Abstract
To accelerate the molecular analysis of behavior in the honey bee (Apis mellifera), we created expressed sequence tag (EST) and cDNA microarray resources for the bee brain. Over 20,000 cDNA clones were partially sequenced from a normalized (and subsequently subtracted) library generated from adult A. mellifera brains. These sequences were processed to identify 15,311 high-quality ESTs representing 8912 putative transcripts. Putative transcripts were functionally annotated (using the Gene Ontology classification system) based on matching gene sequences in Drosophila melanogaster. The brain ESTs represent a broad range of molecular functions and biological processes, with neurobiological classifications particularly well represented. Roughly half of Drosophila genes currently implicated in synaptic transmission and/or behavior are represented in the Apis EST set. Of Apis sequences with open reading frames of at least 450 bp, 24% are highly diverged with no matches to known protein sequences. Additionally, over 100 Apis transcript sequences conserved with other organisms appear to have been lost from the Drosophila genome. DNA microarrays were fabricated with over 7000 EST cDNA clones putatively representing different transcripts. Using probe derived from single bee brain mRNA, microarrays detected gene expression for 90% of Apis cDNAs two standard deviations greater than exogenous control cDNAs.
[The sequence data described in this paper have been submitted to Genbank data library under accession nos. BI502708–BI517278. The sequences are also available at http://titan.biotec.uiuc.edu/bee/honeybee_project.htm.]
The honey bee (Apis mellifera) is an important model for studies of neural and behavioral plasticity, particularly with respect to social behavior, learning, and memory (Fahrbach and Robinson 1995; Robinson 1998; Menzel 2001; Maleszka et al. 2000). The neuroanatomy, neurophysiology, and neurochemistry of the honey bee brain have been studied extensively, and several functions have been mapped to particular brain regions (e.g., Menzel 2001; Fahrbach and Robinson 1995). Honey bees also have been used extensively to study the genetic underpinnings of behavior (Rothenbuhler 1967; Page and Robinson 1991). In the past few years, these lines of inquiry have been extended to the discovery of quantitative trait loci (Hunt et al. 1995, 1998) and analyses of expression levels of genes in the brain (Kucharski et al. 1998, 2000; Fiala et al. 1999; Toma et al. 2000; Shapira et al. 2001; Kucharski and Maleszka 2002).
One strong advantage of working with honey bees is that it is possible to study behavior under both laboratory and natural conditions. The natural social life of honey bees, though arguably as complex as in many vertebrate societies, can be extensively manipulated with precision. Insights gained from both lab and field studies ultimately will enable information on genes influencing neural and behavioral plasticity to be interpreted from ecological and evolutionary perspectives, contributing to a more comprehensive understanding of genes, brain, and behavior (Robinson 1999).
Molecular analyses in the honey bee have been constrained by the high investment required to identify and clone individual genes and the need to have an a priori hypothesis about each gene. The public databases contained only about 101 complete or near-complete A. mellifera gene sequences (nonredundant entries in SWISS-PROT and TrEMBL, as of December 2001) and, prior to this study, a total of 800 nucleotide sequences, most of them expressed sequence tags (ESTs) from antennae (H.M.R., unpubl.) or larvae (Evans and Wheeler 2001). The value of studying many genes simultaneously in the honey bee was demonstrated by Evans and Wheeler (2001) who identified gene expression profiles that were characteristic for worker/queen caste differentiation. This study involved the initial identification of 158 candidate clones using subtractive methods, and was thus limited by the small number of genes analyzed. Current DNA microarray technologies allow expression studies of many thousands of genes at the same time (Schena et al. 1995; DeRisi et al. 1997). ESTs provide an economical approach to identifying large numbers of genes that can be used in gene expression and other genomic studies (reviewed by Gerhold and Caskey 1996; see also Dimopoulos et al. 2000 and Porcel et al. 2000).
Here, we describe a collection of more than 20,000 ESTs generated from the A. mellifera brain, putatively representing 8912 different transcripts after sequence assembly. To facilitate gene identification and functional genomic studies in the honey bee, the brain EST set has been annotated using the structured vocabulary provided by the Gene Ontology Consortium (2001), based on molecular studies of gene function in Drosophila melanogaster. We describe a DNA microarray resource composed of over 7000 EST cDNA clones putatively representing different transcripts. We demonstrate the utility of this resource by reporting on gene expression measured in single honey bee brains. Additionally, comparative genomics approaches were used to predict or improve predictions for 122 genes in Drosophila, as well as to identify 126 genes conserved between Apis and other organisms that apparently have been lost from the Drosophila genome.
RESULTS AND DISCUSSION
Generation and Assembly of Brain ESTs
A normalized, unidirectional cDNA library was generated from dissected honey bee brains. An initial 7968 clones were sequenced from the 5′ end. The library was then subtracted, and 12,288 more clones were sequenced (also from the 5′ end). An additional 1152 sequences (3′ and duplicate 5′ ends) were obtained from previously sequenced clones. Thus, the EST set represents 20,256 cDNA clones and 21,408 total sequences. The 21,408 sequences were trimmed of vector and low-quality sequence and filtered for minimum length (200 bp), identifying 15,311 high-quality ESTs of 494 bp average length (Table 1). The estimated number of ESTs per putative transcript was initially 1.2 when sequencing was initiated and rose to 1.7 at the time sequencing was terminated (based on phrap analyses of high-quality ESTs after each batch of sequences; see below).
Table 1.
Honey Bee Brain EST Summary
| Total sequences | 21408 |
| cDNA clones sequenced (5′ end) | 20256 |
| Normalized library | 7968 |
| Normalized/subtracted library | 12288 |
| Redundant 5′ end sequences | 960 |
| 3′ end sequences | 192 |
| Total high-quality sequences | 15311 |
EST, expressed sequence tag.
The 15,311 high-quality ESTs were analyzed with the CAP3 assembly program to identify those that represent redundant transcripts (Table 2; see Table 8 for all program references). A total of 9481 ESTs were assembled into 3136 contiguous sequences (contigs). The remaining 5830 ESTs did not assemble into contigs (referred to as singlets). Thus, the combined set of contigs and singlets included 8966 sequences (hereafter referred to as “assembled sequences”), putatively representing different transcripts. Only 40 contig sequences contained more than 10 ESTs, and the largest number of ESTs assembled into one contig was 44.
Table 2.
EST Assembly Results
| PHRAPa | CAP3b | CAP3c | |
|---|---|---|---|
| Total sequences analyzed | 14642 | 14642 | 15311 |
| Number of ESTs in contigs | 8464 | 8357 | 9481 |
| Number of contigs | 3119 | 2910 | 3138 |
| Number of singlets | 6178 | 6285 | 5830 |
| Number of putative transcripts (assembled sequences) | 9297 | 9196 | 8966 |
| Number of contigs containing: | |||
| 2–4 ESTs | N/A | 2626 | 2762 |
| 5–10 ESTs | N/A | 255 | 334 |
| 11–20 ESTs | N/A | 28 | 33 |
| 21–40 ESTs | N/A | 2 | 6 |
| >40 ESTs | N/A | 1 | 1 |
Default settings
High-quality ESTs assembled using high-quality, vector-trimmed sequence only. Default settings were used except minimum overlap was 40 bp and 95% identity (default is 30 bp, 75% identity).
High-quality ESTs assembled using high- and low-quality, vector-trimmed sequence, 3′ and reductant 5′ ESTs were included (these were treated as independent clone sequences to avoid error resulting from manual clone picking). Default settings were used except minimum overlap was 40 bp. These assembly results were used for all analyses in this study, except where noted.
EST, expressed sequence tag.
Table 8.
Databases and software used
| Version and/or date downloaded | Source | |
|---|---|---|
| Sequence analysis and assembly | ||
| phred | 0.000925.c | 1 |
| Cross match | 0.990319 | 1 |
| Qualtrim | September, 2000 | 2 |
| Simpletrim | July, 2000 | 2 |
| RepeatMasker | July, 2000 | 1 |
| phrap | 0.990319 | 1 |
| CAP3 | July, 2000 | 3 |
| Flip | 2.0 | 4 |
| Sequence similarity searches | ||
| Stand-alone BLAST | Oct., 01 and later | 5 |
| nr | Aug., 2001 | 5 |
| nt | May, 2001 | 5 |
| EST_Human | May, 2001 | 5 |
| EST_Mouse | May, 2001 | 5 |
| EST_Other | May, 2001 | 5 |
| aa_gadfly.dros.RELEASE2 | RELEASE2 | 6 |
| na_arms.dros.RELEASE2 | RELEASE2 | 6 |
| na_EST.dros | May, 2001 | 6 |
| nr_Arthropoda | Aug., 2001 | 7 |
| nr_Chordata | Aug., 2001 | 7 |
| Functional annotation | ||
| function.ontology | 2.99; Sep., 2001 | 8 |
| process.ontology | 2.88; Sep., 2001 | 8 |
| gene_association.fb | 1.29; Sep., 2001 | 8 |
University of Washington Genome Center; http://www.genome.washington.edu/UWGC
Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana-Champaign; http://www.biotech.uiuc.edu/keck.htm
Organelle Genome Megasequencing Project, University of Montreal; http://megasun.bch.umontreal.ca/ogmpproj.html
National Center for Biotechnology Information (NCBI); http://www.ncbi.nim.nih.gov
Berkeley Drosophila Genome Project (BDGP); http://www.fruitfly.org
Sub-databases were extracted from nr using NCBI gene identification (gi) numbers for each taxonomic group.
We separately processed the high-quality ESTs using PHRAP and CAP3 using different levels of stringency (Table 2). These different assemblies produced very similar results, and we retained the CAP3 results for further analyses. Fifty-four assembled sequences were removed from the database (sequencing artifacts and/or exogenous contaminants; see Methods), leaving 8912 assembled sequences used in subsequent analyses.
EST Quality Analysis and Sequence Survey
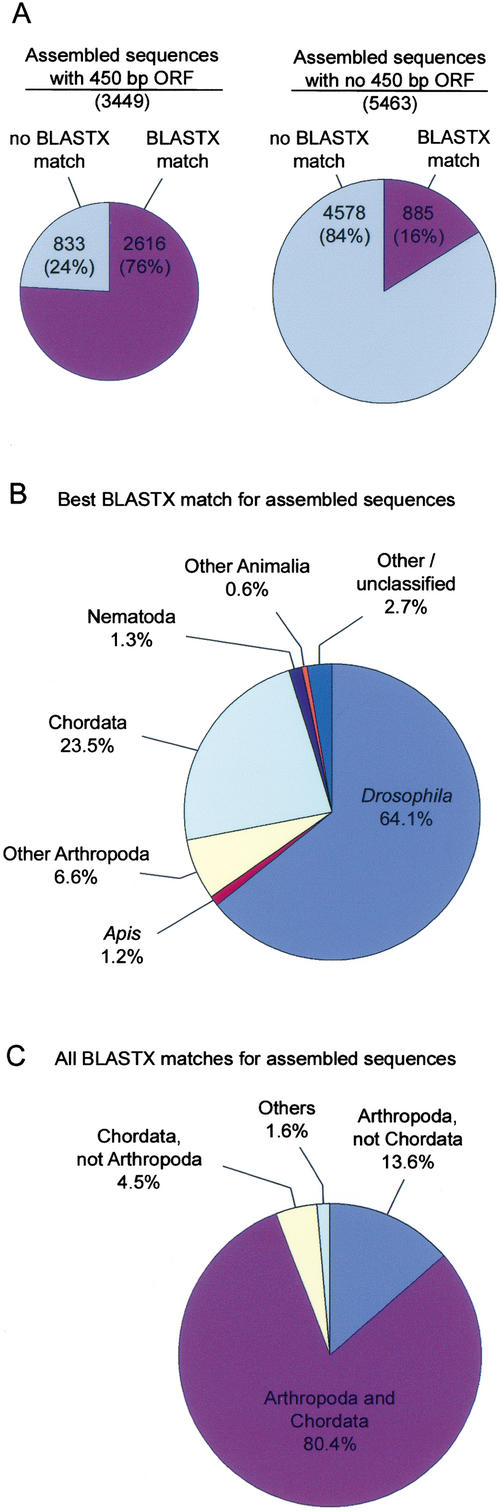
Of the 8912 assembled sequences, 3501 (39%) were similar to known protein sequences in the Non-Redundant Protein (nr) database (BLASTX; E ≤10−5). To estimate the proportion of transcript sequences that represent truly novel genes, the assembled sequences were screened to identify only those with clear protein coding capacity. A total of 3449 assembled sequences have an open reading frame (ORF) of at least 450 bp. Of these, 2616 (76%) had matches in the nr database and 833 (24%) had no matches (Fig. 1A). This result indicates that perhaps 24% of the protein-encoding genes expressed in the honey bee brain are highly diverged in primary structure. A total of 5463 assembled sequences did not have an ORF of at least 450 bp; of these, 885 (16%) had matches in the nr database and 4578 (84%) had no matches. Many assembled sequences did not have an ORF of 450 bp because they were too short (916 assembled sequences were <450 bp long). Other assembled sequences may have lacked an ORF for a variety of reasons, including frame shift errors, 5′ truncation of cDNA clones (causing ESTs to consist mostly or entirely of 3′ untranslated region [UTR]) or ESTs that were not derived from mRNA. Microarray hybridization results indicated that the vast majority of ESTs were derived from legitimate transcripts (see below). To assess 5′ truncation of cDNA clones, we examined sequence alignments of 130 ESTs (5′) that had matches to A. mellifera full-length cDNA sequences in GenBank (matches defined as ≥98% identity over at least 200 bp). Nine of these clones were in a backwards orientation (see below). Of the 121 ESTs in a forward orientation, 56 (46%) had 5′ sequences that corresponded to the 5′ end of the full-length cDNA sequence. The remaining 65 ESTs (54%) were derived from 5′ truncated cDNA inserts. This result suggests that a large fraction of noncoding ESTs may have been derived from severely truncated cDNAs consisting mostly or entirely of 3′ UTR.
Figure 1.
Open reading frame (ORF) and BLASTX results. (A) The proportion of assembled sequences with and without BLASTX matches in the Non-Redundant Protein (nr) database (E ≤10−5) is indicated for assembled sequences with and without an identified 450 bp ORF. Relative area of pie charts indicates number of sequences. (B) Apis sequences with matches in the nr database (3501 total) were classified by the organism of the “best hit” protein sequence. (C) Apis sequences with matches in the nr database (3501 total) were separately analyzed for matches in Arthropoda and Chordata protein databases (see Table 8 for sub-database creation).
ESTs were analyzed to identify a variety of other possible artifacts (see Methods). We estimated that 10% of the clones in the library are at least partially unspliced (often resulting from priming of the oligo(dT) primer within an unspliced AT-rich intron). Approximately 18% of the cDNA clones appear to be inserted in a reverse orientation. Finally, a single chimeric clone was identified that contained linker sequence within an EST flanked by back-to-back poly(A)+ sequences. No chimeras were identified by comparing BLASTX matches for 3′ and 5′ ESTs corresponding to the same cDNA clones (68 clones with 3′ and 5′ BLASTX matches were tested).
Figure 1B summarizes the top hits (matches with lowest E value) for each of the 3501 assembled sequences that had matches in the nr database. As expected, the majority (2245; 64%) were most similar to predicted protein sequence from Drosophila. Only 41 (1.2%) were most similar to predicted protein sequence from Apis (because of the small number of Apis gene sequences in the database). An additional 230 (6.6%) were most similar to sequence from a variety of other Arthropoda, including the insects Bobyx mori (28 best matches) and Manduca sexta (27 best matches). A surprisingly large number, 823 (24%), were most similar to sequence from Chordata (see Comparative Genomics, below). Others were most similar to proteins from Nematoda (47) or other Animalia (22). Twenty had best matches to various bacterial proteins with amino acid identities ranging from 42% to 92% (specifically, Mycobacterium [15], Caulobacter [4], and Agrobacterium [1]). We suggest that these 20 sequences were derived from unknown bacterial infections or contamination of bee brains or associated tissues. Two sequences appeared to be derived from an uncharacterized virus, having 24% and 39% amino-acid identity to different regions of the 2858 amino-acid polyprotein of the honey bee sacbrood virus.
Separate BLASTX searches of Arthropoda and Chordata protein databases revealed that the majority of assembled sequences with matches (80%) were similar to predicted protein sequences from both Arthropoda and Chordata (Fig. 1C). Others were similar to sequences from Arthropoda but not Chordata (13.6%), from Chordata but not Arthropoda (4.5%), or from non-Arthropoda and non-Chordata organisms only (1.6%). The implications of these findings for Drosophila were investigated further (see Comparative Genomics, below).
The assembled EST database was searched for simple sequence repeats using BLASTN and a database of simple sequence repeats of one to four bases (excluding (A)n repeat). This search identified simple sequence repeats in 767 of the assembled sequences using a highest scoring pair (HSP) cutoff value of 50, and 76 sequences using an HSP cutoff value of 100. These HSP cutoff values roughly correspond to 25 and 50 bp of perfect match, respectively (note that identified repeats are not necessarily contiguous because default BLAST parameters allow gaps in alignment). Repeat sequences are likely to reside primarily in EST noncoding sequence (which constitute a large fraction of the ESTs, see above).
Gene Number
EST assembly is expected to generate an overestimation of the actual number of genes represented, as failure of ESTs to assemble can result from nonoverlapping ESTs, alternate splicing, sequence polymorphism, and sequencing errors. Assuming approximately one-to-one correspondence between genes in Apis and Drosophila, the level of redundancy can be estimated based on BLASTX searches of Drosophila predicted proteins. A total of 3362 Apis assembled sequences had “best hits” to 2672 different Drosophila sequences, suggesting 19.6% redundancy in the Apis assembled sequence set. Similar levels of redundancy after EST assembly have been estimated in other large EST collections (e.g., roughly 20% in a large mouse cDNA set; see Kawai et al. 2001). Taking 20% as an estimate of redundancy in the 8912 assembled Apis sequences, the EST set may represent a total of 7100 genes expressed in the honey bee brain. If Apis has about the same number of genes as does Drosophila, this would represent roughly 50% of the total number of genes in the Apis genome.
A similar estimate of representation was provided by comparison of the 8912 assembled sequences with a set of 101 full- or near-full–length cDNA sequences obtained from an independent honey bee brain library (sequences kindly provided by R. Maleszka). A total of 55 assembled sequences from the EST set matched 54 different cDNA sequences from the independent brain library (match defined as ≥98% nucleotide identity over 200 bp). This result suggested that (based on this small sample set of 101 brain expressed cDNA sequences) the chance of finding a gene in the EST set was about 54%.
Functional Annotation of Bee Brain ESTs
We characterized the A. mellifera EST sequences with respect to functionally annotated genes in Drosophila melanogaster, taking advantage of the fact that this insect genome has been sequenced and extensively annotated (Adams et al. 2000). Each Apis assembled sequence was tentatively assigned Gene Ontology (GO) classification based on annotation of the single “best hit” match in BLASTX searches of Drosophila predicted proteins (E ≤10−5). Functional assignments of Apis ESTs described here are at the “inferred from electronic annotation” (IEA) level of evidence (see The Gene Ontology Consortium 2001). We take a conservative approach and avoid using Drosophila annotations that are, themselves, assigned at the IEA level of evidence. We do not exclude Drosophila annotations that are assigned at the “inferred from sequence similarity” (ISS) level of evidence (which requires human judgment and is therefore a higher level of evidence than IEA).
Tables 3 and 4 summarize assignments of Apis sequences to major molecular functions and biological processes, respectively. A broad range of functions and processes are represented in the brain ESTs. Table 5 lists Apis sequences that match Drosophila genes implicated in synaptic transmission (GO:0007268). Fifty-four (out of 116) Drosophila genes implicated in synaptic transmission were “best hit” for at least one Apis-assembled sequence. Table 6 lists Apis sequences that match Drosophila genes implicated in behavior. Note that current GO annotation for Drosophila includes only 42 genes implicated in behavior (as of December 2001). To provide information for comparative analysis, we generated a list of 106 genes directly implicated in behavior based on mutant analysis and/or transgenic experiments in Drosophila (compiled from FlyBase and J. Hall, pers. comm.). Genes were listed if at least one mutant allele or transgene affected a specific aspect of behavior, such as rhythmicity, mating, feeding, or learning and memory. (Global locomotor effects such as paralysis, uncoordinated movement, or shaking were not considered in this analysis, although many of the genes listed do exhibit global locomotor or lethal phenotypes when mutated to the null state.) Using this criteria, 47 (out of 106) Drosophila behavior genes were “best hit” for at least one Apis-assembled sequence. Annotation of Apis EST sequences with respect to all GO terms for molecular function, biological process, and cellular component are regularly updated and can be accessed at http://titan.biotec.uiuc.edu/bee/honeybee_project.htm.
Table 3.
Molecular Function
| Gene ontology term | Drosophila genesa | Apis assembled sequencesb | Drosophila genes representedc |
|---|---|---|---|
| All molecular function terms | 6260 | 1958 | 1509 |
| nucleic acid binding | 1052 | 353 | 269 |
| DNA binding | 696 | 186 | 144 |
| transcription factor | 495 | 132 | 104 |
| RNA binding | 265 | 125 | 92 |
| translation factor | 72 | 40 | 32 |
| transcription factor binding | 37 | 18 | 12 |
| cell-cycle regulator | 17 | 6 | 6 |
| chaperone | 114 | 50 | 35 |
| motor | 74 | 31 | 22 |
| microtubule binding | 83 | 41 | 28 |
| defense/immunity protein | 46 | 8 | 6 |
| enzyme | 2916 | 949 | 752 |
| GTPase | 92 | 51 | 42 |
| kinase | 355 | 133 | 108 |
| phosphatase | 171 | 52 | 41 |
| peptidase | 491 | 110 | 93 |
| enzyme activator | 61 | 21 | 20 |
| enzyme inhibitor | 74 | 7 | 7 |
| apoptosis activator | 3 | 1 | 1 |
| apoptosis inhibitor | 10 | 3 | 3 |
| signal transducer | 677 | 193 | 141 |
| receptor | 443 | 107 | 70 |
| ligand-dependent nuclear receptor | 22 | 9 | 7 |
| transmembrane receptor | 400 | 79 | 55 |
| G-protein coupled receptor | 207 | 21 | 17 |
| olfactory receptor | 58 | 1 | 1 |
| receptor signaling protein | 161 | 76 | 61 |
| ligand | 71 | 9 | 9 |
| cell adhesion | 53 | 27 | 17 |
| storage protein | 7 | 0 | 0 |
| structural protein | 354 | 106 | 92 |
| transporter | 792 | 296 | 195 |
| ion channel | 141 | 72 | 46 |
| voltage-gated ion channel | 43 | 29 | 16 |
| neurotransmitter transporter | 13 | 14 | 6 |
| ligand binding or carrier | 1095 | 451 | 335 |
| protein tagging | 6 | 5 | 3 |
| antioxidant | 8 | 7 | 5 |
Classification is hierarchical: indented terms are children of parent terms listed above.
Genes may be assigned to more than one term. Also note that child terms may have more than one parent term (e.g., “ligand-dependent nuclear receptor” is a child of both “receptor” and “transcription factor”) (see The Gene Ontology Consortium 2001).
Total number of Drosophila genes assigned to each Gene Ontology term (from databases listed in Table 8).
Number of Apis assembled sequences that match Drosophila genes assigned to each term in (a). Match means that the Drosophila gene was “best hit” for the Apis sequence (and e-value ≤10−5).
Number of different Drosophila genes matched by Apis sequences.
Table 4.
Biological Process
| Gene Ontology term | Drosophila genes | Apis assembled sequences | Drosophila genes represented |
|---|---|---|---|
| All biological process terms | 2746 | 906 | 696 |
| cell growth and maintenance | 2102 | 766 | 597 |
| metabolism | 1493 | 531 | 424 |
| protein metabolism and modification | 887 | 300 | 239 |
| ion homeostasis | 7 | 9 | 6 |
| intracellular protein traffic | 158 | 88 | 67 |
| vesicle transport | 150 | 83 | 62 |
| synaptic vesicle transport | 108 | 67 | 48 |
| stress response | 120 | 22 | 16 |
| response to external stimulus | 390 | 73 | 51 |
| cell organization and biogenesis | 253 | 108 | 79 |
| cell cycle | 187 | 42 | 34 |
| apoptosis | 29 | 2 | 2 |
| cell communication | 772 | 257 | 178 |
| cell adhesion | 46 | 25 | 15 |
| cell recognition | 43 | 33 | 20 |
| neuronal cell recognition | 14 | 3 | 3 |
| synaptic target recognition | 5 | 2 | 2 |
| signal transduction | 274 | 84 | 67 |
| cell-cell signaling | 117 | 79 | 54 |
| synaptic transmission | 116 | 79 | 54 |
| neurotransmitter release | 111 | 71 | 50 |
| developmental processes | 406 | 153 | 106 |
| embryogenesis and morphogenesis | 227 | 99 | 67 |
| neurogenesis | 95 | 59 | 36 |
| imaginal discs development | 63 | 35 | 27 |
| sex determination | 8 | 4 | 3 |
| dosage compensation | 8 | 6 | 4 |
| metamorphosis | 6 | 3 | 2 |
| physiological processes | 30 | 6 | 3 |
| perception external stimulus | 196 | 31 | 23 |
| behavior | 42 | 18 | 15 |
See notes for Table 3.
Table 5.
Apis Matches to Drosophila Synaptic Transmission Genes
| Apis sequence | Drosophila gene | Alignment length (aa) | HSP | e-value | Identities |
|---|---|---|---|---|---|
| Contig276 | amphiphysin | 295 | 390 | 1E-108 | 65% |
| BB160003A10D03 | AP-47 | 251 | 411 | 1E-115 | 82% |
| BB170001B10H02 | AP-50 | 234 | 407 | 1E-115 | 88% |
| BB160013B20B02 | Arf51F | 172 | 349 | 1E-96 | 97% |
| Contig 1946 | Arf72A | 179 | 340 | 8E-94 | 93% |
| BB160006B10F12 | Arf79F | 171 | 344 | 2E-95 | 98% |
| BB170032A10C06 | BcDNA:LD23336 | 200 | 95 | 3E-20 | 30% |
| BB170005A10D08 | CaMKII | 59 | 123 | 1E-28 | 96% |
| BB160014B20D10 | Caps | 225 | 372 | 1E-103 | 77% |
| Contig2785 | Cdk5 | 264 | 443 | 1E-125 | 80% |
| BB160022A10H03 | CG10617 | 93 | 61 | 6E-10 | 40% |
| BB16000BA20A11 | CG1107 | 121 | 137 | 6E-33 | 55% |
| Contig 1704 | CG14296 | 178 | 335 | 2E-92 | 91% |
| BB170005B20G04 | CG15694 | 149 | 212 | 2E-55 | 63% |
| Contig1152 | CG17762 | 186 | 92 | 3E-19 | 33% |
| Contig 1768 | CG2381 | 201 | 399 | 1E-111 | 91% |
| Contig2868 | CG2903 | 51 | 62 | 3E-10 | 47% |
| BB170016A20B10 | CG3020 | 38 | 53 | 2E-09 | 57% |
| Contig2061 | CG3029 | 210 | 340 | 1E-93 | 81% |
| Contig190 | CG5014 | 89 | 101 | 2E-38 | 58% |
| BB160022A20H05 | CG5627 | 220 | 263 | 7E-71 | 60% |
| BB170011A20D07 | CG5678 | 164 | 287 | 3E-78 | 85% |
| BB160024A20D05 | CG7034 | 199 | 202 | 2E-52 | 51% |
| BB160003B20B01 | CG7127 | 235 | 130 | 8E-31 | 37% |
| Contig2640 | CG7321 | 213 | 192 | 2E-49 | 49% |
| BB160020B10H05 | CG7736 | 208 | 115 | 1E-42 | 37% |
| BB160009A10E12 | CG8608 | 131 | 198 | 4E-51 | 70% |
| Contig1193 | Chc | 295 | 446 | 1E-126 | 74% |
| Contig924 | Csp | 237 | 218 | 4E-57 | 51% |
| BB160022A10D06 | Dap160 | 202 | 155 | 3E-38 | 46% |
| BB170019A20D08 | dlg1 | 266 | 377 | 1E-105 | 72% |
| Contig1272 | gammaSnap | 247 | 283 | 8E-77 | 55% |
| BB160010B20F08 | Gdi | 109 | 181 | 3E-46 | 75% |
| BB160015B20E03 | I(2)gl | 193 | 96 | 1E-20 | 33% |
| Contig1207 | Iqf | 102 | 66 | 3E-11 | 45% |
| BB170026B10H11 | Nrx | 142 | 215 | 9E-57 | 67% |
| Contig1277 | Nsf2 | 108 | 148 | 9E-37 | 66% |
| BB170025B20H08 | n-syb | 73 | 135 | 2E-32 | 91% |
| Contig1852 | Rab3 | 198 | 367 | 1E-102 | 89% |
| Contig2442 | Rop | 147 | 222 | 8E-59 | 70% |
| Contig1960 | Sed5 | 150 | 141 | 4E-34 | 50% |
| Contig734 | Snap | 279 | 427 | 1E-120 | 72% |
| BB170015A10F09 | Snap24 | 32 | 51 | 4E-07 | 78% |
| BB160017B20C04 | Stam | 219 | 253 | 5E-68 | 57% |
| Contig2134 | syt | 330 | 559 | 1E-160 | 85% |
| Contig80 | SytIV | 190 | 274 | 6E-74 | 70% |
Synaptic transmission (GO:0007268)
Table 6.
Apis Matches to Drosophila Behavior Genes
| Apis sequence | Drosophila gene | Alignment length (aa) | HSP | e-value | Identities |
|---|---|---|---|---|---|
| Contig3015 | 14-3-3zeta | 246 | 452 | 1E-127 | 91% |
| BB170004B20H03 | acj6 | 45 | 92 | 2E-19 | 97% |
| BB160024A10A12 | Adar | 169 | 185 | 3E-47 | 54% |
| Contig 1753 | ap | 83 | 143 | 2E-34 | 81% |
| Contig467 | ari-1 | 55 | 103 | 1E-22 | 81% |
| BB170007B20D03 | Atpalpha | 97 | 177 | 3E-45 | 89% |
| BB160016A10E02 | CadN | 126 | 117 | 4E-27 | 47% |
| Contig2335 | Cam | 145 | 228 | 5E-60 | 74% |
| BB170005A10D08 | CaMKII | 59 | 123 | 1E-28 | 96% |
| BB170029A10F02 | Cha | 87 | 55 | 4E-08 | 36% |
| BB170016A10D05 | chp | 32 | 47 | 5E-06 | 62% |
| BB160005A10C01 | CoVa | 144 | 151 | 3E-37 | 53% |
| Contig924 | Csp | 237 | 218 | 4E-57 | 51% |
| BB170022A10F08 | dare | 155 | 101 | 1E-32 | 38% |
| Contig377 | Dat | 167 | 113 | 9E-26 | 39% |
| BB170029B10C01 | dnc | 66 | 120 | 8E-29 | 84% |
| BB160007B10G06 | dsx | 47 | 50 | 2E-06 | 46% |
| Contig1083 | e | 147 | 86 | 2E-17 | 36% |
| BB160017B10B10 | Fas2 | 199 | 189 | 1E-48 | 47% |
| BB160010A10H12 | for | 193 | 291 | 2E-79 | 75% |
| BB170016A20G06 | fru | 103 | 168 | 3E-42 | 76% |
| BB160008B10H11 | G-salpha60A | 234 | 357 | 4E-99 | 73% |
| BB170024A20D07 | Hk | 28 | 52 | 4E-07 | 78% |
| Contig12 | lark | 367 | 380 | 1E-106 | 58% |
| BB170007B10A02 | mas | 167 | 340 | 4E-94 | 94% |
| Contig923 | mle | 375 | 355 | 3E-98 | 50% |
| Contig362 | nbA | 159 | 70 | 2E-12 | 33% |
| BB160015B10F09 | Nf1 | 210 | 226 | 1E-59 | 57% |
| BB170031B10G07 | ninaA | 172 | 212 | 2E-55 | 55% |
| BB170001A10G09 | ninaE | 163 | 239 | 1E-63 | 69% |
| BB170022A10G08 | nompC | 161 | 101 | 4E-22 | 40% |
| BB160020B20C08 | para | 34 | 76 | 1E-14 | 97% |
| Contig730 | Pka-C1 | 197 | 402 | 1E-112 | 95% |
| BB160004A20E12 | plx | 53 | 82 | 3E-16 | 71% |
| Contig397 | Pp1-87B | 141 | 282 | 1E-76 | 94% |
| BB170012A20E10 | rdgB | 160 | 58 | 4E-09 | 28% |
| Contig2777 | Reg-5 | 90 | 69 | 3E-12 | 40% |
| BB160003A20A12 | Rya-r44F | 197 | 316 | 2E-89 | 73% |
| BB160013A20B02 | sbb | 53 | 52 | 2E-06 | 54% |
| BB160004A10F11 | sd | 260 | 360 | 1E-100 | 68% |
| Contig830 | sgg | 285 | 522 | 1E-148 | 87% |
| Contig3064 | Shab | 88 | 98 | 3E-21 | 59% |
| Contig 1958 | Shal | 72 | 95 | 3E-20 | 58% |
| Contig2624 | slo | 90 | 184 | 5E-47 | 95% |
| Contig2399 | tipE | 64 | 107 | 7E-24 | 78% |
| Contig1139 | vri | 247 | 99 | 2E-21 | 30% |
| Contig2819 | w | 68 | 108 | 2E-56 | 73% |
Behavior genes defined in text. Drosophila genes tested but not found: Ace, Acp70A, Adf1, amn, bi, Btk29A, Caki, Ca-alpha1D, clk, CrebB-17A, Crg-1, crl, cry, cyc, Cyp4e2, dco, Ddc, disco, Dr, dsf, dy, eag, gk, G-oalpha65A, Hdc, inaC, lat, lio, lush, lz, mnb, mud, mys, ninaC, nompA, nompB, nonA, norpA, ogre, otu, Pdf. per, Pka-R1, ppl, qtc, rb, rut, scb, Sh, Shaw, shi, sol, spin, sws, tim, to, tutl, Ubc47D, W.
We expect that ongoing improvements in GO annotation for Drosophila, human, mouse, and Caenorhabditis elegans will lead to significant improvements in Apis gene annotation in the near future. The current annotation of Apis sequences, based solely on matches to Drosophila proteins, allowed useful comparative analyses but had several drawbacks. We often found Apis sequences that clearly encoded members of important gene families of known function, but nevertheless were not annotated. In every case examined, this occurred because the “best hit” gene in Drosophila was not yet assigned GO annotation. Conversely, Apis sequences sometimes were assigned function based on fairly weak matches (i.e., close to the E-value cutoff of 10−5), resulting from the short length of the Apis EST. Annotation also was limited by a high proportion of ESTs in this project that contain transcript noncoding sequence (e.g., 3′ UTR). Additional ESTs, especially from full-length, enriched, normalized, and subtracted libraries (e.g., Carninci et al. 2000), would enhance Apis gene annotation by allowing more ESTs to be assembled into larger contig sequences.
Honey Bee Brain Microarray
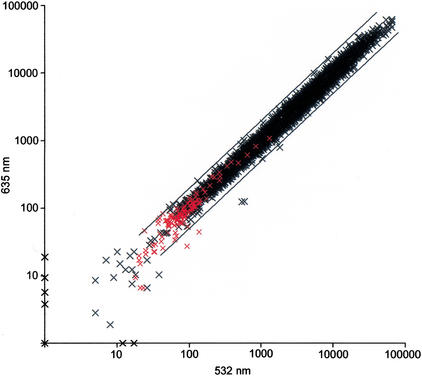
To allow functional genomic studies of brain and behavior in the honey bee, we generated cDNA microarrays from the annotated EST set described above. A total of 7329 cDNAs (putatively representing different transcripts) were successfully amplified as “single-band” PCR product and spotted on the microarray. Pilot studies indicated that fluorescent probe derived from single-brain mRNA (amplified by in vitro transcription; see Methods) could be used to label the vast majority of Apis cDNA spots on the microarray. Data obtained from one microarray experiment are presented in Table 7 and Figure 2. In this experiment, two dissected adult bee brains were combined and mixed during homogenization, then split into two equal samples. Each of the two samples was used to generate an independent probe (one Cy5-labeled probe [635 nm] and one Cy3-labeled probe [532 nm]). The two probes were combined and hybridized to a single microarray. A total of 7300 and 7305 cDNAs produced hybridization signal at least two standard deviations (SD) greater than background at 635 and 532 nm, respectively. To determine whether this hybridization signal was specific, we compared signal produced by Apis cDNA spots with exogenous negative control cDNA spots on the microarray (derived from vertebrate and plant genes). A total of 6647 (91%) and 6631 (90%) of the Apis cDNAs produced signal at least two standard deviations greater than exogenous control cDNAs at 635 and 532 nm, respectively. Signal intensities between 635 and 532 nm were highly correlated in this experiment (r = 0.9926) indicating that technical variation (from RNA isolation, mRNA amplification by in vitro transcription, and fluorescent labeling of probe) is very low. Results from additional microarrays were qualitatively similar using different bee brains as source material (data not shown). These results indicate that genomic scale gene expression profiling is feasible in single honey bee brains using the microarrays and protocols described here.
Table 7.
Signal analysis of an example microarray
| Number of spots | 635 nm | 532 nm | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Feature | Background | Feature | Background | ||||||
| Avg. | SD | Avg. | SD | Avg. | SD | Avg. | SD | ||
| Apis cDNAs | 7329 | 3.42 | 0.47 | 2.23 | 0.05 | 3.43 | 0.48 | 2.15 | 0.04 |
| Exogenous control 1 | 16 | 2.40 | 0.07 | 2.24 | 0.05 | 2.35 | 0.07 | 2.15 | 0.04 |
| Exogenous control 2 | 16 | 2.43 | 0.07 | 2.23 | 0.04 | 2.41 | 0.06 | 2.15 | 0.04 |
| Exogenous control 3 | 16 | 2.32 | 0.08 | 2.23 | 0.05 | 2.26 | 0.08 | 2.15 | 0.04 |
| Exogenous control 4 | 16 | 2.38 | 0.06 | 2.24 | 0.04 | 2.32 | 0.05 | 2.15 | 0.04 |
| Exogenous controls 5–48 | 43 | 2.49 | 0.20 | 2.23 | 0.05 | 2.47 | 0.22 | 2.15 | 0.05 |
| All controls spots | 107 | 2.43 | 0.15 | 2.23 | 0.05 | 2.39 | 0.17 | 2.15 | 0.04 |
| 635 nm | 532 nm | |
|---|---|---|
| # Apis cDNAs > background + 1 SD | 7308 | 7311 |
| # Apis cDNAs > background + 2 SD | 7300 | 7305 |
| # Apis cDNAs > control spots + 1 SD | 7029 (96%) | 7035 (96%) |
| # Apis cDNAs > control spots + 2 SD | 6647 (91%) | 6631 (90%) |
All inteneity values were log10 transformed. Feature and background readings are indicated for each cDNA spot (based on median pixel intensity) for 635 and 532 nm wavelengths. Average (Avg.) and standard deviation (SD) are indicated. Exogenous controls are described in Methods.
Figure 2.
Signal intensities from an example microarray. Values plotted are feature minus background intensity at 635 and 532 nm wavelengths for each cDNA spot (see Methods). Values were normalized such that the median ratio (635:532 nm) equals 1.0. Apis cDNAs are shown as black x's, exogenous negative control cDNAs are shown as red x's. Cy3-labeled probe (532 nm) and Cy5-labeled probe (635 nm) were independently derived from the same starting sample (using in vitro transcription to amplify starting mRNA; see Methods). The starting sample consisted of a mixture of two dissected adult bee brains (one bee observed foraging and one bee observed caring for brood). The coefficient of correlation (r) between 635 and 532 nm values was 0.9926 (based on log-transformed values). Divergence of values from the diagonal (ratio = 1) reflects technical variation introduced during RNA isolation, mRNA amplification by in vitro transcription, and fluorescent labeling of probe. The two diagonal bars indicate ratios (635:532 nm) equal to 0.5 and 2.0.
Microarray hybridization data have been used for the validation of gene sequences (e.g., Andrews et al. 2000; Shoemaker et al. 2001). The results presented above indicate that the vast majority of bee ESTs were derived from legitimate brain-expressed gene transcripts.
Comparative Genomics in Apis and Drosophila
A total of 823 of the assembled sequences (24% of those with matches) were most similar to protein sequence from Chordata (Fig. 1B). The high level of Apis “best hits” to Chordata could arise from a high rate of sequence divergence or gene loss in Drosophila and/or be related to deficiencies in Drosophila gene prediction. To distinguish between these possibilities, Drosophila genome sequence and EST databases were searched for matches to Apis-assembled sequences using TBLASTX. Matches were screened individually to identify true gene alignments based on plausible exon structure and amino-acid composition. In 99 cases, predicted proteins in Drosophila were missing one or more exons (predicted by alignment between Apis ESTs and Drosophila genome sequence). This caused a weak or no match to the Drosophila-predicted protein sequence and a misleading “best hit” to Chordata. In 23 cases, genes were identified in Drosophila genomic sequence (based on alignment with Apis sequence) that were not represented in Drosophila predicted protein or EST databases. Suggestions for annotations of these 122 Drosophila genes have been communicated to FlyBase.
Of the 701 remaining cases where the best match for the Apis sequence was to Chordata, 574 (16% of Apis-assembled sequences with matches) had likely orthologs in Drosophila, but these Drosophila genes were so diverged that better matches for the Apis sequences were identified in human, mouse and/or other non-Arthropoda. In 126 cases (3.6% of Apis assembled sequences with matches), the Apis sequence had significant and clear matches to proteins from human, mouse and/or other organisms, but no plausible ortholog was identified in searches of Drosophila-predicted protein, genome, or EST databases. These Apis sequences appear to define genes that have been lost from the Drosophila genome. Detailed analysis of these highly diverged genes and gene loss events in Drosophila will be presented in a subsequent manuscript.
Future Prospects
The relationship between genes and behavior is complex and is only beginning to be understood. Honey bees exhibit a wide variety of behavioral phenomena that are not observed in Drosophila, such as kin recognition, complex communication via the dance language, socially regulated division of labor, and a larger variety of forms of learning. The honey bee also is haplodiploid and has the highest known recombination rate of any animal (Hunt and Page 1995), traits that can facilitate genetic analyses of behavior. A wide range of naturally variable behavior traits has been described in honey bees, including defensive behavior (Hunt et al. 1998), foraging preferences (Hunt et al. 1995), and differences in socially regulated division of labor (Robinson 1992; see also Brillet et al. 2001). A comprehensive, web-based atlas of the bee brain currently in development (see http://www.neurobiologie.fu-berlin.de/Menzel.html) also will be helpful in providing a stronger neurobiological foundation for the study of genes and behavior in the honey bee. Early efforts to develop transgenic bees (Omholt et al. 1995; Ronglin et al. 1997; K. Robinson et al. 2000) suggest that there are no barriers to harnessing this technology. The work described here provides additional resources that should contribute to molecular analyses of honey bee behavior, using candidate gene studies, positional cloning, and functional genomic approaches.
METHODS
Bees
Approximately 600 adult workers were collected from a typical field colony at the University of Illinois Bee Research Facility. The colony had about 40,000 adult bees and was derived from a naturally mated queen. The bees in this area are a mixture of various races of European honey bees, predominantly Apis mellifera ligustica (Pellett 1938). Bees were collected when they were 1, 5, 10, 15, 20, 25, and 30 days old, which spans the typical lifespan during the active season (Winston 1987). This collection scheme ensured a broad representation of behavioral states, because bees specialize on different tasks at different ages (Robinson 1992). To obtain bees of known age, frames of pupae were removed from the colony and placed in an incubator (33°C). About 3500 one-day-old bees were marked with a spot of paint (Testor's Pla) on the thorax and then returned to their natal colony. We supplemented these age-based collections with samples of bees taking preforaging orientation flights (Capaldi et al. 2000) and foragers returning with either pollen or nectar loads. Collections were made both in the early morning and late in the afternoon. Bees were collected directly into liquid nitrogen (Toma et al. 2000) to minimize the possible effects of collection on gene expression. Brains were dissected on dry ice.
Brain cDNA Libraries
Total RNA was isolated from 400 bee brains (ca. 500 μg) with Rneasy total RNA isolation kit (Qiagen) followed by treatment with Dnase (1 unit RQ1 Dnase; Promega). Poly(A)+ RNA was purified and cDNA was synthesized and directionally cloned into NotI and EcoRI digested pT7T3-Pac phagemid vector as in Bonaldo et al. (1996). cDNA inserts are flanked by linker sequences 5′-NotI-GTTGC-3′ (library specific, 3′ linker) and 5′-EcoRI-GGCACGAGG-3′ (5′ linker). The library was normalized and (subsequently) subtracted as in Bonaldo et al. (1996).
Sequencing and Sequence Analysis
Plasmid DNA was extracted and sequenced using ABI 377 and 3700 sequencers. The sequencing primer used was 5′-AGCGGATAACAATTTCACACACAGGA-3′. Base-calling was performed with phred (see Table 8 for all programs and databases used). Vector sequences were trimmed using Cross-match. Low-quality bases (quality score <20) were trimmed from both ends of sequences using Qualtrim and Simpletrim. Those ESTs having a length of more than 200 bp after both vector and quality trimming were considered “high-quality” ESTs. The repeat sequences in these ESTs then were masked by RepeatMasker program using Drosophila repeat sequences as reference. The masked sequences were further screened for bacterial chromosomal DNA, RNA, insect viral DNA, rRNA, and mitochondrial DNA using BLASTN. Further screens for possible contaminants were conducted by BLASTN searches of the Non-Redundant Nucleotide Sequences (nt), EST_human, EST_mouse, and EST_others databases. Eighty-one ESTs were removed that corresponded to clear contaminants likely derived from other library and/or sequencing projects (from mouse or rat [49], cattle [9], human [6], pig [2], undetermined vertebrate [2], and various non-Escherichia coli bacteria [9]). No other ESTs were found to be ≥90% identical (over any 100 bp span) to nucleotide sequence from any non-Apis species, suggesting that the EST set did not include contamination from Drosophila or other sources not identified here. An additional 101 ESTs were removed as informatic artifacts (e.g., sequencing lanes that should not have produced sequence). Some EST screening was conducted after assembly, resulting in 54 contig sequences that were composed of contaminant or artifact ESTs. These 54 sequences were removed from the “assembled sequence” database and did not affect analyses presented here.
ESTs were analyzed to identify chimeric, backward, or unspliced inserts. Chimeric clones could be indicated by back-to-back poly(A)+ tails or vector linker sequences within ESTs. BLASTN searches for these instances identified only one chimera (out of all 21,408 ESTs). In this instance the 3′ linker sequence was found in the middle of an EST, flanked by back-to-back poly(A)+ tails from two different transcripts. Furthermore, in all cases where 3′ ESTs had BLASTX matches (E ≤10−20) to a Drosophila predicted protein (68 cases), 5′ ESTs from the same cDNA matched the same Drosophila protein. To estimate the total number of backward cDNA inserts, singlet ESTs with BLASTX matches to Drosophila-predicted proteins were analyzed. Out of 1919 singlet EST matches, 364 (19%) had a negative reading frame, indicating a backward cDNA insert. Of 720 individually analyzed ESTs with BLASTX matches to proteins from other organisms, 72 (10%) had clear instances of unspliced intron sequence (based on alignment with putative orthologs, ORF analysis, and identification of putative splice junctions); many of these clones appear to have resulted from priming of the oligo(dT) primer within an unspliced AT-rich intron.
ESTs were assembled using CAP3 and phrap (see Table 2 for settings).
ORFs were identified using FLIP with the minimum length set to 150 amino acids (450 bp). All BLAST searches were conducted on a desktop PC or local server using stand-alone BLAST software and sequence databases indicated in Table 8. All E-value cutoffs were 10−5, except where indicated otherwise. GO databases were installed on a local server. A GO browser was designed and implemented at the W.M. Keck Center for Comparative and Functional Genomics (University of Illinois at Urbana-Champaign) and used for functional annotation of the assembled EST sequences.
Microarray Fabrication
A single EST cDNA clone was selected to represent each assembled sequence (putatively unique transcript). For contigs with multiple ESTs, the rule followed was to select the 3′-most EST that had at least 300 bp of high-quality sequence. This procedure biases the cDNAs on the microarray toward the 3′ end but ensures that at least 300 bp of cDNA is spotted on the array. A total of 8872 cDNA clones were selected. These clones were picked from the library stock plates (384-well bacteria clones) and rearrayed to a new set of 384-well plates. These clones were grown overnight followed by sequence verification (see Clone Tracking, below).
Creation of the microarrays was essentially as described by Brown and Botstein (1999). Bacteria clones were inoculated to 96-well plates with LB and Amp and grown overnight. Plasmid inserts were amplified by PCR using 1 μL of the overnight bacteria inoculant and modified M13 (5′-CCAGTCACGACGTTGTAAAACGAC-3′) and M13 reverse (5′-GTGTGGAATTGTGAGCGGATAACAA-3′) primers in 50 μL volume reactions. Amplifications were performed in a MJ PTC-200 thermocycler (MJ Research). PCR reaction mixes contained 5 μL 10x reaction buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl), 2.0 mM MgCl2, 100 μM dNTPs, 0.2 μM each primer, and 1U Amplitaq Gold (Perkin Elmer). An initial 9-min denaturation was followed by 35 cycles of 40 sec denaturation at 94°C, 40 sec annealing at 65°C, and 3.5 min elongation at 72°C. The reaction ended with an additional incubation of 5 min at 72°C. Products were cleaned using Sephadex G-50 columns. Five microliters of each clean PCR product was analyzed on a 1% agarose gel. cDNA amplification products were visually examined and subjectively classified as follows: “strong single band” (86%), “weak or absent band” (13%), or “multiple bands” (1%). Only cDNAs that were amplified as “single strong band” and successfully spotted on the array (see below) were used in subsequent data analysis (7329 total).
PCR products were dried and resuspended in 8 μL 3x SSC, 1.5 M betaine. Betaine was used as in Diehl et al. (2001) to improve spot homogeneity and to increase hybridization signal on the microarray. All cDNAs were printed as single spots on Telechem Superamine slides (Arrayit) using a Cartesian Technologies spotter. Exogenous control cDNAs derived from cattle (phosphoglycerate kinase 1 and β-2-microglobulin) and soy (rubisco small chain 1 and chlorophyll ab binding protein) were spotted on the array 16 times each, such that they were represented on each of the 16 subgrids on the microarray (“exogenous controls 1–4”, respectively, in Table 7). An additional 43 vertebrate-derived cDNAs (singly spotted at random positions throughout the microarray) were used as control spots (“exogenous controls 5–48” in Table 7).
Spot and printing quality were assessed visually after printing. cDNA spots do not fully evaporate after arraying (as a result of 1.5 M betaine) allowing inspection of spot morphology under a dissecting scope. A few slides (about one in every five) exhibited minor defects (e.g., a single spot missing or several spots damaged by dust or lint particles). The majority of slides exhibited no defects (no spots missing, no spots joined, and all spots uniform in size).
DNA was crosslinked to slides by baking at 80°C for 1 h. Slides were blocked in 0.2% SDS for 4 min, followed by two washes in water. Slides were denatured in boiling water for 2 min, spun dry, and stored.
Microarray Hybridization, Scanning, and Data Analyses
Frozen brains were dissected from bees of known age and behavioral state as above. mRNA was amplified exactly as in Baugh et al. (2001), using only one round of in vitro transcription. Amplified RNA (aRNA) was analyzed by spectrophotometer and gel electrophoresis. Negative control reactions (no template and genomic DNA only) conducted in parallel produced no aRNA. aRNA was labeled by reverse transcription as follows: 5 μg of aRNA was mixed with 5 μg of random primer (Roche) (10 μL volume), denatured at 70°C for 4 min, and placed on ice. Labeling reaction (6 μL of 5x 1st Strand Buffer [Gibco]; 3 μL of 100 mM DTT; 6 μL of low T dNTPs [2.5 mM each dATP, dCTP, dGTP and 1.0 mM dTTP] (Sigma), 3 μL of 1 mM Cy3– or Cy5-dUTP [Amersham Pharmacia] and 2 μL of 200 U/μL SuperScript II [Gibco]) was prepared on ice, mixed with aRNA and primer, then incubated at 42°C for 1 h. One microliter of SuperScript II was added and the reaction was incubated at 42°C for an additional hour. RNA was removed by adding 1 μL of 0.25 mg/mL RNAse A (NEB) and 0.5 μL of 2 U/μL RNAse H (Stratagene) and incubating at 37°C for 30 min. Labeled cDNA was purified using the Qiagen PCR Purification Kit.
Thirty microliters of purified, labeled cDNA was mixed with blocking oligos dT-T7 (20 μg; see Baugh et al. (2001)) and dT30 (40 μg), boiled for 3 min, allowed to anneal at 60°C for 10 min and then room temperature for 10 min, mixed with an equal volume of 2x hybridization buffer (50% formamide, 10x SSC, and 0.2% SDS), and then hybridized to microarray at 42°C overnight. Excess probe was removed by a series of 4 min washes in 1x SSC, 0.2% SDS at 42°C; 0.1x SSC, 0.2% SDS at room temperature; and 0.1x SSC at room temperature. Slides were scanned using an Axon 4000B scanner, and images were analyzed with GenePix software.
All data analyses were conducted using log-transformed values (median pixel intensities) generated by the GenePix software.
Clone Tracking
To identify and correct possible errors in clone tracking, 420 cDNA clones (of the initial set of 20,256) were resequenced from the stock bacterial 384-well plates. Two clones were selected from different positions from each 96-well quadrant (there are four quadrants per 384-well plate). These sequences were tested against existing EST sequences in the database. A PERL script was used to identify expected matches, possible lane-tracking errors, quadrant or plate swaps, or errors in quadrant or plate orientation. In the majority of cases, one or two sequences were obtained from each quadrant and matched expected database sequences, thus confirming tracking accuracy. In cases where a sequence was not obtained or did not match the expected sequence, two additional clones were grown and sequenced. Tracking errors affecting whole quadrants were indicated for 16 (of 212 total) quadrants, including quadrant swaps, duplicate sequencing of quadrants, and quadrants in which database sequences were in an upside-down orientation with respect to the actual clones. The exact nature of each quadrant error was determined (in all cases, the initial determination was confirmed by additional sequencing) and corresponding sequence entrees in the database were corrected to reflect their true plate positions. Lane-tracking errors (i.e., ABI 377 generated sequences that drift from one lane into a neighboring lane) were not observed.
After rearraying the 8872 clones to be used for the microarray, an additional 192 cDNA clones were regrown and sequenced to verify tracking integrity (two clones were picked from each 96-well quadrant, as above). From these, 136 high-quality sequences were obtained and tested for identity with the expected EST. Only one sequence of the 136 tested did not match the expected EST, suggesting that clone tracking was close to 99% accurate at this stage.
WEB SITE REFERENCES
http://titan.biotec.uiuc.edu/bee/honeybee_project.htm.
http://www.fruitfly.org; Berkeley Drosophila Genome Project (BDGP).
http://www.geneontology.org; The Gene Ontology Consortium (2001).
http://www.genome.washington.edu/UWGC; University of Washington Genome Center.
http://megasun.bch.umontreal.ca/ogmpproj.html; Organelle Genome Megasequencing Project, University of Montreal.
http://www.ncbi.nim.nih.gov; National Center for Biotechnology Information (NCBI).
Acknowledgments
We thank L. Hood and D. Smoller for helpful discussions; A.J. Ross, S. O'Brien, and A. Cziko for bee collections; S. O'Brien for bee brain dissections; D. Toma for RNA extraction; M. Rebeiz for assistance with PERL programming; A. Cziko for assistance in microarray fabrication; and R. Hoskins, S. Clough, and members of the Robinson lab for reviewing the manuscript. Special thanks to H.A. Lewin, Director of the Keck Center, for excellent advice throughout the project and his tireless and creative efforts to facilitate genomics research on this campus. This research was supported by an NSF Postdoctoral Fellowship in Bioinformatics (C.W.W.) and grants from the University of Illinois Critical Research Initiatives Program and the Burroughs Wellcome Trust (G.E.R.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL generobi@life.uiuc.edu; FAX (217) 244-3499.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.5302.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Andrews J, Bouffard GG, Cheadle C, Lu J, Becker KG, Oliver B. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 2000;10:2030–2043. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Brown EL, Hunter CP. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 2001;29:E29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Brillet, C., Robinson, G.E., Bues, R., and Le Conte, Y. 2001. Racial differences in division of labor in colonies of the honey bee, Apis Mellifera. Ethology 2002. In press.
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Capaldi EA, Smith AD, Osborne JL, Fahrbach SE, Farris SM, Reynolds DR, Edwards AS, Martin A, Robinson GE, Poppy GM, et al. Ontogeny of orientation flight in the honeybee revealed by harmonic radar. Nature. 2000;403:537–540. doi: 10.1038/35000564. [DOI] [PubMed] [Google Scholar]
- Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. Normalization and subtraction of cap-trapper–selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000;10:1617–1630. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Diehl F, Grahlmann S, Beier M, Hoheisel JD. Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res. 2001;29:E38. doi: 10.1093/nar/29.7.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G, Casavant TL, Chang S, Scheetz T, Roberts C, Donohue M, Schultz J, Benes V, Bork P, Ansorge W, et al. Anopheles gambiae pilot gene discovery project: Identification of mosquito innate immunity genes from expressed sequence tags generated from immune-competent cell lines. Proc Natl Acad SciUSA. 2000;97:6619–6624. doi: 10.1073/pnas.97.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Wheeler DE. Expression profiles during honeybee caste determination. Genome Biol. 2001;2:research0001.1–0001.6. doi: 10.1186/gb-2000-2-1-research0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrbach SE, Robinson GE. Behavioral development in the honey bee: Toward the study of learning under natural conditions. Learn Mem. 1995;2:199–224. doi: 10.1101/lm.2.5.199. [DOI] [PubMed] [Google Scholar]
- Fiala A, Muller U, Menzel R. Reversible downregulation of protein kinase A during olfactory learning using antisense technique impairs long-term memory formation in the honeybee, Apis mellifera. J Neurosci. 1999;19:10125–10134. doi: 10.1523/JNEUROSCI.19-22-10125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Creating the gene ontology resource: Design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold D, Caskey CT. It's the genes! EST access to human genome content. Bioessays. 1996;18:973–981. doi: 10.1002/bies.950181207. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt GJ, Guzman-Novoa E, Fondrk MK, Page RE., Jr Quantitative trait loci for honey bee stinging behavior and body size. Genetics. 1998;148:1203–1213. doi: 10.1093/genetics/148.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt GJ, Page RE., Jr Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics. 1995;139:1371–1382. doi: 10.1093/genetics/139.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt GJ, Page RE, Jr, Fondrk MK, Dullum CJ. Major quantitative trait loci affecting honey bee foraging behavior. Genetics. 1995;141:1537–1545. doi: 10.1093/genetics/141.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Ball EE, Hayward DC, Maleszka R. Molecular cloning and expression analysis of a cDNA encoding a glutamate transporter in the honey bee brain. Gene. 2000;242:399–405. doi: 10.1016/s0378-1119(99)00503-x. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka R. Evaluation of differential gene expression during behavioral development in the honeybee using microarrays and northern blots. Genome Biol. 2002;3:research0007.1–0007.9. doi: 10.1186/gb-2002-3-2-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka R, Hayward DC, Ball EE. A royal jelly protein is expressed in a subset of Kenyon cells in the mushroom bodies of the honey bee brain. Naturwissenschaften. 1998;85:343–346. doi: 10.1007/s001140050512. [DOI] [PubMed] [Google Scholar]
- Maleszka R, Helliwell P, Kucharski R. Pharmacological interference with glutamate re-uptake impairs long-term memory in the honeybee, Apis mellifera. Behav Brain Res. 2000;115:49–53. doi: 10.1016/s0166-4328(00)00235-7. [DOI] [PubMed] [Google Scholar]
- Menzel R. Searching for the memory trace in a mini-brain, the honeybee. Learn Mem. 2001;8:53–62. doi: 10.1101/lm.38801. [DOI] [PubMed] [Google Scholar]
- Omholt SW, Rishovd S, Elmholdt O, Dalsgard B, Fromm S. Successful production of chimerical honeybee larvae. J Exp Zool. 1995;272:410–412. [Google Scholar]
- Page RE, Robinson GE. The genetics of division of labour in honey bee colonies. Adv Insect Physiol. 1991;23:117–171. [Google Scholar]
- Pellett FC. History of American beekeeping. Ames, Iowa: Collegiate Press; 1938. [Google Scholar]
- Porcel BM, Tran AN, Tammi M, Nyarady Z, Rydaker M, Urmenyi TP, Rondinelli E, Pettersson U, Andersson B, Aslund L. Gene survey of the pathogenic protozoan Trypanosoma cruzi. Genome Res. 2000;10:1103–1107. doi: 10.1101/gr.10.8.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GE. The regulation of division of labor in insect societies. Annu Rev Entomol. 1992;37:637–665. doi: 10.1146/annurev.en.37.010192.003225. [DOI] [PubMed] [Google Scholar]
- Robinson GE. From society to genes with the honey bee. Amer Sci. 1998;86:456–462. [Google Scholar]
- Robinson GE. Integrative animal behaviour and sociogenomics. Trends Ecol Evol. 1999;14:202–205. doi: 10.1016/s0169-5347(98)01536-5. [DOI] [PubMed] [Google Scholar]
- Robinson KO, Ferguson HJ, Cobey S, Vaessin H, Smith BH. Sperm-mediated transformation of the honey bee, Apis mellifera. Insect Mol Biol. 2000;9:625–634. doi: 10.1046/j.1365-2583.2000.00225.x. [DOI] [PubMed] [Google Scholar]
- Ronglin Y, Hagen A, Omholt SW. Cryopreservation of totipotent nuclei from honeybee (Apis mellifera) embryos by rapid freezing. Cryobiology. 1997;35:41–45. [Google Scholar]
- Rothenbuhler WC. Genetic and evolutionary considerations of social behavior of honeybees and some related insects. In: Hirsch J, editor. Behavior-genetic analysis. New York: McGraw-Hill; 1967. pp. 61–106. [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Shapira M, Thompson CK, Soreq H, Robinson GE. Changes in neuronal acetylcholinesterase gene expression and division of labor in honey bee colonies. J Mol Neurosci. 2001;17:1–12. doi: 10.1385/JMN:17:1:1. [DOI] [PubMed] [Google Scholar]
- Shoemaker DD, Schadt EE, Armour CD, He YD, Garrett-Engele P, McDonagh PD, Loerch PM, Leonardson A, Lum PY, Cavet G, et al. Experimental annotation of the human genome using microarray technology. Nature. 2001;409:922–927. doi: 10.1038/35057141. [DOI] [PubMed] [Google Scholar]
- Toma DP, Bloch G, Moore D, Robinson GE. Changes in period mRNA levels in the brain and division of labor in honey bee colonies. Proc Natl Acad Sci USA. 2000;97:6914–6919. doi: 10.1073/pnas.97.12.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston ML. The biology of the honey bee. Cambridge, Massachusetts: Harvard University Press; 1987. [Google Scholar]