Abstract
Our goal is to construct an improved Escherichia coli to serve both as a better model organism and as a more useful technological tool for genome science. We developed techniques for precise genomic surgery and applied them to deleting the largest K-islands of E. coli, identified by comparative genomics as recent horizontal acquisitions to the genome. They are loaded with cryptic prophages, transposons, damaged genes, and genes of unknown function. Our method leaves no scars or markers behind and can be applied sequentially. Twelve K-islands were successfully deleted, resulting in an 8.1% reduced genome size, a 9.3% reduction of gene count, and elimination of 24 of the 44 transposable elements of E. coli. These are particularly detrimental because they can mutagenize the genome or transpose into clones being propagated for sequencing, as happened in 18 places of the draft human genome sequence. We found no change in the growth rate on minimal medium, confirming the nonessential nature of these islands. This demonstration of feasibility opens the way for constructing a maximally reduced strain, which will provide a clean background for functional genomics studies, a more efficient background for use in biotechnology applications, and a unique tool for studies of genome stability and evolution.
[Sequence data described in this paper have been submitted to the DNA Data Bank of Japan, European Molecular Biology Laboratory, and GenBank databases under accession nos. AF402780, AF402779, and AF406953, respectively.]
Escherichia coli is both a tool and object of study of genome science. As the primary model organism for bacteria, it was used to define the genetic code and elucidate mechanisms of central importance such as transcription, translation, restriction, replication, and much of basic metabolism. Its annotations are regularly transferred to other organisms as their genome sequences are completed, so the accuracy of E. coli functional genomics is especially important for the overall genomics enterprise. It is also used extensively as a genomics tool to propagate DNA subclones for sequencing and for a variety of functional studies in many species. Industrially, E. coli is used to produce hormones, enzymes, and antibiotics.
E. coli is a facultative anaerobe and a metabolic opportunist, spending part of its natural life cycle living anaerobically in the intestinal tracts of animals, part living aerobically in diverse natural environments such as rivers or soil, and, in the case of pathogens, part invading animal or human hosts (Schaechter and Neidhardt 1987). Individual strains of E. coli vary in their preferences for niches/hosts, and these variations are reflected in their gene contents. It has been estimated that the genomes of natural isolates of E. coli range from 4.5 to 5.5 megabases (Bergthorsson and Ochman 1998).
Two strains of E. coli have been fully sequenced: (1) the lab K-12 strain MG1655 with 4,639,221 bp (Blattner et al. 1997) and (2) two isolates of the enteric pathogen O157:H7 of 5,528,445 bp (Hayashi et al. 2001; Perna et al. 2001). K-12 and O157:H7 are phylogenetically distant relatives within the E. coli species (Reid et al. 2000; Perna et al. 2001).
Comparison between them revealed a startling pattern in which hundreds of strain-specific “islands” are found inserted into a common “backbone” that is highly conserved (98% sequence identity) between the two strains. A third strain, the uroseptic E. coli strain CFT073, nearing completion in our laboratory, confirms and extends this pattern. It contains virtually the same backbone peppered with an almost completely different set of islands (data not shown). The strain-specific islands are termed K-islands, O-islands, and C-islands after the strain from which they were sequenced. The size of the backbone genome is ∼3.7 Mbp, and the total of K-islands amounts to 0.9 Mbp. Thus, removal of all the K-islands from MG1655 would result in a 20% reduction of the length of the genome.
Gene loss and horizontal gene transfer were the major genetic processes that shaped the ancestral E. coli genome, resulting in the spectrum of divergent present-day strains that possess very different arrays of genes (McClelland et al. 2000; Ochman and Jones 2000; Riley and Serres 2000; Perna et al. 2001). The genes contained in the backbone regions generally include basic core functions of E. coli that are necessary regardless of environmental niche. Islands contain a disproportionate share of genes that are of unknown function, as well as toxins, virulence factors, and metabolic capabilities that may be of advantage in the niche to which the strain is adapted. Islands also contain many transposable elements, phages, cryptic prophages, pseudogenes, gene remnants, and damaged operons.
Clearly, E. coli possesses many dispensable functions that would not be needed in a strain designed for laboratory purposes. Comparative genomics provide the working hypothesis that genes and gene islands that are present in MG1655 and not in other strains may be deleted. A second approach is to eliminate genes that are not expressed in any of a wide variety of conditions. Based on microarray data, ∼25% of E. coli genes are clearly expressed in log phase growth in minimal medium, and 50% appear not to be; the remainder is probably expressed at a low level. (Richmond et al. 1999; data not shown).
A minimal strain consisting just of the backbone would be interesting to study. A backbone-only strain would have ∼3700 genes, many more than the minimal gene set defined by Koonin (2000) to be “sufficient to sustain cellular life under the most favorable conditions” and which may be as small as 150 genes. Instead, a backbone-only strain would constitute a robust set that has stood the test of time in the evolution of the Enterobacteriaceae and would include considerable redundant functions.
Many questions of bacterial physiology, genetics, and ecology could be addressed experimentally by using a simplified “clean” E. coli strain. Would such a strain be healthier than its parent MG1655? Would it have a more stable genome? What horizontal transmissions would it pick up from the environment? Does “selfish DNA” extract a cost?
Potential advantages of a deleted host in biotechnology are also readily apparent. Any unnecessary gene product that is expressed in a production host represents a potential contaminant that could drive up the cost of product purification. There is also metabolic waste entailed in producing unwanted products. Some sideproducts are detrimental, even in tiny quantities, when drugs or vaccines must pass certification. Deletion of the gene is by far the most reliable and effective way to ensure the complete absense of an unwanted component in a biotechnological product.
The genetic instability of E. coli owing to transposable elements is certainly a problem that can be addressed by genomic deletion. E coli MG1655 includes 44 transposable elements of 10 kinds and 6 copies of Rhs, a repeated sequence that resembles a transposon but for which there are no data indicating that it can move. The genome also contains 8 prophage or phage remnants. Transposases associated with the transposable elements are induced by stress, such as heat and cold shock, and might be activated by procedures such as electroporation. Genome rearrangements and mutations associated with the activity of transposons are very common in E. coli, including the preponderance of spontaneous null mutations (Kitamura et al. 1995) and inversions.
Many examples show that transposon hopping can happen inadvertently during laboratory handling. A closely related lab strain, W3110, sequenced in Japan, is almost identical except for a large inversion and differences in the distributions of transposable elements, indicating evolution that must have happened on the time scale of <50 years. Even more striking, we have found ∼18 instances of transposable elements in the draft human genome sequence (mostly IS186, IS2, and IS5) and many more in the GenBank database. These presumably happened when a mobile element hopped from the E. coli production host onto a bacterial artificial chromosome clone before the DNA was isolated for sequencing on a time scale of a year. Clearly a stable bacterium would be desirable for a production process and for many scientific experiments.
Host Modifications
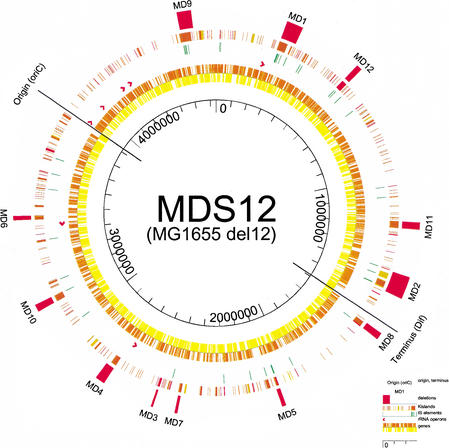
Figure 1 shows MG1655 with the open reading frames (ORFs) in the inner ring (rightward transcribed genes are shown in yellow, leftward in orange). Ribosomal RNA operons are shown as red arrows. The next ring shows portions of the K12 genome that are absent in O157H7, either cleanly deleted (red) or substituted with completely unrelated sequences (brown) or with highly variable (yellow) sequences. The outer ring shows the positions of the deleted regions.
Figure 1.
Representation of deletions MD1 through MD12 on the circular map of the MG1655 genome. Concentric rings denote the position of (from outside to inside) (1) deletions, (2) K-islands, (3) insertion sequence elements, (4) rRNA operons, and (5, 6) genes, by orientation. The replication origin and terminus are indicated.
RESULTS
To construct targeted deletions, a rapid and straightforward method was developed. A polymerase chain reaction (PCR)–generated DNA fragment was inserted into the genome by λ red-type homologous recombination, followed by a double-strand break (DSB)–stimulated recombinational repair process, resulting in a scarless deletion.
The procedure is based on a modified version of the λ red-type or RecET-mediated recombination methods (Zhang et al. 1998; Datsenko and Wanner 2000; Yu et al. 2000) and on the DNA repair activities of the cell stimulated by a DSB in the chromosome (Pósfai et al. 1999). The method is rapid and efficient and produces scarless deletions.
Outline of the Deletion Method
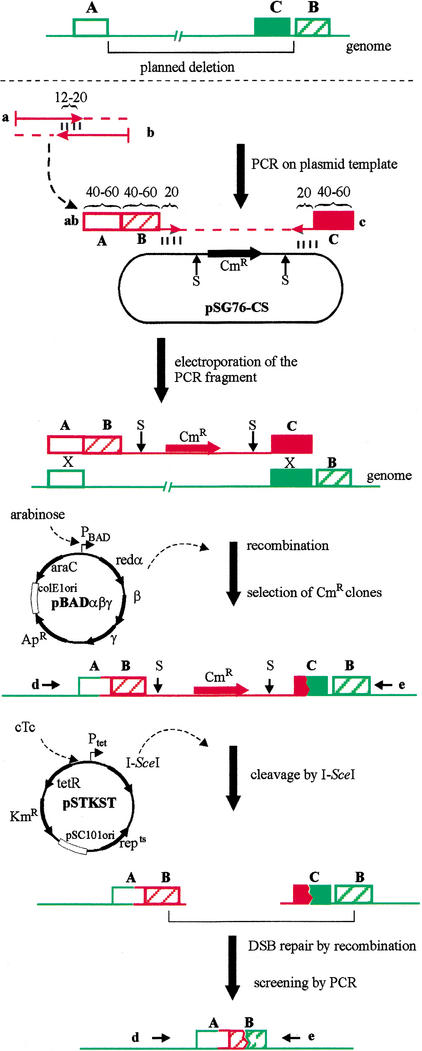
Steps of the method are depicted in Figure 2 and detailed in the Methods section. To delete the chromosomal region between arbitrarily chosen segments (indicated as box A and B), a linear DNA molecule is generated by PCR on the template plasmid pSG76-CS. The resulting fragment, carrying a selectable marker (chloramphenicol resistance [CmR]) flanked by two I-SceI meganuclease sites, is electroporated into the target cell, where it can replace a segment of the chromosome. This replacement requires a double crossover and involves short (40 to 60 bp) terminal segments (homology boxes A and C) of the DNA fragment. The helper plasmid pBADαβγ (Muyrers et al. 1999) provides the arabinose-inducible recombinase functions (Redαβγ) necessary for the integration event. Cells that integrated the linear fragment into their chromosome are then selected by their CmR (hereafter referred to as intermediate clones). Correct-site insertions are confirmed by PCR screening using primers d and e. The procedure is similar to published protocols (Zhang et al. 1998; Datsenko and Wanner 2000; Yu et al. 2000). However, in contrast to the original λ red recombination method, the integrated fragment carries a third homology region (box B) fused to box A as part of the PCR primer. Because synthesis of very long primers is difficult, the fusion is produced in a PCR-like filling-in reaction of two partially complementary oligonucleotides (primers a and b), resulting in the composite primer ab, which in turn is used to initiate the generation of the linear DNA fragment. Note that the bulk of the deletion is generated by the integration of the fragment. Moreover, the sequence corresponding to box B is duplicated in the chromosome at this stage.
Figure 2.
Overview of the deletion procedure. A, B, and C represent arbitrarily chosen DNA segments (homology boxes). Numbers refer to the length of oligonucleotides in basepairs. Polymerase chain reaction primers are labeled by lower case letters in bold (a, b, c, d, e, ab). S indicates an I-SceI cleavage site. Step-by-step description of the procedure can be found in the article.
Next, I-SceI meganuclease expression is induced by derepressing the tet promoter on the helper plasmid pSTKST. (Alternatively, pKSUC1, a plasmid constitutively expressing I-SceI, can be transformed into the cell.) As a result, the chromosome is cleaved at the 18-bp I-SceI sites (Monteilhet et al. 1990) present on the integrated fragment. The broken chromosomal ends are then subjected to RecA-mediated DSB repair by intramolecular recombination. Because the broken ends carry short homologous regions (box B) close to their termini, recombinational repair is likely to proceed via these homologous segments. Surviving colonies are screened by PCR using primers d and e. Correct-size PCR fragments confirm the generation of the desired scarless deletion.
Features of the Deletion Method
Over a dozen large deletions have been constructed in our laboratory using the new method (for the subset presented in this study, see Table 1). Total length of the targeting linear DNA fragment was 1.7 kb. Sizes of the homology boxes A, B, and C were in the range of 40 to 80, 40 to 60, and 40 to 45 bp, respectively. Typically a 200- to 500-ng DNA fragment was electroporated into MG1655, and 10 to 200 recombinant (CmR) colonies were obtained, 5% to 90% of which contained an insertion in the desired locus. In our hands, this relatively low number of correct insertions was the limiting factor in the procedure. There were anecdotal reports that internal regions of the fragment homologous to chromosomal sequences cause a drop in the integration efficiency. The template plasmid pSG76-CS carries a 78-bp piece of IS1 that is copied into the targeting DNA fragment. Because IS1 is present in seven copies in the MG1655 genome, it seemed likely to interfere with the integration process. However, when the 78-bp internal homology was deleted from the plasmid and the resulting pSG76-CSH was used as a template to generate the targeting fragment, no increase was observed in the number of integrants.
Table 1.
Genomic Deletions in MDS12
| Deletion | Endpointsa | Size (bp) | Descriptionb |
|---|---|---|---|
| MD1 | 263080, 324632 | 61553 | b0246–b0310; includes K-islands Nos. 16–18, CP4-6, eaeH |
| MD2 | 1398351, 1480278 | 81928 | b1336–b1411; includes K-island No. 83, Rac |
| MD3 | 2556711, 2563500 | 6790 | b2441–b2450; includes K-island No. 128, CP-Eut |
| MD4 | 2754180, 2789270 | 35091 | b2622–b2660; includes K-island No. 137, CP4-57, ileY |
| MD5 | 2064327, 2078613 | 14287 | b1994–b2008; includes K-islands Nos. 94–96, CP4-44 |
| MD6 | 3451565, 3467490 | 15926 | b3323–b3338; includes K-islands Nos. 164 and 165 |
| MD7 | 2464565, 2474198 | 9634 | b2349–b2363; includes K-island No. 121 |
| MD8 | 1625542, 1650785 | 25244 | b1539–b1579; includes K-island No. 77, Qln |
| MD9 | 4494243, 4547279 | 53037 | b4271–b4320; includes K-island No. 225, tec operon, fim operon |
| MD10 | 3108697, 3134392 | 25696 | b2968–b2987; includes K-island No. 153, gic operon |
| MD11 | 1196360, 1222299 | 25940 | b1137–b1172; includes K-island No. 71, e14 |
| MD12 | 564278, 585331 | 21054 | b0538–b0565; includes K-island No. 37, DLP12 |
Endpoints indicate the first and last nucleotides in the genome of MG1655 that were removed in each deletion.
Descriptions of the deletions include the span of genes deleted (indicated by b-number; Blattner et al. 1997), the MG1655-specific regions (K-islands; Perna et al. 2001), and specific elements such as genes, operons, and cryptic prophages.
Sizes of the deletions were in the range of 7 to 82 kb. No significant correlation was found between the efficiency of integration and the size of the deletion it caused. In the case of deletion MD2, two different targeting fragments were constructed. The DNA fragment replacing a 2-kb segment of the chromosome produced five times more integrants (53 colonies) than the fragment replacing an 82-kb segment (10 colonies). However, we note that the results are not fully comparable, because the box C segments were from different genomic regions in the two constructs.
The second step of the procedure, DSB-stimulated recombinational repair, proceeded efficiently. After induction of I-SceI expression, 10% to 100% of the surviving cells carried the desired deletion. However, our initial experiments showed that positioning the broken chromosomal ends relatively close to box B segments is essential, otherwise recombinational gap repair can proceed via other randomly occurring short homologies. Therefore, it is advisable to select a box C sequence close to box B on the chromosome. For similar reasons, in an experiment involving deletion MD2, placing two I-SceI sites on the targeting fragment (as shown in Fig. 2) resulted in an increased level (15%) of recombinational repair proceeding via box B, compared with having a single I-SceI site on the fragment (5%).
Deletions were created in the recombination-proficient strain MG1655. A number of recombinational activities are thought to be involved in DSB repair (for review, see Kowalczykowski 2000), with RecA playing a central role. Because recombination of short (40 to 60 bp) repeats, atypical substrates for the recombinase, is involved in the deletion process, we tested the role of RecA. It was found that deletion of the recA gene practically abolished recombinational repair when pUC19RP12 (Pósfai et al. 1999), a plasmid constitutively expressing I-SceI, was transformed into MG1655 cells carrying the insertion corresponding to the intermediate construct for deletion MD12 (data not shown).
Deletion Target Selection
Chromosomal regions marked for deletion were chosen primarily on the basis of comparing the genomes of MG1655 and EDL933. We hypothesized that the regions present only in one of the two strains are unlikely to encode essential functions under common culturing conditions. To maximize the amount of DNA removed, 12 large genomic regions, including the 11 largest and six smaller K-islands, were selected for deletion (Table 1; Fig. 1). Candidate regions were checked for the presence of potentially essential genes by comparing the inferred amino acid sequences of all included ORFs to 24 bacterial proteomes in reciprocal BLAST searches. Except for phage integrase genes, none of the predicted proteins matched homologs in more than nine of the 24 genomes, indicating the absence of widely conserved orthologs. In four cases (MD1, MD2, MD4, and MD9), relatively large regions (6 to 15 kb) adjacent to the K-islands and encoding several genes of unknown functions were included in the target. Generally, deletion endpoints were placed in noncoding regions, next to the nearest ORFs to be deleted. Because of primer design considerations, in a few cases the deletion endpoint was shifted into coding sequences, leaving the 3′-end region of the deleted ORF in the genome. No new fusion protein was created by the deletions. Eight of the 12 deleted regions carried cryptic prophages or phage remnants. All together, the segments marked for deletion possessed a higher than average ratio (50% versus 38%) of ORFs classified as unknown (Blattner et al. 1997).
Deletions
A total of 12 regions (MD1 to MD12) were sequentially deleted from the MG1655 genome. The strain carrying all 12 deletions was designated MDS12. Coordinates of the deletions are shown in Table 1. A total of 376,180 nucleotides were deleted, resulting in an 8.1% reduction in the size of the genome. All together, 409 protein-coding ORFs and 2 stable RNA genes were removed, 9.3% of the total gene count.
Deletion MD1 was created using a suicide plasmid–based, FLP recombinase-assisted method, described earlier (Posfai et al. 1997). As a consequence, a 114-bp vector fragment, including a frt site, remained in the chromosome. Because the frt sequence could serve as a convenient entry site for various insertions, no attempt was made to reengineer this deletion to remove the extra nucleotides.
Deletions MD2 through MD12 were constructed by the scarless method. However, as the work progressed, several variations of the procedure were tested. MD2 through MD6 were made in the same strain sequentially, and the incompatible helper plasmids pBADαβγ and pKSUC1, possessing different antibiotic resistance markers, were introduced into the strain by repeated alternating transformations to replace each other in a cyclic fashion. To construct deletions MD7 through MD12, I-SceI was expressed from pSTKST. Some insertion intermediates were first constructed in wild-type MG1655 carrying pBADαβγ. The inserted regions were then transduced by P1 phage (Miller 1992) to the strain carrying all previous deletions, and the second step of the procedure, DSB-stimulated recombinational repair, was performed in this final host.
All 12 new chromosomal joints, formed by the deletions, were sequenced across. Test primer pairs (d, e) were used to generate a PCR fragment spanning the deletion, and the fragment was sequenced from the ends. A minimum of 60-bp sequences were determined both upstream and downstream of the joint. In most cases, results matched the predicted sequence. In three cases, however, small unintended alterations (4-, 3-, and 2-bp deletions at MD11, MD2, and MD10, respectively) occurred at the 3′-ends of primers a or b, and were likely caused by misannealing of the primer ends in the filling-in reaction.
Detailed data, including nucleotide sequences of the primers, additional methodological descriptions, and actual sequences of the joints can be found on our Web page (http://www.szbk.u-szeged.hu/~posfaigy/).
Characterization of MDS12
Growth rates of MDS12 and the parental strain MG1655 were compared in LB and in M9/glucose minimal medium with and without casamino acids (Fig. 3). Cultures were grown in duplicate, and experiments were repeated twice. The doubling times of the two strains proved to be nearly identical at exponential growth phase (37 min in LB, 42 min in minimal medium supplemented with casamino acids, and 56 min in minimal medium). A small but detectable difference in culture densities was observed in stationary phase: In all three media, MDS12 reached an optical density ∼10% higher than that of MG1655.
Figure 3.
Growth curves of MG1655 and MDS12. Cultures were grown in duplicate in either LB, M9/glucose minimal medium supplemented with 0.5% casamino acids, or M9/glucose minimal medium. One hundred milliliters of liquid medium was inoculated with 1 mL overnight starter culture and grown at 37°C with moderate aeration. The optical density (OD) of the culture was measured at 540 nm.
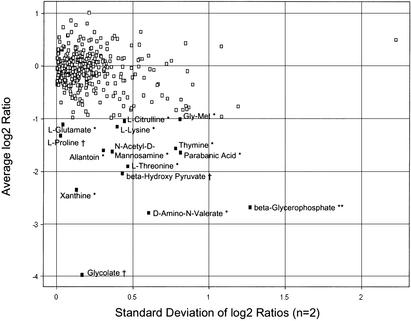
The two strains were analyzed for differences in growth phenotype using Biolog Phenotype Microarrays (Bochner et al. 2001). Cells from both strains were inoculated in parallel into 379 different growth media arrayed in 96-well plates, including 190 different carbon sources, 95 nitrogen sources, 59 phosphorus sources, and 35 sulfur sources. Growth was indicated by production of color from a tetrazolium dye and was measured in a microplate reader. MG1655 showed detectable growth in 202 of the 379 different media. Of these, 15 showed endpoint growth reduced twofold or more for MDS12 (Fig. 4). Three such differences were observed in the use of carbon sources, one difference in phosphate sources, and 11 differences in nitrogen sources.
Figure 4.
Biolog Phenotype Microarray analysis. MG1655 and MDS12 were inoculated in parallel into Biolog microtiter plates containing 379 different growth media, and endpoint growth was measured by absorbance. The value for MDS12 was divided by that of MG1655, and the log2 value was calculated. The average of two replicates and the standard deviation are shown. Media on which the mutant grew more than twofold less than weight are indicated, with symbols denoting tests for sources of nitrogen (*), phosphorous (**), and carbon (†).
Transformability of MDS12 and MG1655 was measured by electroporating a small supercoiled plasmid, pTZ18U (Bio-Rad), into the cells. Electroporation efficiencies for the two strains were identical (4 × 108/μg plasmid DNA). Similarly, chemical transformation (Sambrook et al. 1989) efficiencies of the two strains were identical (5 × 105/μg plasmid DNA).
DISCUSSION
Up to now, there has not been a concerted study to improve the genome of E. coli by large-scale reduction. Early work by Squires and co-workers (Asai et al. 1999) used genome deletion of E. coli to reduce the number of ribosomal RNA genes for study of their individual functions, if any. In Bacillus subtilis, Itaya and Tanaka (1997) subdivided the genome into two chromosomes.
A number of recently developed methods for genomic engineering use bacteriophage recombinases and are based on homologous recombination of a PCR-generated linear DNA fragment carrying a selectable marker gene into the genome (Zhang et al. 1998; Datsenko and Wanner 2000; Murphy et al. 2000; Yu et al. 2000). In applications aimed at deletion construction, the inserted fragment replaces the chromosomal region marked for deletion. The method is fast; however, removal of the marker gene requires a second step. One option is to use a frt- or loxP-flanked selection marker gene that can subsequently be removed by site-specific recombinases. The disadvantage is that there still remain some foreign sequences (including frt or loxP) in the genome, and they may interfere with gene expression or with subsequent rounds of deletions. Another solution is to replace the inserted sequences by a PCR-generated marker-less fragment in a second round of recombination. This procedure, however, requires that the original insertion carries a counter-selectable marker that is usually strain-specific and culturing condition–specific.
The method presented here combines the speed of λ red-type recombination procedures with precise removal of all foreign sequences. The bulk of the chromosomal sequences to be deleted is replaced by inserting a PCR-generated fragment into the genome by λ red-type recombination. The novelty of this step is that the inserted fragment carries the actual joint of the final deletion in its synthetic primer part, and consequently, 40- to 60-bp direct repeats, separated by ∼1.7 kb, are created on the chromosome. Intramolecular recombination between the duplicated sequences then results in the loss of the selectable marker gene and all foreign sequences, producing a clean scarless deletion. However, spontaneous recombination of the short duplicated sequences is a rare event. To stimulate recombination, in the second step of the procedure I-SceI meganuclease is expressed, causing DSBs at the two I-SceI sites uniquely found on the inserted fragment. In surviving cells, DSBs are repaired primarily by the desired intramolecular homologous recombination of the duplicated sequences positioned close to the broken ends. Meganuclease cleavage thus has a dual role: (1) It stimulates the intended recombination by producing free DNA ends, and (2) it provides selection pressure for the loss of the inserted sequences responsible for deleterious chromosomal breaks. We note that in some cases, intramolecular recombinational repair of the DSBs can proceed via other randomly occurring short homologies flanking the site of damage. In fact, in some cases the majority of the surviving cells displayed such undesired deletions. A PCR-based screening of the candidate colonies using test primers flanking the site of deletion is thus necessary.
All attempted deletions were successful; however, insertion efficiency varied widely and was generally lower than that reported by others using λ red-type recombination. A possible explanation for the generally low number of integrants is the relatively low electroporation efficiency achieved in MG1655. Site-to-site variations in integration efficiencies are likely to be caused by the particular nucleotide sequence context of the insertion sites. We note that several of the difficult deletions were positioned next to tRNA genes with potential secondary structure.
This method is suitable to construct single or multiple deletions in a wide range of sizes. The largest deletion we constructed covered 82 kb, and the smallest possible deletion is ∼40 bp, as topological limits are imposed by the duplicated sequences in the latter case.
For the insertion step, pBADαβγ helper plasmid was used in all cases. Phage recombinases responsible for homologous recombination are expressed from the plasmid on transient induction by arabinose. For expression of I-SceI, two alternatives, based on two different plasmids, were presented. The high-copy-number pKSUC1 expresses I-SceI constitutively and can be used at 37°C, but curing the cell of it at the end of the process is difficult. On the other hand, pSTKST is easily curable at 37°C to 43°C, but the procedure is more time consuming, because cells must be cultured at 30°C, and an additional step, derepression of the tet promoter is needed to express I-SceI. Introducing a DSB into the genome induces the SOS response (Walker 1996). This might lead to error-prone DNA synthesis, causing incorporation of mutant nucleotides into the chromosome at damaged sites. However, we did not detect (Pósfai et al. 1999; T. Fehér unpubl.) a significant change in the mutation rate when the method was applied. This might be because a single DNA lesion is introduced into the chromosome, and there are no other damaged sites serving as potential mutational hotspots.
Repair of a DSB by recombination of duplicated sequences requires a number of repair activities (Kowalczykowski 2000). Limits of primer synthesis dictated the use of short homologies (40 to 60 bp) as substrates for recombination. RecA is thought to play a minor role in recombination of homologous sequences <100 bp (Lloyd and Low 1996). However, it was found that presence of RecA was an absolute requirement in this repair process.
The 4.263-Mb genome of MDS12 is smaller than the chromosome of any E. coli strain for which data are available (4.4 to 5.5 Mb; Bergthorsson and Ochman 1995, 1998). The 12 deletions reduced the genome by removing most of the potentially “selfish” DNA (cryptic prophages, phage remnants, insertion sequences [ISs]) and a large fraction of genes of tentative or unknown functions. All together, of the 409 ORFs and 2 stable RNA genes deleted, 114 possessed tentative functional classification and 204 were marked as unknown (Blattner et al. 1997).
Transposons and ISs are thought to be the major source of mutations in E. coli (Chalmers and Blot 1999). Over half of these mobile genetic elements (24 of the 44 transposons of MG1655) have been removed from the genome of MDS12, presumably increasing the genetic stability. Further deletions aimed at removing all ISs from the genome are underway to test this hypothesis.
Chromosomal regions identified as K-islands can theoretically carry replacements of essential genes preserved in EDL933 but lost from MG1655. The fact that all 12 regions chosen for deletion could be removed from the MG1655 genome indicates that such nonorthologous gene displacements must be rare events, at least for genes essential under laboratory growth conditions. It may also reflect the high level of paralogy among E. coli genes (Riley and Serres 2000) and the robustness of the E. coli metabolic network (Edwards and Palsson 2000).
The two oppositely replicating halves (replichores) of the E. coli genome are nearly identical in size (Blattner et al. 1997). Large rearrangements significantly affecting this size balance can have deleterious effects on replication (Hill and Gray 1988). The 12 deletions presented here are nearly evenly distributed along the genome, but size reduction of replichore 1 is 121 kb larger than that of replichore 2. However, this relatively small size difference was well tolerated by the cell.
Partial characterization of MDS12 revealed few phenotypic changes compared with those of the parental strain. Growth characteristics and transformability of MDS12 were essentially identical to those of MG1655. The use of 379 carbon, nitrogen, phosphorous, and sulfur sources was compared using Biolog Phenotype Microarray plates, and the deletion mutant showed more than twofold reduced growth on 15 compounds. Of the observed phenotypes, the one showing the most marked change can be conclusively attributed to a deleted gene. That is the use of glycolate substrate, which is normally metabolized to glyoxylate by the product of the deleted glcD gene. Most of the other phenotypes are for compounds with known catabolic pathways (KEGG database, 2001; http://www.genome.ad.jp/kegg/) that are left intact in MDS12. Some of the deleted genes—such as tynA, argF, and gabT (the putative promoter of which was eliminated)—code for products involved in possible alternate or intersecting pathways related to the observed phenotype, but a causal connection is not apparent. In the case of argF, the alternative enzyme argI is still present in MDS12. A few of the growth defects were observed for compounds that have not been studied in E. coli (5-aminovalerate, β-glycerolphosphate, and parabanic acid). This supports the idea that many unknown ORFs in E. coli encode proteins to metabolize uncommon compounds.
Successful size reduction of the MG1655 genome by 8.1% underlines the feasibility of a rational targeted approach in constructing modified bacterial genomes. MDS12, a strain with a DNA content approaching the core E. coli genome, appears to retain E. coli capabilities important under standard laboratory culturing conditions. Further refinements and deletions could eventually result in a greatly simplified cell that displays fully predictable reactions and programmable functions.
METHODS
Plasmids
Sequence data for plasmids pSG76-CS, pKSUC1, and pSTKST have been submitted to the DNA Data Bank of Japan, European Molecular Biology Laboratory, and GenBank databases under accession Nos. AF402780, AF402779, and AF406953, respectively. The three plasmids were engineered from parental plasmids described previously (Pósfai et al. 1997, 1999). pSG76-CS serves as template plasmid to generate linear targeting fragments by PCR. It contains the CmR gene and two I-SceI sites and was obtained by the PCR-mediated insertion of a second I-SceI recognition site into pSG76-C, downstream of the NotI site. The two I-SceI sites are in opposite orientation. pKSUC1 is a high-copy-number plasmid expressing I-SceI constitutively. It was assembled by ligating the XbaI-NotI fragment of pSG76-K, blunted at the NotI-end and carrying the kanamycin resistance (KmR) gene, to the XbaI-DraI fragment of pUC19RP12, carrying the I-SceI gene and the pUC origin of replication. pSTKST is a low-copy-number plasmid expressing I-SceI under the control of the tet promoter-operator. It was derived by ligating the XbaI-PstI fragment of pUC19RP12, carrying the I-SceI gene, to the large XbaI-PstI fragment of pFT-K, carrying the gene for KmR. Replication of the plasmid is temperature sensitive and is abolished at 37°C to 43°C. pSG76-CSH was constructed by cleaving pSG76-CS by EcoRI and HhaI, blunting the ends by DNA polymerase I large fragment and recircularizing the plasmid by ligation. pBADαβγ has been described (Zhang et al. 1998) and was a gift of A.F. Stewart (EMBL, Heidelberg).
Strains and Media
Plasmids were generally prepared from DH5α (Woodcock et al. 1989). Deletions were engineered in the strain K-12 MG1655 (Blattner et al. 1997). Standard laboratory media (LB, SOC, and M9/glucose minimal medium) and agar plates were used (Sambrook et al. 1989). Casamino acids were added to 0.5% when used as a medium supplement. Antibiotics were applied at the following concentrations: ampicillin (Ap) 50 μg/mL, chloramphenicol (Cm) 25 μg/mL, and kanamycin (Km) 25 μg/mL. Heat-treated chlor-tetracycline (cTc) was used to inactivate the Tet repressor. Preparation of the inducer stock solution has been described (Pósfai et al. 1999).
Deletion Procedure
Construction of a Linear Targeting Fragment by PCR
To generate primer ab (Fig. 2), 20 pmole of primer a was mixed with 20 pmole of primer b, and PCR was performed in a total volume of 50 μL. Cycle parameters were 15 × (94°C 40 sec/57°C or lower [depending on the extent of overlap between primers a and b] 40 sec/72°C 15 sec). Next, 1 μL of this PCR product was mixed with 20 pmole of primers a and c (Fig. 1) each and 50 ng of pSG76-CS template, and a second round of PCR was performed in a volume of 2 × 50 μL. Cycle parameters were 28 × (94°C 40 sec/57°C 40 sec/72°C 80 sec). The resulting PCR-generated linear DNA-fragment was purified by Promega Wizard PCR purification kit and suspended in 20 μL water. Elimination of the template plasmid (e.g., by DpnI digestion) is not needed.
Replacement of a Genomic Region by Insertion of the DNA Fragment
The target strain carrying pBADαβγ was grown at 37°C in the presence of Ap, and electrocompetent cells were prepared as described (Pósfai et al. 1999), except that 0.1% l-arabinose was added to the culture 0.5 to 1 h before harvesting the cells; 4 μL targeting DNA-fragment (200 to 500 ng) was electroporated into 40 μL of electrocompetent cells. Cells were plated on Cm + Ap plates and incubated at 37°C. A total of 10 to several hundred colonies were usually obtained after 12 to 24 h of incubation. Typically 12 colonies were checked for correct-site insertion of the fragment by colony PCR using primers d and e (Fig. 2).
Deletion of the Inserted Sequences
A colony harboring the desired insertion was grown in LB + Cm, competent cells were prepared for chemical transformation, and pSTKST was introduced into them by standard procedures (Sambrook et al. 1989). Cells were spread on LB + Km plates and incubated at 30°C. A colony from this plate was inoculated into 10 mL of LB + Km supplemented with heat-treated inducer cTc (final concentration, 25 μg/mL) and grown for 24 h at 30°C. Dilutions of the culture were plated on LB + Km plates and incubated overnight at 30°C. (Alternatively, instead of pSTKST, pKSUC1 can be used and the cTc induction step can be omitted. The transformation mixture is spread on LB + Km plates and incubated overnight at 37°C.) Typically six colonies were screened by PCR using primers d and e. A fragment size matching the predicted length indicated the presence of the desired scarless deletion. Helper plasmids can be eliminated from the cells by growing the culture at 37°C to 43°C in LB. Replication of pSTKST is inhibited at this temperature, and pBADαβγ is rapidly lost from the cells under nonselective conditions.
For additional information see our Web page (http://www.szbk.u-szeged.hu/~posfaigy).
Biolog Phenotype Microarray Analysis
The strains MG1655 and MDS12 were streaked from frozen glycerol stocks on M9/glucose minimal medium and grown overnight at 37°C. Colonies were transferred into 1 mL sterile 0.85% NaCl, and 100 μL was spread on R2A plates and grown overnight at 37°C. Cells were then inoculated into Phenotype Microarrays PM1–4 (Biolog; Bochner et al. 2001) and grown according to the manufacturer's instructions. Growth was quantitated by measuring absorbance at 620 nm in either a Labsystems Multiskan Ascent or Tecan SpectraFluor Plus plate reader, and results were confirmed visually. The comparison of MG1655 and MDS12 was performed twice.
WEB SITE REFERENCES
http://www.szbk.u-szeged.hu/~posfaigy/; nucleotide sequences of the primers, additional methodological descriptions, and actual sequences of the joints.
http://www.genome.ad.jp/kegg/; KEGG database.
Acknowledgments
We thank Agnes Szalkanovics for technical assistance and Vincent Starai for assistance with Biolog plates. We thank Valerie Burland and Antal Kiss for critical reading of the manuscript. This work was supported by grants OTKA T030136, ETT24280, and HHMI 75195542102 to G.P. and National Institutes of Health grant GM35682 to F.R.B. C.D.H. was supported by National Institutes of Health grant 5-T32-GM08349.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL: fred@genome.wisc.edu.; FAX (608) 262-2976.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.217202.
REFERENCES
- Asai T, Zaporojets D, Squires C, Squires CL. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Ochman H. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J Bacteriol. 1995;177:5784–5789. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R, Blot M. Insertion sequences and transposons. In: Charlebois RL, editor. Organization of the prokaryotic genome. Washington, DC.: ASM Press; 1999. pp. 151–169. [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Palsson BO. Robustness analysis of the Escherichia coli metabolic network. Biotechnol Prog. 2000;16:927–939. doi: 10.1021/bp0000712. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Hill CW, Gray JA. Effects of chromosomal inversion on cell fitness in Escherichia coli K-12. Genetics. 1988;119:771–778. doi: 10.1093/genetics/119.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya M, Tanaka T. Experimental surgery to create subgenomes of Bacillus subtilis 168. Proc Natl Acad Sci. 1997;94:5378–5382. doi: 10.1073/pnas.94.10.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Torii Y, Matsuoka C, Yamamoto K. DNA sequence changes in mutations in the tonB gene on the chromosome of Escherichia coli K12: Insertion elements dominate the spontaneous spectra. Jpn J Genet. 1995;70:35–46. doi: 10.1266/jjg.70.35. [DOI] [PubMed] [Google Scholar]
- Koonin EV. How many genes can make a cell: The minimal-gene-set concept. Annu Rev Genomics Hum Genet. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- Lloyd RG, Low KB. Homologous recombination. In: Neidhart FC, et al., editors. Escherichia coli and Salmonella. Washington, DC.: ASM Press; 1996. pp. 2236–2255. [Google Scholar]
- McClelland M, Florea L, Sanderson K, Clifton SW, Parkhill J, Churcher C, Dougan G, Wilson RK, Miller W. Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res. 2000;28:4974–4986. doi: 10.1093/nar/28.24.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 271–273. [Google Scholar]
- Monteilhet C, Perrin A, Thierry A, Colleaux L, Dujon B. Purification and characterization of the in vitro activity of I-SceI, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res. 1990;18:1407–1413. doi: 10.1093/nar/18.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Campellone KG, Poteete AR. PCR-mediated gene replacement in Escherichia coli. Gene. 2000;246:321–330. doi: 10.1016/s0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET- recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Jones IB. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 2000;19:6637–6643. doi: 10.1093/emboj/19.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Pósfai G, Koob MD, Kirkpatrick HA, Blattner FR. Versatile insertion plasmids for targeted genome manipulations in bacteria: Isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Serres MH. Interim report on genomics of Escherichia coli. Annu Rev Microbiol. 2000;54:341–411. doi: 10.1146/annurev.micro.54.1.341. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 1989. pp. 1.82–1.84. , A.1–A.2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schaechter M, Neidhardt FC. Introduction. In: Neidhart FC, et al., editors. Escherichia coli and Salmonella. Washington, DC.: ASM Press; 1987. pp. 1–2. [Google Scholar]
- Walker GC. The SOS response of Escherichia coli. In: Neidhart FC, et al., editors. Escherichia coli and Salmonella. Washington, DC.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]