic mice promotes tumorigenesis (Pierce et al. 1998), implicating E2F-1 functions as both an oncogene and a tumor suppressor. Furthermore, deletion of E2F-1 in mice causes testicular atrophy, exocrine gland dysplasia, and defects in thymocyte development (Field et al. 1996; Yamasaki et al. 1996). The molecular mechanism for E2F-driven transcription activation is unclear at present. However, the transactivation domain of E2F-1 is known to interact with general transcription factors such as TATA-box-binding protein (TBP; Hagemeier et al. 1993; Pearson and Greenblatt 1997), TFIIA/TFIIB (Ross et al. 1999), and the coactivator p300/CBP (Trouche and Kouzarides 1996; Trouche et al. 1996; Fry et al. 1999).
The Rb tumor suppressor plays a pivotal role in negative regulation of cell proliferation (for review, see Weinberg 1995), and such growth-inhibition function depends, at least in part, on its ability to repress E2F transcription activity. In early-to-mid-G1 phase, the Rb protein is sequentially phosphorylated and inactivated by G1-cyclin-dependent kinase complexes (DeCaprio et al. 1989; Ludlow et al. 1990; Harbour et al. 1999). This event releases E2F from the Rb complex, and E2F can then mediate cell proliferation. The importance of the Rb–E2F interaction is underscored by observations that (1) E2F activity is aberrantly controlled in most human tumor cells through mutation of genes required to control the Rb–E2F interaction, including Rb, cyclin D, and p16 (Flemington et al. 1993; Zhang et al. 1999; Nevins 2001); (2) viral oncoproteins inactivate the tumor-suppressor properties of Rb by interacting with Rb in such a way that E2F is liberated from the Rb–E2F complex (Chellappan et al. 1992); (3) the formation of some tumor types in Rb+/− mice is reduced or delayed in animals deficient in E2F-1 (Yamasaki et al. 1998).
Two models have been proposed to explain how inhibition of E2F transcription by Rb might affect cell cycle progression. The “direct binding and masking” model proposes that an underphosphorylated, active form of the Rb tumor suppressor directly binds to the transactivation domain of E2F and, by blocking the ability of E2F to activate transcription, leads to cell cycle arrest (Hiebert et al. 1992; Flemington et al. 1993; Helin et al. 1993a; Dynlacht 1997). In the second model, the Rb protein binds to the promotor of certain cell cycle genes as a complex with E2F and recruits chromatin-modifying enzyme complexes (Weintraub et al. 1995; Brehm et al. 1998; Zhang et al. 2000; Nielsen et al. 2001). The histone-modifying enzymes then actively repress transcription through changes in chromatin structure. In this “active repression” model, E2F repression is promotor-dependent as certain promoters are insensitive to histone deacetylase-1-mediated inhibition (Luo et al. 1998). Consistent with this observation, Rb represses transcription in a reconstituted transcription system lacking histones (Dynlacht et al. 1994). Although both models are well supported by a number of studies, the relative importance of these two distinct mechanisms is unclear. Nevertheless, recognition of the E2F transactivation domain by Rb is a primary step for the repression of E2F in both models.
In humans, six E2F members (E2F-1–E2F-6) and two of their partners, DP-1 and DP-2, have been identified (for review, see Dyson 1998). Each E2F subunit has a DNA-binding domain and a dimerization domain, and, except for E2F-6, all E2F subunits have a C-terminal transactivation domain that provides an interaction interface for Rb and two other pocket-protein family members, p107 and p130 (Bagchi et al. 1991; Kaelin et al. 1992; Cress et al. 1993; Helin et al. 1993b; Krek et al. 1993). The pocket region of Rb (amino acid residues 380–785), formed from the A-box, a spacer, and the B-box, is essential for the biological function of Rb. It is the most frequently mutated region in Rb-related cancers and is essential for growth suppression (Onadim et al. 1992; Kratzke et al. 1994). Furthermore, it is the minimal interaction region for several Rb-binding viral oncoproteins and cellular proteins, including E2F (Hu et al. 1990; Huang et al. 1990; Kaelin et al. 1990, 1992; Helin et al. 1992). Evidence from several investigations suggests that an 18-amino-acid residue sequence (residues 410–427) in the transactivation domain of E2F is essential and sufficient for the interaction of E2F with the Rb-pocket region (Kaelin et al. 1992; Cress et al. 1993; Hagemeier et al. 1993; Shan et al. 1996; Lee et al. 1998).
Recently, the crystal structures of the Rb pocket bound to the human papilloma virus (HPV) E7 peptide or the N-terminal fragment of SV40 large T antigen have been determined (Lee et al. 1998; Kim et al. 2001). The structures reveal that the Rb pocket is composed of two cyclin boxes with additional helices and a hairpin. Also, the structures show how the LxCxE motif within the E7 peptide or the SV40 large T antigen binds to the conserved groove in the B-box of the Rb pocket. However, despite the central importance of the Rb–E2F interaction in cell cycle regulation, it remains unclear how Rb binds to E2F and blocks its transcription activity at the molecular level. To understand the basis of E2F-transactivation-domain recognition by the Rb tumor suppressor, we have determined the 2.2-Å crystal structure of the Rb pocket bound to an 18-residue peptide of the E2F-2 transactivation domain (Fig. 1a; Table 1). The results reveal that an L-shaped E2F-2 peptide binds to the highly conserved central interface in the Rb pocket. We show that five conserved residues in the E2F transactivation peptide are critical for the transcription activity of E2F, and that Rb conceals these residues upon binding to E2F. Our studies support the importance of the “direct binding and masking” model in Rb-mediated E2F inhibition and provide the structural basis for how the E2F transactivation domain is recognized by the Rb tumor suppressor.
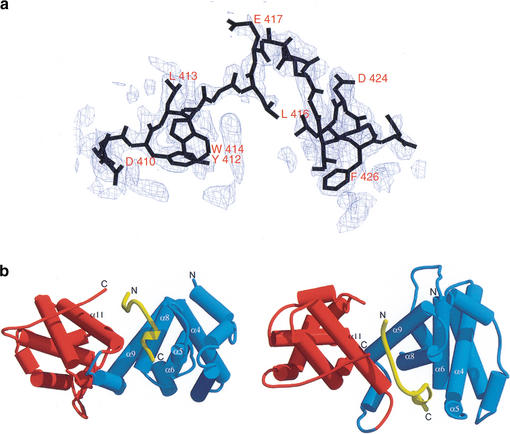
Figure 1.
(a) Electron density map for the bound E2F-2 peptide. The difference map was calculated using phases derived from the model after omitting the entire E2F-2 peptide and subjecting the model to simulated annealing refinement from 2000 K using CNS to remove model bias. The map was calculated at 2.2-Å resolution and contoured at 2.5 ς. (b) Overall structure of the Rb pocket in complex with the E2F-2 transactivation-domain peptide. The schematic representation of the right panel is related to the one on the left by a 90° rotation around the horizontal axis. (Blue) A-box in Rb; (red) B-box in Rb; (yellow) E2F-2. The secondary structure in the binding region in the Rb pocket is labeled.
Table 1.
Statistics from crystallographic analysis
| Data collection Parameter
|
Data
|
|---|---|
| Resolution (Å) | 50.0–2.2 |
| Total observations | 93798 |
| Unique observations | 37599 |
| Data coverage (%) (last shell, 2.28–2.20) | 86.3 (70.3) |
| Rsym (last shell, 2.28–2.20) | 8.2 (23.7) |
| Refinement | |
| Resolution range (Å) | 20–2.2 |
| Number of reflections (I/ς > 1) | 37466 |
| Rworking/Rfree (%) (last shell, 2.28–2.20) | 22.4/28.6 (28.7/32.8) |
| Number of atoms | 5981 |
| Number of water molecules | 267 |
| RMSD bond length (Å) | 0.009 |
| RMSD bond angles (°) | 1.38 |
| RMSD B-factor (Å2) for main-chain atoms | 1.48 |
| RMSD B-factor (Å2) for side-chain atoms | 2.06 |
Rsym = ∑h∑i|Ih,i − Ih|I∑h∑iIh,i, where Ih is the mean intensity of the i observations of symmetry-related reflections of h. R = ∑|Fobs − Fcalc|∑ Fobs, where Fobs = Fp, and Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections). RMSD in bond lengths and angles are the deviations from ideal values, and the RMSD in B factors is calculated between bonded atoms.
Results
Overall structure of the complex
Because our initial attempt to obtain crystals using the full Rb pocket (residues 380–785) and the E2F-2 peptide was not successful, we removed the flexible spacer region (residues 582–642) to aid the crystallization. Previous biochemical studies showed that the deletion of a spacer region did not affect the interaction between the Rb pocket and an 18 amino-acid residue peptide of the E2F-1 transactivation domain, or between the Rb pocket and viral oncoproteins containing an LxCxE motif (Lee et al. 1998; Kim et al. 2001).
Within the asymmetric unit of each crystal, there are two complexes of the Rb pocket bound to the E2F-2-transactivation-domain peptide plus an extra E2F-2 peptide. The extra E2F-2 peptide, located between the B-boxes of the two Rb pockets, makes protein–protein contacts that stabilize the crystal packing. The two molecules in the asymmetric unit have similar conformations; the root-mean-squared deviations (RMSDs) of the Cα atoms between the two molecules are 0.65 Å for Rb and 0.43 Å for the E2F-2 peptide.
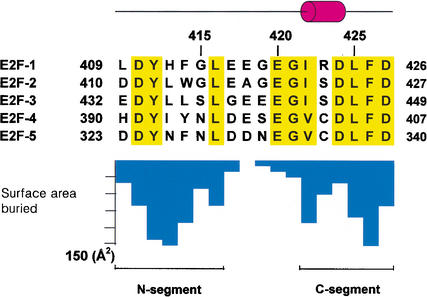
The structure shows that the 18-amino-acid peptide of E2F-2 binds Rb in a bipartite manner, with the two E2F-2 segments interacting with different sites on the Rb pocket (Fig. 1b). The seven-residue N-terminal segment (N-segment, residues 410–416) adopts an extended β-strand-like conformation, and the six-residue C-terminal segment (C-segment, residues 422–427) forms a helical shape containing a 310 helix (Fig. 1b). The two segments are separated by a bend formed from the five amino acid residues in the middle of the peptide. The binding between Rb and E2F-2 buries 2333 Å2 of surface area, the interface involving the N-segment and the C-segment in E2F-2 contributing ∼50% and ∼42% respectively, to the surface (Fig. 2). The conserved residues in the N-segment and C-segment that are biochemically known to affect binding of E2F to Rb make a major contribution to the buried surface area upon complex formation (Hagemeier et al. 1993; Shan et al. 1996).
Figure 2.
Sequence of the E2F family showing secondary structure elements. Conserved residues are highlighted in yellow. The bar graph shows the buried surface area per E2F-2 peptide residue upon complex formation.
The binding does not involve significant conformational changes to the main chain of the Rb pocket (RMSDs of Cα atoms are 1.23 Å between the HPV E7–Rb-pocket complex and the Rb pocket bound to the E2F-2 peptide). Some rearrangements are observed in the C-terminal region (residues 773–776) compared with the Rb pocket structure bound to the E7 peptide, but the alterations are distant from the binding site for the E2F-2 peptide.
N-segment of E2F-2 and Rb-pocket interface
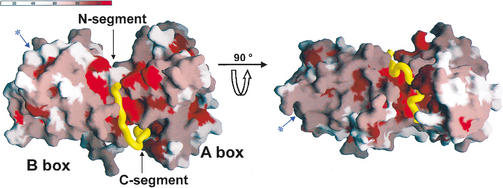
The Rb pocket is formed from the A-box and B-box and the extended C-terminal tail of Rb. Upon packing of the A-box and B-box, a highly conserved groove is formed that runs halfway around the interface (Fig. 3). This groove is ∼35 Å distant from the shallow groove in the B-box where viral oncoproteins containing the LxCxE motif bind (Fig. 3). Helices α8, α9, and α10 from the A-box and α11, α12, α13, and α14 from the B-box participate in the formation of this groove. The binding site for the N-segment of the E2F-2 peptide is formed from three helices at this groove in the A–B interface: α8 and α9 from the A-box form one side, and α11 from the B-box forms the other side of the binding site. The top of the binding site is lined with residues from the extended C-terminal loop of the Rb pocket. Here, several exposed polar residues, together with a partly exposed hydrophobic patch that formed upon A–B packing, complete the binding site for the N-segment of the E2F-2 peptide.
Figure 3.
Surface representation of the Rb pocket colored according to the degree of conservation in the Rb orthologs of five species and p107 and p130 family members (Lee et al. 1998). The A–B interface and the A-box where E2F binds are shown. The conserved groove in the B-box where the LxCxE motif binds is in the opposite face (marked with an arrow and an asterisk shown in blue) and cannot be seen in this view.
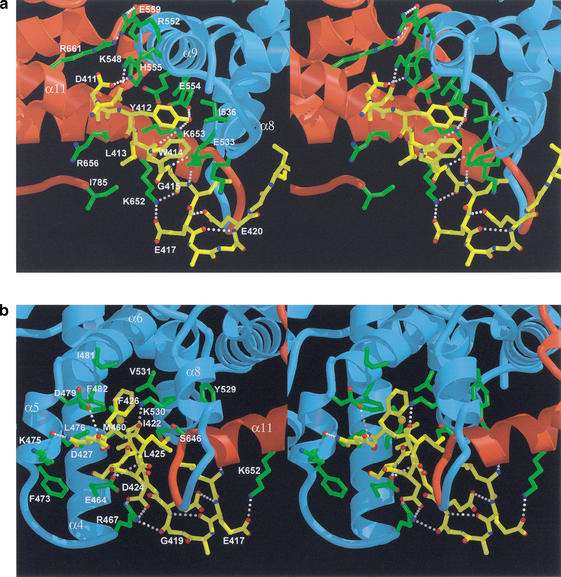
In the extended β-strand-like N-segment, the side chains of Tyr 412, Trp 414, and Leu 416 of E2F-2 are directed toward the A-box, where they make inter- and intramolecular contacts. In contrast, Leu 413 and Gly 415 make contacts with the B-box. The intermolecular H-bonds and van der Waals contacts made by this part of E2F-2 are uniformly distributed among A- and B-box residues. This area of the interface contains eight intermolecular H-bonds made by two backbone amide groups (Gly 415 and Leu 416), two carbonyl groups (Leu 413 and Gly 415), and the side chains of Asp 410, Asp 411, and Tyr 412 of the E2F-2 peptide (Fig. 4a). Mutations in the conserved Tyr 411 prevent E2F-1 (Tyr 412 from E2F-2) from binding to Rb (Shan et al. 1996), and in our structure, Tyr 412 is almost entirely buried, with 99% of its accessible surface area (in a hypothetical free peptide) covered by residues from the Rb pocket and the E2F-2 peptide. The hydroxyl group is inserted deep within the partly exposed polar region of the A–B interface of the pocket, making H-bonds with the side chains of Glu 554 from the A-box and Lys 653 from the B-box, both of which are conserved in pocket-protein family members (Lee et al. 1998; Kim et al. 2001). The side chains of Glu 554 and Lys 653, which form H-bonds with each other, are partly exposed (6% and 20%, respectively) but become entirely buried upon E2F binding. The side-chain ring of Tyr 412 of E2F-2 is surrounded by the side chains of Glu 533 and Ile 536 from helix α8, by Glu 551 from α9 of the Rb pocket, and by the indole ring of Trp 414 from the E2F-2 peptide. The side chain of Asp 411 of E2F-2 makes an H-bond to His 555 in helix α9 of the A-box. In the same helix, Glu 559 forms a salt bridge with Arg 661, a hot spot for tumorigenic mutations of the Rb protein. Another tumorigenic hot spot, Ala 562, makes van der Waals contacts with surrounding residues (Onadim et al. 1992; Kratzke et al. 1994). Thus, our structure indicates that tumorigenic mutations at these sites are likely to disturb the local interactions between Rb and E2F. The side chain of Leu 413 forms van der Waals interactions with Ile 785 from the C-terminal end of the Rb pocket and the aliphatic side chains of Lys 652, Lys 653, and Arg 656 from helix α11. Finally, while making no intermolecular contacts, the side chain of the conserved residue Leu 416 of E2F-2 is directed toward and makes van der Waals contacts with residues Gly 421, Ile 422, and Leu 425 from the C-segment of the E2F-2 peptide, helping to stabilize the peptide conformation within the Rb pocket.
Figure 4.
Schematic representation of interactions between the Rb pocket and the E2F-2 transactivation-domain peptide. (a) A closeup view of the interactions between the N-segment of the E2F-2 peptide and helices α8 and α9 from the A-box and α11 from the B-box in the A–B interface of the Rb pocket. (Yellow) E2F-2 peptide; (blue) A-box; (red) B-box; (yellow) interacting side chains in E2f-2; (green) interacting side chains in Rb; (red) O atoms; (blue) N atoms. In addition to the van der Waals contacts at the interface, there are eight intermolecular H-bonds: Lys 548 NZ to Asp 410 OD2, His 555 NE2 to Asp 411 OD2, Glu 554 OE2 to Tyr 412 OH, Lys 653 NZ to Tyr 412 OH, Lys 653 NZ to Leu 413 carbonyl, Glu 533 OE1 to Gly 414 amide, Lys 652 NZ to Gly 414 carbonyl, and Glu 533 OE2 to Leu 415 amide. (b) A closeup view of the interactions between the C-segment of E2F-2 and helices α4, α5, α6, α8, and α11 of the Rb pocket. The conserved residues, Ile 422, Leu 425, and Phe 426, of E2F-2 form multiple van der Waals contacts with residues from the Rb pocket. In the Rb pocket, the interacting residues are Met 460 on helix α4; Leu 476 on helix α5; Ile 481 and Phe 482 on helix α6; Tyr 529, Lys 530, Val 531, and Ser 534 on helix α8; and Ser 646 and Leu 649 on helix α11. The color-coding scheme is as in a.
C-segment of E2F-2 and Rb-pocket interface
The C-segment of the E2F-2 peptide binds to the A-box of the Rb pocket. The groove formed at this interface is continuous with the groove in the A–B interface, but in contrast to the N-segment, in which the contacts are made with both the A-box and the B-box, the C-segment of the E2F-2 peptide binds to helices almost exclusively from the A-box. The structural framework of the interface is formed by helices α4, α5, α6, and α8 from the A-box and helix α11 from the B-box. Except for α5, all these helices are from the cyclin fold.
The C-segment forms an amphipathic helical shape (Figs. 1b, 4b). In this region of E2F-2, the interacting residues, including Ile 422, Leu 425, and Phe 426, form an extensive network of van der Waals contacts with the surface residues of the Rb pocket (Fig. 4b). In the Rb pocket, the interacting residues are Met 460 on helix α4; Leu 476 on helix α5; Ile 481 and Phe 482 on helix α6; Tyr 529, Lys 530, Val 531, and Ser 534 on helix α8; and Ser 646 and Leu 649 on helix α11. Residues Leu 476, Phe 482, and Lys 530 are all highly conserved between Rb and other pocket family members. Notably, the interactions between the hydrophobic face of E2F-2, including Ile 422 and Phe 426, and the residues in the groove of the Rb pocket show strong steric complementarity. The significance of this steric complementarity is supported by a study in which a mutation of Phe 426 to Leu 426 in E2F-1 disrupted the interaction between E2F-1 and Rb (Shan et al. 1996).
In addition to van der Waals contacts, this part of the Rb–E2F-2 interface contains five intermolecular Hbonds, including one backbone-to-backbone contact between the amide of Asp 428 of E2F-2 and Lys 475 in the Rb pocket, and four backbone-to-side-chain contacts. In particular, the carbonyl atom of Leu 425 of E2F-2 accepts an H-bond from the side chain of the conserved Lys 530 in Rb, and the backbone amide of Ser 423 makes an H-bond to the side chain of the conserved Glu 464 in Rb. Importantly, the side chain of Asp 424 in the E2F-2 peptide forms a salt bridge with the Arg 467 in the loop connecting helices α4 and α5 of the Rb A-box.
Flexibility of the E2F-2 peptide
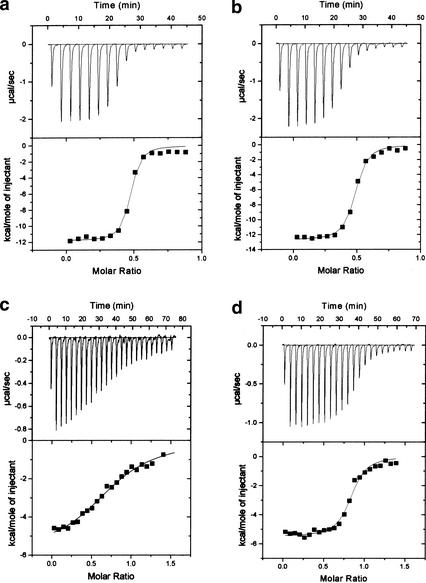
A sharp bend is formed between the N-segment and the C-segment of the E2F-2 peptide of ∼90° (Figs. 1b, 3). This is stabilized by an intramolecular H-bond from the backbone amide group of Glu 417 of the E2F-2 peptide to the side chain of Glu 420. In addition, Gly 421 of E2F-2 confers flexibility at this point of the peptide by adopting an unusual main-chain dihedral angle, the φ and ψ dihedrals being 50° and 160°, respectively. The bend in this region of E2F-2 seems to be essential for the tight interaction of the E2F-2 peptide with the Rb pocket in that it provides the flexibility necessary to permit the two segments of the E2F-2 peptide to make bipartite interactions on two different Rb-pocket binding sites. The significance of this bend is further highlighted by the absolute conservation of Glu 420 and Gly 421, neither of which makes intermolecular contacts, in E2F family members (Fig. 1b). To further investigate the relative contribution of Glu 420 and Gly 421 in the Rb–E2F interaction and the correlation between peptide bending at this position and the strength of E2F binding to Rb, we used isothermal titration calorimetry to measure the binding affinities of wild-type and mutant E2F-2 peptides to the Rb pocket (Fig. 5; Table 2). The wild-type peptide bound the Rb pocket with a dissociation constant of 0.18 μM, whereas the mutant peptides, Glu420Ala and Gly421Ser, bound the Rb pocket with dissociation constants of 0.32 μM and 10.3 μM, respectively. The twofold decrease in the binding affinity of the Glu420Ala mutant agrees with a mutational analysis in which a Glu 419 (Glu 420 in E2F-2) to Lys mutation in E2F-1 resulted in a reduced but significant binding affinity of the peptide to Rb (Shan et al. 1996), suggesting that this conserved acidic residue is not absolutely required for the interaction of E2F with Rb but presumably has other roles such as an interaction with factors that are involved in transcription. However, the decreased affinity between the Gly421Ser mutant and Rb when compared with wild-type E2F-2 indicates that E2F-2 must have the flexibility necessary to achieve the observed peptide bending if the E2F-2 peptide is going to interact with the Rb pocket, a result that supports our structure.
Figure 5.
ITC titration curves for E2F-2 peptide binding to the Rb pocket. The interactions between the Rb pocket and intact E2F-2 peptide (a); a Glu420Ala mutant (b); a Gly421Ala mutant (c); and intact E2F-5 peptide (d) are measured.
Table 2.
Thermodynamic parameters of E2F-2 peptide binding to Rb pocket
| Parameter
|
ΔH (kcal/mole)
|
TΔS (kcal/mole)
|
ΔG (kcal/mole)
|
Kd (μM)
|
|---|---|---|---|---|
| Intact E2F-2 | −11.8 ± 0.2 | −2.86 | −8.94 ± 0.2 | 0.19 ± 0.04 |
| Glu420Ala | −12.7 ± 0.1 | −4.01 | −8.69 ± 0.1 | 0.32 ± 0.04 |
| Gly421Ser | −5.8 ± 0.1 | 0.81 | −6.61 ± 0.1 | 10.3 ± 1.4 |
| Intact E2F-5 | −10.9 ± 0.1 | −2.72 | −8.21 ± 0.1 | 0.69 ± 0.1 |
Mutational analysis of the E2F-binding site within the Rb pocket
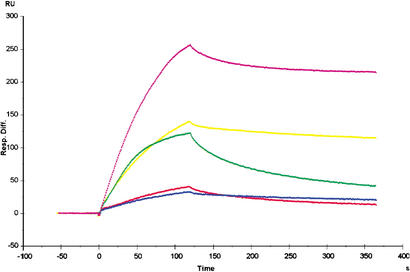
Previous work has shown that the Rb pocket structure contains two highly conserved regions, namely, the A–B interface and a shallow groove in the B-box (Lee et al. 1998). Viral oncoproteins containing an LxCxE motif bind to the shallow groove in the B-box, but the available data indicate that E2F binds to a different region on Rb (Lee et al. 1998; Kim et al. 2001). This suggests that the A–B interface of the Rb pocket is the possible E2F-binding site. Our structural data demonstrate that the A–B interface is, indeed, the region for E2F binding in Rb. To further investigate the interaction between the E2F peptide and residues in the A–B interface of the Rb pocket observed in our complex structure, we used surface plasmon resonance spectroscopy to study the binding of wild-type and mutant Rb pocket to E2F-1 protein containing the whole transactivation domain (Fig. 6; Table 3). For these studies, two residues in the N-segment binding site of Rb, Glu 554 and Lys 653, and two residues in the C-segment binding site, Arg 467 and Phe 482, were replaced by alanine. The wild-type Rb pocket bound to the E2F-1 protein with a dissociation constant of ∼3 nM, whereas the mutant Rb proteins exhibit a 6- to 35-fold decreased binding affinity for the E2F-1 protein. Thus, our mutational analyses demonstrate the importance of H-bonds between Tyr 412 of E2F-2 and Glu 554/Lys 653 of Rb, a salt bridge between Asp 424 of E2F-2 and Arg 467 of Rb, and the hydrophobic network mediated by Phe 482 of Rb in complex formation of Rb–E2F, providing biochemical confirmation that the E2F-2 peptide binding site within the Rb pocket is as shown in our structure.
Figure 6.
Surface plasmon resonance experiments. Sensograms of E2F-1 binding to wild-type and mutant Rb proteins that were used to calculate the kinetic binding constants are shown. The sensogram data at 100 nM for each Rb protein are superimposed and displayed. (Magenta) wild-type Rb pocket; (yellow) Arg467Ala mutant; (red) Phe482Ala mutant; (blue) Glu554Ala mutant; (green) Lys653Ala mutant.
Table 3.
Binding kinetics of wild-type or mutant Rb proteins to E2F-1 transcription factor as assayed by surface plasmon resonance
| Ligand
|
ka (q104 M−1 sec−1)
|
kd (×10−3 sec−1)
|
KD (×10−8 M)
|
|---|---|---|---|
| Wild-type Rb | 97.7 ± 7.3 | 5.85 ± 0.34 | 0.6 ± 0.01 |
| Wild-type GST-Rb | 3.62 ± 0.06 | 0.27 ± 0.03 | 0.74 ± 0.07 |
| GST-Rb (R467A) | 4.31 ± 1.34 | 1.77 ± 0.72 | 4.02 ± 0.27 |
| GST-Rb (F482A) | 2.65 ± 0.74 | 4.06 ± 0.96 | 15.5 ± 0.90 |
| GST-Rb (E554A) | 7.43 ± 0.04 | 1.95 ± 0.11 | 26.2 ± 1.70 |
| GST-Rb (K653A) | 6.99 ± 0.22 | 3.24 ± 0.10 | 4.64 ± 0.05 |
ka (on-rate) and kd (off-rate) are the kinetic association and dissociation constants, respectively. The equilibrium dissociation constant (affinity constant) is KD = kd/ka.
E2F regulation by Rb phosphorylation
E2F release upon Rb phosphorylation is a critical regulation mechanism in cell proliferation. Sequential phosphorylation events in the C-terminal region of Rb induce conformational changes in this region, triggering intramolecular interactions between the C-terminal region of the Rb protein and the lysine patch on the B-box in the pocket (Harbour et al. 1999). These changes would allow Cdk2–cyclin E to access and phosphorylate the buried Ser 567 of Rb and subsequently release E2F from the complex (Harbour et al. 1999). In our structure, the side chain of Ser 567 makes an H-bond with the backbone carbonyl of Ala 562 in helix α9, which forms part of the E2F-binding site. Thus, the movement of this helix upon Ser 567 phosphorylation would perturb the binding site in the A–B interface, and this alteration would release the E2F from the Rb, supporting the proposed experimental model (Ezhevsky et al. 1997; Lundberg and Weinberg 1998; Harbour et al. 1999). The fact that a missense tumorigenic mutation that occurs in Ser 567 disrupts the E2F interaction with Rb protein highlights the significant role of Rb–E2F interactions in Rb-related tumorigenesis (Templeton et al. 1991).
E2F-binding specificity on Rb
Each member of the E2F family is bound and regulated preferentially by specific Rb family members. E2F-1, E2F-2, and E2F-3 interact almost exclusively with Rb; E2F-5 binds to p130; whereas E2F-4 associates with Rb, p130, and p107 (Lee et al. 1993; Hijmans et al. 1995; Ikeda et al. 1996; Moberg et al. 1996). To investigate whether this specificity comes from differences in key contact residues for the interaction between the different Rb family members and the 18 residues within the Rb-binding regions in E2F family members, we have measured the binding constants for Rb–E2F-1 and Rb–E2F-5 peptide using isothermal titration calorimetry (Fig. 5). As shown in Table 2, the Rb pocket binds to E2F-1 with fourfold higher affinity than to E2F-5, indicating that the 18 residues within the E2F transactivation domain do contribute to the specificity in the Rb–E2F interaction. However, the difference in binding affinity is not as significant as expected, suggesting that other factors such as (1) regions other than the 18 residues within the transactivation domain of E2F family members and/or (2) additional proteins are likely to contribute to the binding specificity of the Rb–E2F complex.
Although our structural analysis of the Rb pocket and E2F-2 cannot fully explain the molecular basis for this specificity, our structure does reveal some notable differences in the E2F-binding sites of Rb family members that may help to resolve the fourfold difference in the binding affinity for Rb–E2F-2 and Rb–E2F-5 complexes (Fig. 2). The side chain of Leu 413 in E2F-2 that makes van der Waals contact with the side chain of Ile 785 is replaced by Asn 326 in E2F-5. This Asn side chain would make a favorable intermolecular interaction with the side chain of Thr 986 (Ile 785 in Rb) in p130. Gly 415 in E2F-2 is replaced by Asn 328 in E2F-5, and the side chain of Asn 328 would form an intramolecular H-bond with the side chain of Asn 326 (Fig. 4a). However, these residues in E2F-5 would be surrounded in the cluster of hydrophobic residues in Rb that is an unfavorable environment for E2F-5. Glu 417 of E2F-2 is replaced by Asp 330 in E2F-5, and such a substitution is short for the formation of a salt bridge with the side chain of Lys 652 in Rb. The change of Ile 422 in E2F-2 to Val 335 in E2F-5 would have a reduced packing effect in the tightly packed hydrophobic core in the Rb–E2F interface (Fig. 4b). The hydroxyl group of Ser 423 in E2F-2 is partly exposed and close to the OE2 group of Glu 464 in Rb. Thus, the change to Cys 336 in E2F-5 would be less favorable in same environment. Taken together, the more favorable electrostatic interactions, H-bonds, and van der Waals interactions between Rb and E2F-2 over Rb and E2F-5 would provide the basis for the binding specificity of the Rb–E2F complex. Conversely, we presume that the binding environment in p130 for the E2F transactivation domain is adjusted to form more stable interactions with E2F-5.
Functional implication for Rb–E2F interaction
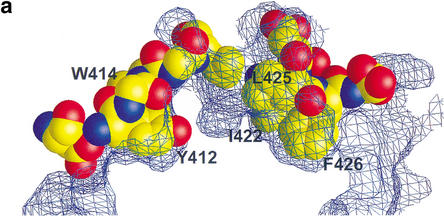
What does the information gained from our structural study tell us about how the Rb–E2F interaction alters E2F-driven transcription? Mutagenesis studies have previously shown that mutation of Tyr 411 or Phe 413 (Tyr 410 or Trp 412 in E2F-2, respectively), which makes contact to Rb in our structure, did not affect the transactivation activity of E2F but disrupted E2F's ability to interact with Rb, enhancing the E2F-1-mediated S-phase entry (Fig. 7a; Shan et al. 1996). However, a double mutant of Tyr411Ala and Phe413Ala had significantly reduced transcription activity, indicating two residues are functionally redundant in transcription (Hagemeier et al. 1993). Our structure also reveals that other conserved hydrophobic residues in the C-segment, Ile 422, Leu 425, and Phe 426, are deeply buried upon Rb binding (Fig. 7a). In previous studies, each of the point mutants on these residues did not affect the transcription activation of E2F (Shan et al. 1996), whereas the deletion mutation studies containing these residues indicate that this region is critical for transactivation (Flemington et al. 1993). Thus, it is likely that these residues may function together in transactivation, and all the residues must be mutated in order to abrogate the transcription activity of E2F as seen in Tyr 411 and Phe 413.
Figure 7.
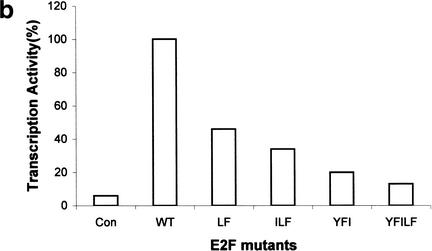
Conserved E2F residues that make contact to Rb are involved in transcription activation. (a) Five conserved residues—Tyr 412, Trp 414, Ile 422, Leu 425, and Phe 426—fit tightly into the A–B interface of the Rb pocket. The Rb surface is represented as a wire mesh, and E2F-2 residues are in space-filling representation. (b) The E2F-dependent transactivation potential for wild-type and mutant E2F-1. Each of wild-type and mutant E2F-1 was transfected into SAOS-2 cells and its ability to activate the expression of luciferase gene was assayed. Double, triple, and multiple E2F mutants were used for the activity measurements (see text).
To investigate whether the key conserved residues in the N- and C-segments in E2F-2 that make contact with Rb are directly involved in the transcription activity, we have created several multiple mutants, cotransfected the plasmids expressing these E2F-1 multiple mutants with DP-1 into an SAOS-1 cell, and measured transcription activity for each mutant (Fig. 7b). A double mutant Leu 425–Phe 426 exhibits twofold decreased transcription activity compared with that of wild type. The triple mutants Ile 421–Leu 424–Phe 425 (Ile 422–Leu 425–Phe 426 for E2F-2) and Tyr 411–Phe 413–Ile 421 show threefold to fivefold decreased activity, respectively. The most dramatic effect is seen in a multiple mutant in which five residues, Tyr 411–Phe 413–Ile 421–Leu 424–Phe 425, that tightly bind to Rb are replaced by Ala; this mutant exhibits 10-fold decreased transcription activity compared with that of wild-type E2F-1. Although weak residual activity is observed in this mutant, our studies suggest that these five residues are most critical for the transactivation of E2F. The fact that triple mutants retain 20%–30% of transcription activity suggests that all five residues have to be concealed to completely inhibit the transcription activity of E2F, and our structure clearly demonstrates that this is what Rb does when it binds to E2F. Thus, our results suggest that the five conserved residues could critically contribute to the interactions with proteins that are required for E2F transcription, and that Rb binding to E2F interferes the physical contact between these residues and cellular proteins such as basal transcription factors and/or coactivators.
The transactivation domain of E2F contains several conserved acidic residues. The residual transcription activity observed in the E2F mutant may contribute from these residues as the residues Glu 416, Glu 417, and Glu 419 in a bending region and Asp 424 and Asp 427 in the C-segment in E2F-1 are involved in transactivation to a certain extent (Hagemeier et al. 1993). However, these acidic residues are only partly exposed to solvent upon Rb binding (Fig. 2), and thus the proteins that are required for E2F transcription, such as components of basal transcription machinery or coactivators, would have limited access to them in the Rb–E2F complex. Taken together, our structure in conjunction with the mutational analysis suggest that the overall steric hindrance imposed by the Rb pocket would prevent the E2F transactivation domain from forming an active complex with the factors that are required for transcription either by directly interfering with their association, or by causing them to assemble in a weakly bound form (Hiebert et al. 1992; Helin et al. 1993a; Ross et al. 1999).
Nonspecific interaction of E2F with LxCxE-motif-binding site
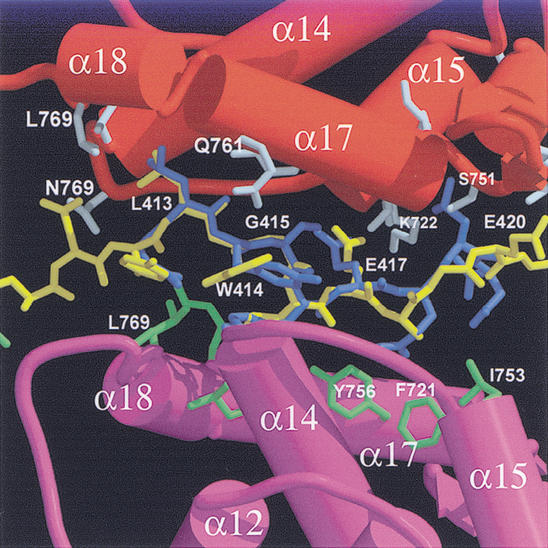
The extra E2F-2 peptide that nonspecifically interacts with the Rb pocket in our crystal binds to the shallow groove formed from helices α14, α15, α16, and α17 in the B-box where the viral peptide LxCxE motif binds (Fig. 8; Lee et al. 1998; Kim et al. 2001). Although this extra E2F-2 peptide forms an extended, β-stranded conformation like that of the E2F-2 that specifically bound to the central groove of Rb, notable conformational differences are observed between these E2F-2 peptides, showing an RMSD of 3.8 Å for 12 Cα atoms.
Figure 8.
Contacts made by the E2F-2 peptide with the conserved B-box in the Rb pocket where the viral protein LxCxE motif binds. The E2F-2 peptide (yellow) is superimposed with a nine-residue HPV E7 peptide containing the LxCxE motif (blue). (Red) Helices in B-box where the conserved Leu, Cys, and Glu in the E7 peptide bind; (magenta) helices from the other B-box in the asymmetric unit.
The E2F-2 peptide contains two LxxxE sequences, LWGLEAGE (the conserved residues are in italic or boldface), and this presumably mimics the binding pattern of the viral LxCxE motif (Fig. 2). The conformation of this region of E2F-2 is similar to that of the LxCxE motif in the E7 peptide (Lee et al. 1998) with an RMSD of 1.2 Å. However, unlike the LxCxE binding to Rb in which alternating x residues are exposed to solvent, the alternating residues in the E2F-2 peptide interact tightly with residues from the equivalent shallow groove formed in the B-box of another Rb pocket, and these interactions contribute to the stable binding of the E2F-2 peptide in this region.
The contacts between Rb and E2F in this region are very similar to those in Rb and the LxCxE motif in HPV E7 or the SV40 large T antigen; in the LWGLE motif, Leu 413 makes van der Waals contacts with side chains of Val 714, Tyr 756, Met 761, Ile 768, and Leu 769 of the B-box. Glu 417 of E2F-2 forms H-bonds to the backbone amide of Phe 721 and Lys 722, respectively. Also most of the H-bonds between the backbone of Rb and the side chain of E2F-2 are conserved. In the LEAGE sequence, Leu 416 and Ala 418 make van der Waals contacts to several residues from the other B-box. Glu 420 in E2F-2 does not make contacts with any surrounding residues. However, unlike the Rb–HPV E7 or SV40 large T antigen interaction in which a conserved bulky hydrophobic residue in the LxCxExΦ (where Φ is a bulky hydrophobic amino acid) motif is involved in binding to Rb (Lee and Cho 2002), no such residue is present in E2F, indicating the significance of the conservation of a large hydrophobic residue as well as Leu, Cys, and Glu in this motif for the tight binding to the groove in the B-box. It is unclear if Rb binds to the E2F in same stoichiometry, in vivo. However, the nonspecific interaction between the E2F-2 peptide and the B-box shown here suggests that the B-box of Rb may associate with the second E2F molecule in weak affinity or in the presence of other proteins, in vivo. Also, it is possible that many proteins that do not contain an LxCxE motif but nevertheless bind to the pocket region may similarly bind to this region of Rb with or without the aid of other cellular proteins. Because Rb has a limited conserved region within the pocket although a large number of cellular proteins bind to the Rb pocket, the nonspecific interaction between Rb and E2F shown here may illustrate the ways to recruit several interacting proteins by the Rb pocket.
Discussion
In this study, we have provided a structural and biochemical basis for the specific recognition of the E2F-2 transactivation domain by Rb, a process that is required for Rb-mediated E2F transcription repression. In the Rb–E2F-2 complex, both the A-box and the B-box of the Rb pocket participate in E2F-2 binding. The conserved groove that runs around the central interface of the Rb pocket shows strong complementarity to the L-shaped transactivation domain of E2F-2, and this allows tight complex formation between the two proteins (Figs. 1b, 3).
Structural analyses of several protein-transactivation domain complexes including MDM2–p53 (Kussie et al. 1996), KIX–pKID (Radhakrishnan et al. 1997), TAF–VP16 (TATA box-binding protein-associated factor; Uesugi et al. 1997), and p300–HIF-1α (Freedman et al. 2002) suggest that the complex formation triggers a folding of the transactivation domain from an unfolded state to an amphipathic helix or helices. It is unclear whether the E2F-2 peptide undergoes a similar structural transition upon binding to Rb. However, the significant conformational difference between the specifically and nonspecifically bound E2F-2 peptides implies structural plasticity of the E2F-2 peptide, which can change its conformation in different environments. Unlike the structures of other transactivation-domain peptides, however, E2F-2 forms an extended β-stranded-like conformation (although a short 310 helix is present), suggesting that more diverse structural motifs can be induced upon complex formation between the protein and the transactivation domain.
In the MDM2–p53 complex, the MDM2 structure has a twofold pseudosymmetry, and the p53 binding is observed at the interface of pseudosymmetry-related domains in MDM2 (Kussie et al. 1996). This feature of intramolecular dimerization and binding of a peptide in the interface cleft is also shared by several other proteins including MHC molecules (Bjorkman et al. 1987) and carmodulin (Meador et al. 1992). Although the A-box and the B-box of the Rb pocket are not related by pseudosymmetry, each box contains an evolutionarily conserved cyclin box that is involved in the binding of the E2F-2 peptide. Thus, the duplication of a cyclin box may be an evolved feature like the intramolecular dimerization observed in several other proteins to accommodate the peptide.
We show that five conserved residues within the E2F transactivation domain are critical for the transcriptional activity of E2F and that they must be simultaneously blocked to inhibit the transactivation activity of E2F. Our structure shows that by forming a tight complex formation with both the N- and C-segments of E2F-2, Rb efficiently masks these five key residues to repress the transcription of E2F.
Our results clearly demonstrate that “direct binding and masking” is an important mechanism in Rb-mediated repression of E2F. However, both the “direct binding and masking” and “active repression” models are well supported by a number of studies, and our data do not reveal the relative importance of the two models. Further studies are required to resolve this matter. Our structure also reveals that the tumorigenic hot spots are observed at or near the E2F binding site, and the missense mutations are likely to affect the local interactions between Rb and E2F. This explains in part the importance of Rb–E2F interactions in tumorigenesis. The structural and biochemical data presented here should serve as a framework for further elucidation of the mechanism of transcriptional repression of E2F by the Rb tumor suppressor, and provide the insight for the design of small molecules or proteins that could regulate E2F transcription activity.
Materials and methods
Protein expression and purification
The recombinant human Rb pocket (residues 380–785) was overexpressed at room temperature in Escherichia coli as a glutathione S-transferase (GST) fusion protein using a pGEX-4T3 vector (Pharmacia). To aid the crystallization, a spacer region between the A-box and B-box (residues 582–642) was deleted. The GST–Rb pocket was purified on glutathione S-sepharose and cleaved from the fusion protein by thrombin. It was purified by cation exchange (Mono-S) and gel filtration (Superdex 200) chromatography. The 18-residue E2F-2 peptide was synthesized chemically with the N and C termini acetylated and amidated, respectively. It was purified by reverse phase chromatography. The sequence of the E2F-2 peptide used in crystallization is given in Figure 2.
Crystallization and structure determination
E2F-2 peptide was mixed with Rb pocket in a 1:1.5 ratio and was concentrated by ultrafiltration to 20 mg/mL in a buffer of 25 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 5 mM dithiothreitol. Crystals were grown at 4°C by hanging-drop vapor diffusion from 1.5 M ammonium sulfate, 100 mM Na-Citrate (pH 5.5). The crystals formed in space group P1 with a = 54.4 Å, b = 65.2 Å, c = 69.3 Å, α = 85.6°, β = 79.5°, and γ = 67.1°, and contained two Rb–E2F complexes and an extra E2F peptide in each asymmetric unit. Diffraction data were collected at −170°C from a crystal flash frozen in crystallization buffer containing 30% glycerol using the B6 beamline in the Pohang Accelerator Laboratory (PAL). Data were processed with the programs DENZO and SCALEPACK (Otwinowski and Minor 1997). The structure was determined by the molecular replacement method. The position of the Rb pocket was determined by molecular replacement with the structure of the Rb–SV40 large T antigen complex (Kim et al. 2001) using the program AMORE (Collaborative Computational Project 4 1994). The correlation coefficient was 0.51 for 10 ∼ 4-Å data. Successive rounds of model building with O (Jones et al. 1991) and simulated annealing refinement with the program CNS (Brunger et al. 1998) clearly showed the density of the E2F-2 peptide and the other region of the Rb pocket (Table 1). The electron density of the portion of the E2F-2 peptide is shown in Figure 1a. The final model consists of residues 380–581 and 643–785 of Rb, residues 55–72 of the E2F-2 peptide, and 269 water molecules. The last six residues at the C-terminal end of an extra E2F-2 peptide are disordered. A total of 85% of the main-chain φ/ψ angles are in the most favored region, and two residues (from Rb pocket) are in the disallowed region of the Ramachandran plot.
Measurement of E2F-2 peptide binding to the Rb pocket
The binding constants of intact and mutant E2F-2 peptides to the Rb pocket (residues 380–785) were measured by isothermal titration calorimetry (ITC) using the Micro Calorimetry System (Microcal Inc.). The Rb pocket was dialyzed against buffer containing 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 5 mM β-mercaptoethanol. E2F-2 peptides were chemically synthesized, purified by reverse phase chromatography, and dissolved in the same buffer. The 18-residue E2F-2 peptide used for crystallization was used in the ITC experiments and also two mutants in which Glu 420 or Gly 421 had been mutated to Ala or Ser, respectively. The ITC measurements were carried out at 18°C, by making 15–25 injections (7 μL each) of the peptide solution into 1.4 mL of Rb protein. The concentrations of the Rb protein and peptides were 0.0625 mM and 0.75 mM, respectively. Curve fitting was done using MicroCal Origin software; the binding curves and parameters are shown in Figure 5.
Surface plasmon resonance spectroscopy
Rb mutants were generated using PCR-based methods and subcloned into the pGEX-4T3 vector. The complete regions encoding Rb mutants were sequenced to ensure no extra mutations had been introduced during PCR. E2F-1 (residues 88–428) was inserted into PET15b and expressed in E. coli BL21. E2F-1 protein was purified to >95% purity by use of the protocol described previously (Kim et al. 2001). All Rb proteins were purified in the GST-fusion form by GST column, ion exchange chromatography and gel filtration chromatography. The CM5 sensor chip (BIAcore) was used to measure the binding of the E2F-1 proteins to Rb. Wild-type and mutant Rb pocket proteins were injected onto the chip at 25, 50, 100, 200, and 400 nM concentrations in 100 mM NaCl for 2 min followed by a 4-min dissociation phase. The binding data were analyzed using the BIAevaluation software (Table 3; Fig. 6).
Transient transfections
Multiple site-specific mutations (to alanine) were carried out on Tyr 411, Phe 413, Ile 421, Leu 424, and Phe 425 of E2F-1 using PCR-based methods. The PCR products were then subcloned into a pcDNA3 vector. Rb-deficient SAOS2 cells were prepared with Dulbecco's modified Eagle's minimal essential medium supplemented with 10% fetal calf serum for the transfection assay. Approximately 1 × 104 cells were transfected with 2 μg of E2F response luciferase reporter construct, 1 μg of effector DNA, and 1 μg DP-1 DNA by calcium phosphate coprecipitation. Cells were harvested 16 h posttransfection and lysates were used for the luciferase assay.
Acknowledgments
We thank J. Bradbury for help with preparation of the manuscript, and the staff of the Pohang Accelerator Light source for help with data collection. This work was supported by the funds from the National Creative Research Initiatives (Ministry of Science and Technology), Frontier 21 program, BK21 program, and the Korean Academy of Science and Technology. Coordinates have been deposited in the RCSB Protein Data Bank under PDB accession no. 1N4M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
EMAIL yunje@postech.ac.kr; FAX (8254) 279-8111.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1046102.
References
- Bagchi S, Weinmann R, Raychaudhuri P. The retinoblastoma protein copurifies with E2F-1, an E1A-regulated inhibitor of the transcription factor E2F. Cell. 1991;65:1063–1072. doi: 10.1016/0092-8674(91)90558-g. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Blake MC, Azizkhan JC. Transcription factor E2F is required for efficient expression of the hamster dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989;9:4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstieve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography & NMR system: A new software suit for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chellappan SF, Kraus VB, Kroger B, Munger K, Howley PM, Phelps WC, Nevins JR. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cress WD, Johnson DG, Nevins JR. A genetic analysis of the E2F1 gene distinguishes regulation by Rb, p107, and adenovirus E4. Mol Cell Biol. 1993;13:6314–6325. doi: 10.1128/mcb.13.10.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S. Cell cycle regulation of the human cdc2 gene. EMBO J. 1992;11:1797–1804. doi: 10.1002/j.1460-2075.1992.tb05231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang CM, Livingston DM. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell. 1989;58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou QP, Markell PJ, Pardee AB. Thymidine kinase transcription is regulated at G1/S phase by a complex that contains retinoblastoma-like protein and a cdc2 kinase. Proc Natl Acad Sci. 1992;89:3256–3260. doi: 10.1073/pnas.89.8.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht BD. Regulation of transcription by proteins that control the cell cycle. Nature. 1997;389:149–152. doi: 10.1038/38225. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Flores O, Lees JA, Harlow E. Differential regulation of E2F trans-activation by cyclin–cdk2 complexes. Genes & Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Ezhevsky SA, Nagahara H, Vocero-Akbani AM, Gius DR, Wei MC, Dowdy SF. Hypo-phosphorylation of the retinoblastoma protein (pRb) by cyclin D:CDK4/6 complexes results in active pRb. Proc Natl Acad Sci. 1997;20:699–704. doi: 10.1073/pnas.94.20.10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, Orkin SH, Greenberg ME. E2F-1 functions in mice to prompt apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- Flemington EK, Speck SH, Kaelin WG. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 α. Proc Natl Acad Sci. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CJ, Pearson A, Malinowski E, Bartley SM, Greenblatt J, Farnham PJ. Activation of the murine dihydrofolate reductase promoter by E2F1. A requirement for CBP recruitment. J Biol Chem. 1999;274:15883–15991. doi: 10.1074/jbc.274.22.15883. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Terui Y, Sakoe K, Ohta M, Saito M. The role of cellular transcription factor E2F in the regulation of cdc2 mRNA expression and cell cycle control of human hematopoietic cells. J Biol Chem. 1994;269:26249–26258. [PubMed] [Google Scholar]
- Guo Z, Yikang S, Yoshida H, Mak TW, Zacksenhaus E. Inactivation of the retinoblastoma tumor suppressor induces apoptosis protease-activating factor-1 dependent and independent apoptotic pathways during embryogenesis. Cancer Res. 2001;61:8395–8400. [PubMed] [Google Scholar]
- Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes & Dev. 2000a;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- ————— Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000b;2:65–67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cell move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- Helin K, Lees JA, Vidal M, Dyson N, Harlow E, Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:33–50. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993a;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Wu CL, Fattaey AR, Lees JA, Dynlacht BD, Ngwu C, Harlow E. Heterodimerization of the transcription factor E2F-1 and DP-1 leads to cooperative trans-activation. Genes & Dev. 1993b;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- Hiebert SW, Lipp M, Nevins JR. E1A-dependent trans-activation of the human MYC prompter is mediated by the E2F factor. Proc Natl Acad Sci. 1989;86:3594–3598. doi: 10.1073/pnas.86.10.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert SW, Chellappan SP, Horowitz JM, Nevins JR. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes & Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- Hijmans EM, Voorhoeve PM, Beijersbergen RL, van'tVeer LJ, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu QJ, Dyson N, Harlow E. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J. 1990;9:1147–1155. doi: 10.1002/j.1460-2075.1990.tb08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wang NP, Tseng BY, Lee WH, Lee EH. Two distinct and frequently mutated regions of retinoblastoma protein are required for binding to SV40 T antigen. EMBO J. 1990;9:1815–1822. doi: 10.1002/j.1460-2075.1990.tb08306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda MA, Jakoi L, Nevins JR. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, et al. Role for p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ewen ME, Livingston DM. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol Cell Biol. 1990;10:3761–3769. doi: 10.1128/mcb.10.7.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Krek W, Sellers WR, DeCaprio JA, Ajchenbaum F, Fuchs CS, Chittenden T, Li Y, Farnham PJ, Blanar MA, et al. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- Kim H-Y, Ahn BY, Cho Y. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 2001;20:295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzke RA, Otterson GA, Hogg A, Coxon AB, Geradts J, Cowell JK, Kaye FJ. Partial inactivation of the RB product in a family with incomplete penetrance of familial retinoblastoma and benign retinal tumors. Oncogene. 1994;9:1321–1326. [PubMed] [Google Scholar]
- Krek W, Livingston DM, Shirodkar S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science. 1993;262:1557–1560. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Lee C, Cho Y. Interactions of SV40 large T antigen and other viral proteins with retinoblastoma tumour suppressor. Rev Med Virol. 2002;12:81–92. doi: 10.1002/rmv.340. [DOI] [PubMed] [Google Scholar]
- Lee JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-O, Russo AA, Pavletich NP. Structure of the retinoblastoma tumor-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- Ludlow JW, Shon J, Pipas JM, Livingston DM, DeCaprio JA. The retinoblastoma susceptibility gene product undergoes cell cycle-dependent dephosphorylation and binding to and release from SV40 large T. Cell. 1990;60:387–396. doi: 10.1016/0092-8674(90)90590-b. [DOI] [PubMed] [Google Scholar]
- Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin–cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 Å structure of a calmodulin–peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- Moberg K, Starz MA, Lees JA. E2F4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudryj M, Hiebert SW, Nevins JR. A role for the adenovirus inducible E2F transcription factor in a proliferation dependent signal transduction pathway. EMBO J. 1990;9:2179–2184. doi: 10.1002/j.1460-2075.1990.tb07387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Gen. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Onadim Z, Hogg A, Baird PN, Cowell JK. Oncogenic point mutations in exon 20 of the RB1 gene in families showing incomplete penetrance and mild expression of the retinoblastoma phenotype. Proc Natl Acad Sci. 1992;89:6177–6181. doi: 10.1073/pnas.89.13.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pearson A, Greenblatt J. Modular organization of the E2F activation domain and its interaction with general transcription factors TBP and TFIIH. Oncogene. 1997;15:2643–2658. doi: 10.1038/sj.onc.1201451. [DOI] [PubMed] [Google Scholar]
- Pearson BE, Nasheuer HP, Wang TS. Human DNA polymerase α gene: Sequences controlling expression in cycling and serum-stimulated cells. Mol Cell Biol. 1991;11:2081–2095. doi: 10.1128/mcb.11.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AM, Gimenz-Conti IB, Schneider-Broussard R, Martinez LA, Conti CJ, Johnson DG. Increased E2F1 activity induces skin tumors in mice heterozygous and nullizygous for p53. Proc Natl Acad Sci. 1998;95:8858–8863. doi: 10.1073/pnas.95.15.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Ross JF, Liu X, Dynlacht BD. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol Cell. 1999;3:195–205. doi: 10.1016/s1097-2765(00)80310-x. [DOI] [PubMed] [Google Scholar]
- Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Durfee T, Lee WH. Disruption of RB/E2F-1 interaction by single point mutations in E2F-1 enhances S-phase entry and apoptosis. Proc Natl Acad Sci. 1996;93:679–684. doi: 10.1073/pnas.93.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton DJ, Park SH, Lanier L, Weinberg RA. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc Natl Acad Sci. 1991;88:3033–3037. doi: 10.1073/pnas.88.8.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmeier K, Synovzik H, Mertz R, Winnacker EL, Lipp M. Nuclear factor E2F mediates basic transcription and trans-activation by E1a of the human MYC promoter. Genes & Dev. 1989;3:527–536. doi: 10.1101/gad.3.4.527. [DOI] [PubMed] [Google Scholar]
- Trouche D, Kouzarides T. E2F1 and E1A 12S have a homologous activation domain regulated by RB and CBP. Proc Natl Acad Sci. 1996;93:1439–1442. doi: 10.1073/pnas.93.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D, Cook A, Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced α helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Weintraub SJ, Chow KN, Luo RX, Zhang SH, He S, Dean DC. Mechanism of active transcriptional repression by the retinoblastoma proteins. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1+/− mice. Nat Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Postigo AA, Dean DC. Active transcriptional repression by the Rb–E2F complex mediates G1 arrest triggered by p16INK4a, TGFβ, and contact inhibition. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC–Rb–hSWI/SNF and Rb–hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]