Abstract
DNA methylation is a covalent modification of the nucleotide cytosine that is stably inherited at the dinucleotide CpG by somatic cells, and 70% of CpG dinucleotides in the genome are methylated. The exception to this pattern of methylation are CpG islands, CpG-rich sequences that are protected from methylation, and generally are thought to be methylated only on the inactive X-chromosome and in tumors, as well as differentially methylated regions (DMRs) in the vicinity of imprinted genes. To identify chromosomal regions that might harbor imprinted genes, we devised a strategy for isolating a library of normally methylated CpG islands. Most of the methylated CpG islands represented high copy number dispersed repeats. However, 62 unique clones in the library were characterized, all of which were methylated and GC-rich, with a GC content >50%. Of these, 43 clones also showed a CpGobs/CpGexp >0.6, of which 30 were studied in detail. These unique methylated CpG islands mapped to 23 chromosomal regions, and 12 were differentially methylated regions in uniparental tissues of germline origin, i.e., hydatidiform moles (paternal origin) and complete ovarian teratomas (maternal origin), even though many apparently were methylated in somatic tissues. We term these sequences gDMRs, for germline differentially methylated regions. At least two gDMRs mapped near imprinted genes, HYMA1 and a novel homolog of Elongin A and Elongin A2, which we term Elongin A3. Surprisingly, 18 of the methylated CpG islands were methylated in germline tissues of both parental origins, representing a previously uncharacterized class of normally methylated CpG islands in the genome, and which we term similarly methylated regions (SMRs). These SMRs, in contrast to the gDMRs, were significantly associated with telomeric band locations (P = .0008), suggesting a potential role for SMRs in chromosome organization. At least 10 of the methylated CpG islands were on average 85% conserved between mouse and human. These sequences will provide a valuable resource in the search for novel imprinted genes, for defining the molecular substrates of the normal methylome, and for identifying novel targets for mammalian chromatin formation.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF484557–AF484583.]
DNA methylation is central to many mammalian processes including embryonic development, X-inactivation, genomic imprinting, regulation of gene expression, and host defense against parasitic sequences, as well as abnormal processes such as carcinogenesis, fragile site expression, and cytosine to thymine transition mutations. DNA methylation in mammals is achieved by the transfer of a methyl group from S-adenosyl-methionine to the C5 position of cytosine. This reaction is catalyzed by DNA methyltransferases and is specific to cytosines in CpG dinucleotides. Seventy percent of all cytosines in CpG dinucleotides in the human genome are methylated and prone to deamination, resulting in a cytosine to thymine transition. This process leads to an overall reduction in the frequency of guanine and cytosine to about 40% of all nucleotides and a further reduction in the frequency of CpG dinucleotides to about a quarter of their expected frequency (Bird 1986). The exception to CpG underrepresentation in the genome is CpG islands, which were first identified as Hpa II tiny fragments (Bird et al. 1985), and were later formally defined as sequences >200 bp in length, with a GC content >0.5, and a CpGobs/CpGexp (observed to expected ratio based on GC content) >0.6 (Gardiner-Garden and Frommer 1987). CpG islands have been estimated to constitute 1%–2% of the mammalian genome (Antequera and Bird 1993), and are found in the promoters of all housekeeping genes, as well as in a less conserved position in 40% of genes showing tissue-specific expression (Larsen et al. 1992). The persistence of CpG dinucleotides in CpG islands is largely attributed to a general lack of methylation of CpG islands, regardless of expression status (reviewed in Cross and Bird 1995).
Although CpG islands are believed to be unmethylated, two exceptions to this rule in normal cells are the inactive X chromosome (Yen et al. 1984) and imprinted genes (Ferguson-Smith et al. 1993; Razin and Cedar 1994; Barlow 1995), both of which are associated with methylated CpG islands. Genomic imprinting is the parental origin-specific differential expression of the two alleles of a gene, and most imprinted genes show differential germline methylation of associated CpG islands (reviewed in Ohlsson et al. 2001). A third exception to the rule of methylation exclusion of CpG islands is aberrant methylation of CpG islands in tumors and in immortalized cultured cells, and such CpG island methylation is thought to contribute to carcinogenesis (Herman et al. 1994; Merlo et al. 1995).
Because of the interest in DNA methylation, genomic imprinting, and cancer, several general approaches have been used to identify CpG islands that are differentially methylated in specific cell types, such as screening tumor-normal pairs for cancer-related methylation changes (Huang et al. 1999; Shiraishi et al. 1999; Toyota et al. 1999), or pronuclear transplantation to examine differential parental origin for imprinted genes (Hayashizaki et. 1994; Plass et al. 1996). However, there has been only one report of a systematic effort to identify normally methylated CpG islands throughout the genome (Brock and Bird 1997), using a methyl-CpG binding column. The resulting sequences were ribosomal DNA and other repeated sequences with no characterization of unique, methylated CpG islands (Brock and Bird 1997). Here, we have taken a different approach, using a restriction enzyme-based library cloning strategy, and we have identified novel unique normally methylated CpG islands.
RESULTS
Isolation of Normally Methylated CpG Islands
We chose a restriction enzyme-based strategy for isolating methylated CpG islands over a PCR-based strategy, to avoid known problems of amplification bias against GC-rich sequences, and to obtain larger clone inserts than would be possible by a PCR-based approach. We used DNA from tissue from a male, to avoid cloning methylated CpG islands from the inactive X chromosome, and to avoid cell culture-induced DNA methylation. The tissue we chose was a Wilms tumor, because this approach would identify either normally methylated CpG islands or those methylated specifically in this tumor, which is of interest to our laboratory. Our plan was to determine after cloning these sequences whether they were methylated in normal cells or in tumors. The first step of our approach (Fig. 1) involved double digestion with Mse I, which recognizes the sequence TTAA and Hpa II, which recognizes the sequence CCGG at unmethylated sites. Mse I digests DNA between CpG islands, and Hpa II digests unmethylated CpG islands into small fragments, as it has a 4-bp recognition sequence. These digestions were followed by gel purification of fragments >1 kb in length. These initial digestions and purification were predicted by computer analysis of GenBank to enrich ∼10-fold for CpG islands, and enrichment of known methylated CpG islands (near imprinted genes) was confirmed by Southern blot hybridization (L.Z. Strichman-Almashanu and A.P. Feinberg, data not shown). At the same time, this step eliminates all unmethylated CpG islands because of the methylcytosine sensitivity of Hpa II. The restriction fragments obtained by this first step then were cloned into the restriction-negative strain XL2-Blue MRF‘ to avoid bacterial digestion of methylated genomic DNA, and the resulting genomic library was termed the “Mse library.” The second cloning step (Fig. 1) involved further enrichment of CpG islands by digesting the purified Mse I library DNA with an infrequently cutting restriction endonucleases (i.e., recognizing ≥ 6 bp CG-rich sequences) specific for sequences common to CpG islands, to isolate relatively large fragments of CpG islands that are normally methylated (i.e., survived the first cloning step), but are now unmethylated in the Mse library and therefore amenable to digestion and subcloning. Most of the work described here was performed by using Eag I (recognition sequence CGGCCG) in this second step, and subcloning Eag I fragments in three size classes separated by agarose gel electrophoresis (100–500 bp, 500–1000 bp, >1000 bp), and the resulting library was termed the Eag library.
Figure 1.
Overall strategy for cloning methylated CpG islands. In step 1, genomic DNA was digested with Mse I (red), which cuts between CpG islands, and Hpa II (blue), which cuts unmethylated CpG islands. Mse I fragments containing methylated CpG islands then are transformed into a bacterial strain that does not cut methylated DNA. However, brief bacterial passage leads to loss of methylation of these previously methylated sequences. In step 2, the library DNA is pooled and digested with Eag I (green), which cuts relatively large fragments within CpG islands, and these fragments are then subcloned.
Methylated CpG Islands within Interspersed Repeats
Our primary goal was to identify unique methylated CpG islands throughout the genome. However, it quickly became apparent that most of the clones in the Eag library represented high copy number methylated CpG islands. The majority of these clones were derived from a sequence termed SVA, which constituted 70% of the Eag I library, and that was not previously known to be methylated. The little-known SVA retroposon contains a GC-rich VNTR region, which embodies a CpG island between an Alu-derived region and an LTR-derived region. Only three such elements had previously been described (Kawajiri et al. 1986; Zhu et al. 1992; Shen 1994), although their methylation has not been characterized. We designed a probe, termed SVA-U, unique to the SVA and present in all of the SVA elements, to analyze copy number and methylation of this sequence in genomic DNA. The copy number was estimated by quantitative Southern hybridization to be 5000 per haploid genome (L.Z. Strichman-Almashanu and A.P. Feinberg, data not shown). The SVA elements were found to be completely methylated in all adult somatic tissues examined, including peripheral blood lymphocytes, kidney, adrenal, liver and lung. A somewhat less abundant high copy repeat, representing an additional 20% of the Eag I library, corresponded to the nontranscribed intergenic spacer of ribosomal DNA, which was a known methylated repetitive sequence (Brock and Bird 1997), suggesting that ribosomal gene methylation may be more extensive than was previously suspected. The focus of the current study was on the unique methylated CpG islands that were identified after excluding these sequences.
Methylation Analysis of Novel Single-Copy CpG Islands
To isolate single-copy clones, we rederived the Mse library, adding restriction endonucleases designed to cleave repeat sequences described above, rendering them unclonable (see Methods). After eliminating redundant clones, 62 unique clones were characterized. All of the sequences were GC-rich, i.e., with a measured (C + G)/N >50%, and they ranged in GC content from 55 to 79%. Forty-three (69%) of the clones showed an observed to expected CpG ratio >0.6, meeting the formal definitional requirement of a CpG island, and they were characterized further. Nevertheless, most of the remaining clones showed an observed to expected CpG ratio >0.5.
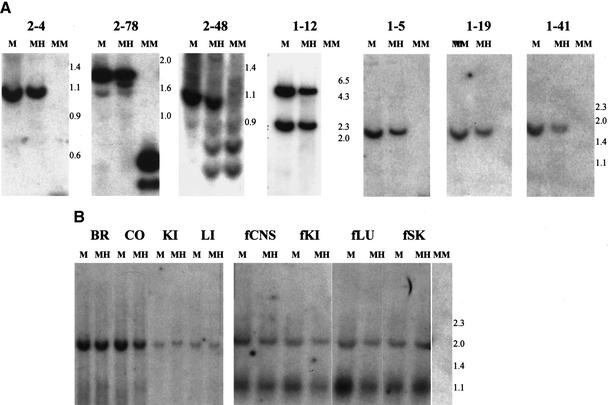
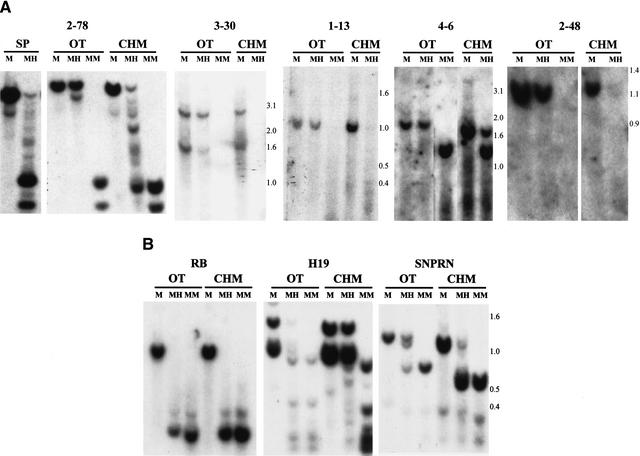
As the original source of DNA was a Wilms tumor, we had no a priori knowledge of the methylation status of these sequences in normal tissue. Surprisingly, all of the sequences were methylated in normal lymphocyte DNA (Fig. 2A). Methylation was not restricted to lymphocyte DNA, as it also was observed in both adult and fetal tissues, including brain, gut, kidney, liver, lung, and skin (Fig. 2B). Thus, these sequences represented normally methylated CpG islands. To determine whether the CpG islands were differentially methylated in the maternal and paternal germline, 30 of the clones were individually hybridized to Southern blots of DNA isolated from ovarian teratomas (OT) and complete hydatidiform moles (CHM), which are of uniparental maternal and paternal origin, respectively (CHM DNA was exhausted at that point). Thirteen clones exhibited methylation in the OT but not or significantly less so in the CHM (Table 1). For example, CpG island 2–78 showed complete digestion after Hpa II treatment of genomic DNA isolated from sperm and CHM, similar to the pattern after Msp I digestion (Fig. 3A). In contrast, 2–78 showed an identical pattern after Mse I + Hpa II digestion, as after Mse I alone, in OT (Fig. 3A). Similarly, Fig. 3A shows OT-specific methylation of CpG islands 3–30, 1–13, 4–6, and 2–48, with relative lack of methylation in CHM. These sequences therefore represent differentially methylated regions, because of their different pattern of methylation in germline tissues of male (sperm and CHM) and female (OT) origin. Because many of these sequences also are methylated in somatic tissues, we refer to them as gDMR’s (germline differentially methylated regions). All of the gDMR sequences were methylated in OT and not CHM. As a negative control, a CpG island associated with the RB gene is unmethylated in both CHM and OT. As a positive control, a CpG island upstream of the imprinted gene H19 is preferentially methylated in CHM, and a CpG island within the imprinted SNRPN gene is methylated in OT (Fig. 3B).
Figure 2.
Methylation of CpG islands in normal human DNA. Genomic DNA from peripheral blood lymphocytes (A) or tissues (B) was digested with Mse I (M), Mse I + Hpa II (MH), or Mse I + Msp I (MM). Fragment sizes are indicated to the right. CpG islands used for Southern blot hybridization are indicated in panel A, and CpG island clone 1–19 was used in panel B. Note that there is an Mse I polymorphism in the fetal tissue that is not in the adult tissue, accounting for the presence of two bands in the fetal tissue Mse I digest. Blots were made in duplicate and one set was hybridized to RB to ensure the presence of DNA in the Msp I lane. BR, brain; CO, colon; KI, kidney; LI, liver; fCNS, fetal CNS; fKI, fetal kidney; fLU, fetal lung; fSK, fetal skin.
Table 1.
Methylated CpG Islands Characterized in Detail
| Clone ID | Methylation pattern | Chromosomal location | Associated genes | |
|---|---|---|---|---|
| Accession no. | Predicted function | |||
| 3-10 | SMR | 1q44 | 14042537a | Similar to Zn finger protein |
| 14423768 | Olfactory receptor 2T1 | |||
| hCG1644736 | Similar to olfactory receptor 2T1 | |||
| hCG1724357 | Similar to olfactory receptor 2T1 | |||
| 3-20 | SMR | 2q36 | 5174481a | Histone deacetylase A |
| hCG1651464 | ||||
| hCG1656118 | ||||
| hCG1651461 | ||||
| hCG1651466 | ||||
| 1-19 | SMR | 4p16 | 3777583a | WFS1 (wolframin) |
| 1-41 | SMR | 4q35 | hCG1788598a | Similar to mouse pair-rule gene ODZ3 |
| hCG1793025a | Hypothetical protein | |||
| hCG1787540 | Similar to mouse pair-rule gene ODZ3 | |||
| 4-8 | SMR | 4q35 | hCG1788598a | Adjacent to but distinct from 1-41 |
| 3-4 | gDMR | 6q24 | hCG1660630a | HYMA1 |
| 4-7 | SMR | 7p22 | 6806913a | Centaurin-α |
| hCG1747708 | ||||
| hCG1790856 | ||||
| hCG1747710 | Cytochrome P450 homolog | |||
| 1-30 | SMR | 7q11.1 | hCG1779529a | Rab5 exchange factor homolog |
| hCG1779527 | Antisense to hCG1779529 | |||
| hCG1789113 | Antisense to hCG1779529 | |||
| 13642872 | 60S ribosomal prot L35 | |||
| 6572672 | Putative transcription factor | |||
| 1-22 | SMR | 7q36 | 11386149a | Tyrosine phosphatase |
| hCG1799787 | BHLF1 protein | |||
| 3-2 | SMR | 8p23 | hCG1659058a | Proline-rich mucin homolog |
| 2-5 | gDMR | 8q21.2 | 17451956 | Similar to antigen GOR |
| hCG1757665 | ||||
| 2-48 | gDMR | 9p13 | hCG1659616 | |
| 1-20 | gDMR | 10q26 | 13325182a | |
| hCG1799063 | ||||
| 3-12 | SMR | 10q26 | 3122245a | Inositol triphosphate phosphatase |
| hCG1654478 | ||||
| 3-30 | gDMR | 11q25 | 17456499a | Hypothetical gene |
| hCG37607 | ||||
| hCG1745526 | Hypothetical protein | |||
| 1-5 | SMR | 13q34 | hCG20146a | |
| 1-21 | gDMR | 14q32 | 8393715a | Heptacellular cancer candidate |
| hCG21408 | Similar to Drosophila CLIP-190 | |||
| 2-42 | gDMR | 16p13.1 | hCG15669a | Ser/Thr protein kinase |
| 3-110 | SMR | 17q25 | 8400736a | β-tubulin cofactor D |
| 10435982 | ||||
| 2-1 | SMR | 17q25 | 1655842a | Sulfamidase |
| 14149793 | Coiled-coil protein | |||
| 1-12 | SMR | 17q25 | hCG1806389 | |
| hCG1796817 | ||||
| 2-78 | gDMR | 18q21 | 13645769a | Elongin A3 |
| 3-8 | gDMR | 18q21 | 13645769a | Adjacent to but distinct from 2-78 |
| 2-3 | SMR | 18q23 | 6912444 | Voltage-dependent K+ channel |
| 1914872 | Choline-binding protein | |||
| 5326898 | RNA Polymerase II CTD phosphatase | |||
| 1-13 | gDMR | 18q23 | hCG1651089a | |
| 6688241 | SALL3 Spalt-like zinc finger protein | |||
| hCG20372 | ATPase | |||
| 1651088 | Mucin1 precursor | |||
| 1-6 | gDMR | 19p13.1 | 14249150a | Hypothetical protein |
| 4-3 | SMR | 19p13.3 | 9665054a | Ser/Thr kinase 11 |
| hCG23965 | Ankyrin repeat protein | |||
| 4506715 | Ribosomal protein S28 | |||
| hCG1794585 | ||||
| 10732648 | Angiopoietin-like protein | |||
| 1-2 | SMR | 19q13.4 | 12053197 | Zinc finger protein |
| 4505329 | NSF attachment protein | |||
| 5901994 | Kaptin-actin binding protein | |||
| 5689511 | Na/Ca exchanger protein | |||
| 7657128 | Glioma tumor suppressor candidate | |||
| 7657054 | EH-Domain containing protein | |||
| 7657130 | Glioma tumor suppressor candidate | |||
| 2-4 | gDMR | 20q12 | 17484155a | Nuclear factor of activated T-cells 2 |
| hCG1800975 | ||||
| hCG1653833 | ||||
| 13378306 | Brain RPTmam4 isoform | |||
| 110743 | Neurofilament triplet H protein | |||
| 7799072 | DNA helicase | |||
Accession numbers correspond to GenBank entries, within 10 kb of the CpG island, unless there is no GenBank entry, in which case they correspond to Celera entries. One additional SMR could not be mapped.
CpG island lies within the transcript.
Figure 3.
Differential methylation of novel gDMRs in uniparental tissues of germline origin. Fragment sizes (kb) are indicated to the right. (A) Sperm (SP), ovarian teratoma (OT), or complete hydatidiform mole (CHM) was digested, and Southern blot hybridization was performed with the gDMRs indicated, as described in the legend to Figure 2. Multiple OT and CHM were examined with similar results, although only one is shown. (B) Similar experiments were performed with an unmethylated CpG island in the retinoblastoma gene (RB), with a CpG island upstream of H19 that shows preferential methylation of the paternal allele, and with a CpG island within the SNRPN gene that shows preferential methylation of the maternal allele.
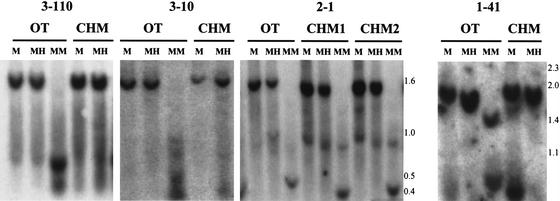
An additional 17 clones identified CpG islands that were methylated equally in OT, CHM, and sperm (Table 1). For example, CpG islands 3–110, 3–10, 2–1, and 1–41 showed an identical pattern after Mse I + Hpa II digestion, as after Mse I alone, in OT and CHM (Fig. 4). We termed these sequences SMRs, to connote their comparable methylation in male and female tissue of germline origin. Like the gDMRs, these SMRs were methylated in cells of somatic origin (Fig. 2A).
Figure 4.
Similar methylation of novel SMRs in uniparental tissues of germline origin. Experiments were performed as described in the legend to Figure 2, using the SMRs indicated. Fragment sizes are indicated to the right.
Chromosomal Location of Methylated CpG Islands and Association with Genes
The methylated CpG islands identified here were distributed throughout the genome. There was a striking localization of SMRs near the ends of chromosomes. Sixteen of 17 SMRs were localized near the ends of chromosomes, either on the last (n = 15) or the penultimate (n = 1) subband of the chromosome on which it resided (Table 2). In contrast, of 12 gDMRs that could be mapped (of the 13 gDMRs studied), only four were localized near the ends of chromosomes (Table 2). This difference was highly statistically significant (P = .0008, Fisher's exact test). The association of SMRs near the ends of chromosomes is consistent with an observation of densely methylated GC-rich sequences near telomeres, although that study did not describe methylated CpG islands (Brock et al. 1999). In addition, there was a segregation of gDMRs and SMRs within compartments of differing genomic composition, i.e., isochores, which are regions of several hundred kilobases of relatively homogeneous GC composition (Bernardi 1995). Approximately 75% of the SMRs fell within high isochore regions (G+C ≥50%), as might be expected from the high GC content of methylated CpG islands. Surprisingly, however, all of the gDMRs fell within low isochore regions (G+C <50%), i.e., of relatively low GC content, despite the high GC content of the gDMRs themselves (L.Z. Strichman-Almashanu and A.P. Feinberg). This difference was statistically significant (P<.01, Fisher's exact test). Thus, the gDMRs and SMRs may lie within distinct chromosomal and/or isochore compartments.
Table 2.
Band Location of Methylated CpG Islands
| CpG Island | Band location | ||
|---|---|---|---|
| Centromeric | Midchromosome | Telomerica | |
| gDMR | 0 | 8 | 4 |
| SMR | 1 | 0 | 16 |
P = 0.0008, Fisher's exact test.
One SMR was subtelomeric.
There were several examples of nonredundant, unique methylated CpG islands localizing to the same chromosomal region. In two cases, two pairs of sequences were adjacent within the genome. Two SMRs on 4q35, 1–41 and 4–8, were adjacent to each other; and two gDMRs on 18q21, 2–78 and 3–8, also were adjacent to each other (Table 1). In addition, 14 methylated CpG islands were located near and on the same chromosomal subband as other methylated CpG islands (Table 1). For example, SMRs 3–110, 2–1, and 1–12 are all on 17q25; two of these sequences, 3–110 and 1–12, lie within 660 kb (data not shown). In some cases, SMRs and gDMRs were found in relatively close proximity. For example, SMR 2–3 and gDMR 1–13 lie within 1 Mb on 18q23. In addition, gDMR 1–20 and SMR 3–12 are both on 10q26 and separated by ∼800 kb (Table 1 and R.S. Lee and A.P. Feinberg, data not shown). All of these data together support the idea that these methylated CpG islands identify specific portions of the genome.
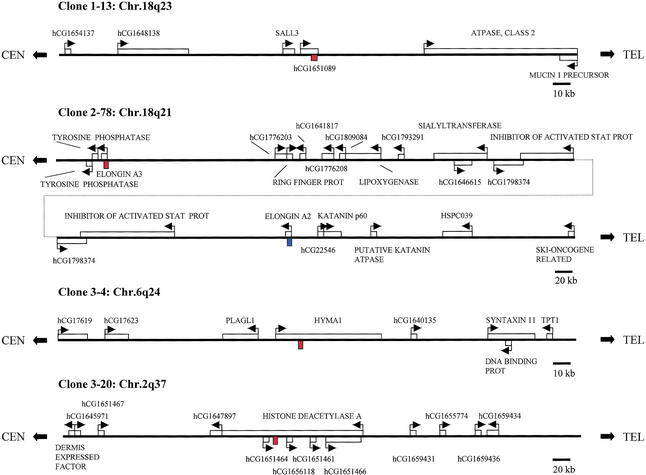
Most of the methylated CpG islands were localized within or near the coding sequence of known genes or of anonymous ESTs within the GenBank or Celera databases. Because of the known ability of DMRs to regulate imprinting over long distances (reviewed in Feinberg 2001), we determined the identity of known or predicted genes within several hundred kilobases of each methylated CpG island. We were particularly intrigued that gDMR 3–4 was located on 6q24 within HYMA1 (Fig. 5), an imprinted gene involved in diabetes mellitus (Arima et al. 2000). This CpG island has been identified independently as a DMR, in a specific analysis of this gene (Arima et al. 2001), and our isolation of this sequence indicates that these methylated CpG islands may identify imprinted gene domains. gDMR 1–13 was located on 18q23, within a predicted gene of unknown function, and near the SALL3 gene (Fig. 5), which encodes a Spalt-like zinc finger protein that is a candidate gene for 18q deletion syndrome (10610715), which involves preferential loss of the paternal allele (Kohlhase et al. 1999). Interestingly, 18q23 also has been implicated in bipolar affective disorder, specifically harboring a predisposing gene transmitted preferentially through the father (Stine et al. 1995; McMahon et al. 1997). Therefore, the localization of this gDMR may serve as a guidepost for identifying candidate imprinted genes for this important disease. SMR 1–2 was located within 19q13.4 (Fig. 5). Even though this sequence is an SMR, 19q13.4 contains the imprinted genes PEG3 and ZIM1 (Kim et al. 1999). Given that SMR 1–2 is ∼10 Mb from these genes, it is unlikely to lie within the same imprinted gene domain. Nevertheless, it will be of interest to examine nearby genes for their imprinting status, including a glioma tumor suppressor candidate gene located 110 kb telomeric to SMR 1–2. Another interesting gene harboring a methylated CpG island was histone deacetylase A (HDAC4), and there were several other predicted genes near this CpG island, SMR 3–20 (Fig. 5). In addition, several antisense transcripts are associated with this CpG island. Given that HDAC4 is itself involved in chromatin remodeling (Wang et al. 2000), methylation of this region could be involved in a feedback loop controlling chromatin structure. Other genes located near methylated CpG islands included the wolframin gene, a transmembrane protein involved in congenital diabetes (Strom et al. 1998); several olfactory receptor genes; several phosphatase and kinase genes likely involved in signal transduction; several genes for DNA-interacting proteins; and the Peutz-Jeghers syndrome gene STK11 (Table 1). A voltage-dependent potassium channel subunit protein was localized only 16 kb from methylated CpG island 2–3 (Table 1), which is of interest given that the voltage-dependent potassium channel KvLQT1 is imprinted (Lee et al. 1997). Finally, in addition to genes directly adjacent to these methylated CpG islands, at least two of the domains flanked by methylated CpG islands harbored several genes within them that may play a role in cancer. For example, contained within the region defined by methylated CpG islands 3–110 and 1–12 are a predicted apoptosis inhibitor, a septin-like cell division gene, a ras homolog, and a predicted translation initiation factor (Table 1 and L.Z. Strichman-Almashanu and A.P. Feinberg, data not shown).
Figure 5.
Chromosomal location and relationship of representative methylated CpG islands to nearby genes. Genes are indicated with boxes, and the arrows show transcriptional orientation. The methylated CpG islands are shown in red. In the case of 2–78, the homologous sequence within Elongin A2 is indicated in blue.
Identification of an Imprinted Gene Homologous to Elongin A
In addition to HYMA1, described above, a DMR within the IGF2R contains an Eag I site, and as predicted, this gene also was found in the Eag library (L.Z. Strichman-Almashanu and A.P. Feinberg, data not shown). We recently have begun to examine genes near methylated CpG islands for allele-specific expression, and we already have found one novel imprinted gene. gDMR 2–78 was localized to 18q21 (Fig. 5) and was completely methylated in all somatic fetal and adult tissues tested (Fig. 2 and L.Z. Strichman-Almashanu and A.P. Feinberg, data not shown). However, this CpG island was unmethylated in CHM and sperm and methylated in OT (Fig. 3A). A BLAST search showed that the CpG island spanned the putative promoter region and body of a gene predicted by GENSCAN (http://genes.mit.edu/GENSCAN), and included 1638 nucleotides encoding 546 amino acids (Fig. 6). BLAST searches of GenBank and Celera databases using the predicted sequences revealed that the predicted gene showed 43% amino acid identity to human transcription elongation factor B (SIII) polypeptide 3 (TCEB3), also known as Elongin A. The novel sequence was even more closely related to a previously identified homolog of Elongin A termed Elongin A2, or TCEB3L, showing 79% amino acid sequence identity to human transcription elongation factor (SIII) Elongin A2 (TCEB3L). To determine whether 2–78 represented a genuine transcript, and if so, whether the gene is imprinted, we designed primers that would amplify 2–78 but not Elongin A2, and amplification products were of the expected size.Sequencing demonstrated that the amplified cDNA corresponded to 2–78 and not Elongin A2, based on sequence differences between the two genes within the PCR product. Analyzing DNA samples from fetal tissues, we then identified a polymorphism at nucleotide 910 (G/A) of 2–78. We found four fetuses heterozygous for this polymorphism, in which maternal decidua DNA was available and homozygous, allowing us to identify parental origin in the fetal samples (Fig. 7). Reverse transcriptase PCR (RT-PCR) analysis of tissues from these fetuses showed that the gene was indeed transcribed. We therefore term this gene Elongin A3. An alternative term is TCEB3L2, but for this term to apply, the nomenclature committee will need to rename TCEB3L (Elongin A2) TCEB3L1.
Figure 6.
Nucleotide and amino acid sequence of Elongin A3. The transcription factor SII similarity motif is shown in red, and the nuclear localization signal is shown in green. The site of the (G/A) polymorphism, at nucleotide 910, used for imprinting analysis is shown in blue, and the PCR primers specific for Elongin A3 are shown in black boldfaced type.
Figure 7.
Tissue-specific imprinting of Elongin A3. The (G/A) polymorphism was used to assess allele-specific expression in four heterozygous fetuses denoted A, B, C, and D. Chromatograms of genomic DNA (gDNA) sequence are included to show heterozygosity, as well as the homozygous maternal decidual DNA indicating parental origin. (A) Monoallelic expression of the maternal allele in lung, central nervous system (CNS), and limbs, and biallelic expression in kidney. (B) Monoallelic expression of the maternal allele in placenta and CNS, and biallelic expression in intestine. (C) Monoallelic expression of the maternal allele in CNS, biallelic expression in kidney and liver. (D) Monoallelic expression of the maternal allele in placenta, and biallelic expression in kidney and liver. Sequencing was done bidirectionally in all cases, and monoallelic expression of the maternal allele did not depend on whether that allele was A or G.
Analysis of allele-specific expression showed monoallelic expression of lung, brain, placenta, and spinal cord, with preferential expression from the maternal allele (Fig. 7A–D). There was incomplete preferential expression from the maternal allele in two of three kidneys (Fig. 7A,C), and absence of imprint-specific gene expression in one kidney and in the intestine or liver (Fig. 7B,C,D). Thus, Elongin A3 shows tissue-specific imprinting, at least in prenatal development. Therefore, the isolation of these novel CpG islands does enable the identification of novel human imprinted genes.
Species Conservation of Methylated CpG Islands
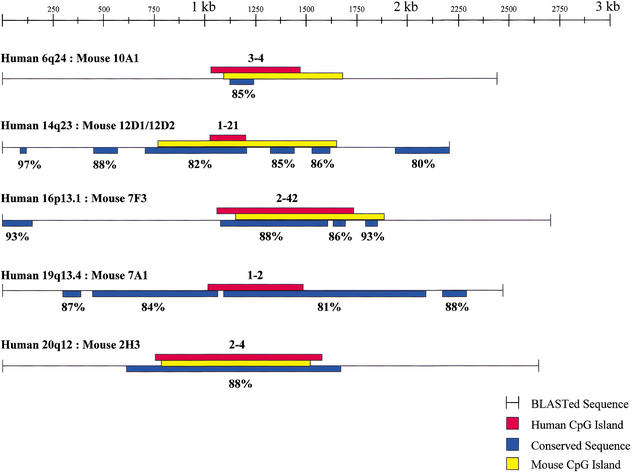
As further confirmation of the importance of the methylated CpG islands we have isolated, we ascertained their sequence conservation in the mouse, using the Celera mouse genome database. Thirteen (46%) of the 30 human noncontiguous methylated CpG islands matched sequences within the mouse genome at 86.9 ± 4.9% identity (Fig. 8 and R.S. Lee and A.P. Feinberg, data not shown). Furthermore, in some cases, the region of conservation extended beyond the CpG island itself. For example, gDMR 1–21 showed, in addition to a 558 bp, 82% conserved region including the CpG island, five additional conserved sequences within 1 kb of the CpG island. These additional sequences varied from 80–97% identity (Fig. 8). Most of the conserved sequences outside of the CpG islands themselves were not predicted genes, and thus may represent conserved regulatory sequences. In all cases in which BLAST analysis of the CpG island and flanking 1 kb on each side was performed, and in which any sequence conservation was found, the CpG island itself was conserved, again supporting the idea that these CpG islands play an important role.
Figure 8.
Sequence conservation of methylated CpG islands between human and mouse. Human methylated CpG islands and ∼1 kb of flanking DNA were compared to mouse sequence, synteny was confirmed, the corresponding mouse CpG islands were identified, and regions of conservation (percentage shown) were determined. In the case of gDMR 1–21, the corresponding mouse sequence, while GC-rich, showed an observed to expected CpG ratio of 0.45–0.50 and therefore was not classified as a CpG island. Colors are as indicated in the figure.
DISCUSSION
The major conclusion of this study is the identification of a subset of unique CpG islands that are methylated in normal tissues, in the first systematic effort to identify such sequences. The experiments were designed to identify CpG islands that are methylated differentially in germline-derived tissues or differentially in cancers. To our surprise, we found no CpG islands methylated specifically in tumors, but slightly more than one half of the unique methylated CpG islands were methylated in germline-derived tissues of both maternal and paternal origin. Conventional wisdom holds that CpG islands are unmethylated, with the exception of the inactive X chromosome, imprinted genes, and tumors. However, rare exceptions to this rule have been described. Some repeated sequences harboring CpG islands have been found to be methylated. We reported methylation of a mouse testis-specific histone H2B gene (Choi et al. 1996), and others have found methylation of some ribosomal gene sequences (Brock and Bird 1997). Indeed, methylation of one of these repeat sequences, the rDNA nontranscribed spacer, previously was found after genomic purification from a methyl-CpG binding protein column (Brock and Bird 1997), and we speculate that the large number of these sequences obscured the identification of unique methylated CpG islands. The methylation of high copy number sequences is not surprising, as it is consistent with the hypothesis that CpG methylation arose as a host defense mechanism (Bestor and Tycko 1996). This is particularly true of the SVA element, which is a high copy number retroposon.
However, the presence of normally methylated unique CpG islands has not been observed systematically. An intriguing exception is the MAGE melanoma gene (Serrano et al. 1996), and it is thought that hypomethylation of this gene leads to its activation in cancer (De Smet et al. 1996). Our results suggest that normally methylated single-copy CpG islands may be more abundant than previously believed. Indeed, the loss of methylation of such sequences may be related to gene activation in cancer, just as the gain of methylation of CpG islands may lead to their silencing. Previous screens for altered CpG island methylation have not been designed to identify normally methylated CpG islands, but it should be noted that the original observation of altered methylation in cancer was widespread loss of methylation (Feinberg and Vogelstein 1983). Furthermore, even in tumors that show increased CpG island methylation, the total methylation content is reduced (Feinberg et al. 1988). DNA methylation serves as an additional layer of genetic information in the genome, which has been termed the methylome (Feinberg 2001), and both increases and decreases may be important in cancer. Our strategy for cloning these sequences can be generalized to secondary libraries in addition to the Eag library, and the identification of additional such sequences thus should enhance our understanding of the methylome.
The second major result of this study was the identification of novel CpG islands that are methylated differentially in OT and CHM. The second (Eag) library would not identify known imprinted genes lacking Eag I sites, but it did contain the DMR of IGF2R, as well as the DMR of the imprinted HYMA1 gene, suggesting that this strategy also can identify novel imprinted gene domains. One such gene was identified to date, a novel homolog of the Elongin A and Elongin A2 genes, which we term Elongin A3. Both Elongin A and Elongin A2 are known to be the active components of the transcription factor B (SIII) complex (Aso et al. 2000), that may compete for other components (Elongin B and C) with the VHL tumor suppressor gene (Kibel et al. 1995). We did not check directly for elongation activity of Elongin A3, but it contains the TFS2N motif as well as a nuclear localization signal, and the predicted protein sequence is 79% identical to that of Elongin A2, so it likely does have such a function.
It should be noted that gDMRs, even the gDMR within this novel imprinted gene, showed variable to complete methylation in somatic tissues. Such a pattern of methylation also is similar to that seen for the promoter of the imprinted gene ZNF127 (Strom et al. 1998), and for at least one methylated CpG island within the 11p15 imprinted gene domain (S. Kane and A. Feinberg, unpubl.). Thus, imprinted gene domains may harbor some methylated CpG islands that show persistent differential methylation in somatic tissues, but also may contain other CpG islands that do not show these differences in somatic tissues. Thus, it is important to compare methylation in sperm or CHM as a representation of the male germline, and OT (as eggs cannot be harvested from humans for this purpose), in the search for imprinted gene domains. The mouse is a useful adjunct and provides access to a greater variety of tissues at varying developmental stages, but there are substantial differences between human and mouse imprinting, both in the identity of the genes themselves, and in their developmental pattern of imprinting.
The localization of these sequences likely will stimulate a great deal of research by many laboratories to identify novel imprinted genes near them. Several of these domains harbor multiple genes that have been implicated in cancer, and that show frequent loss of heterozygosity, including 4p16, 4q35, 10q26, 18q21, and 19p13. An imprinted tumor suppressor gene in one or more of these regions might not show conventional mutations in tumors, and thus identifying imprinted genes is an important part of tumor suppressor gene identification within these regions. The same region of 18q also has shown linkage in bipolar affective disorder, with preferential transmission through the paternal allele (McMahon et al. 1997). Furthermore, these domains appear to harbor both SMRs and gDMRs, suggesting that both types of methylated CpG islands may be useful for identifying imprinted gene domains.
What is the function of these normally methylated CpG islands? CpG islands normally must be under selective pressure for their maintenance, as methylation leads to deamination and loss of cytosine. This is especially true in the case of the SMRs we have described, as they are methylated even in sperm DNA. In the case of gDMRs, their methylation in somatic tissues and oocyte-derived cells may be critical for suppression of nearby gene expression in spermatocyte progenitor cells. This may be particularly important for genes involved in establishing epigenetic states and in epigenetic reprogramming, as the chromatin of spermatocyte differs markedly from oocytes and somatic cells.
It also is possible that normally methylated CpG islands are involved directly in chromatin formation. For example, they could serve as chromatin insulators separating enhancers from promoters. If that is so, then we would expect to find their loss of methylation in specific tissues at specific developmental stages, which would be consistent with the observation that imprinted genes can show developmental (tissue- and timing-specific) imprinting (Lee et al. 1997). Support for this idea also comes from our observation that SMRs were more frequently localized near the ends of chromosomes. Given that chromosomal ends are associated with the nuclear lamina in interphase (Cockell and Gasser 1999), the relative proximity of SMRs to the ends of chromosomes might permit their association with the nuclear lamina and chromatin proteins found within it.
Normally methylated CpG islands also might promote chromatin formation. In an intriguing review, Pardo-Manuel de Villena et al. (2000) suggest that imprinting involving differences among homologous chromosomes arose under selective pressure to facilitate pairing and distinguish homologous chromosomes during meiosis. We suggest that SMRs also might enhance pairing and recombination by recruiting chromatin factors to specific locations along a given chromosome and allowing those factors to interact between homologous chromosomes. A prediction of our suggestion is that recombination frequencies in meiosis or even mitosis might be enhanced near normally methylated CpG islands. Methylated CpG islands also may play a role intrachromosomal compartmentalization. For example, the gDMRs lay within regions of comparatively lower CpG content (GC-poor isochores). Consistent with this idea, we have noted that most known imprinted genes also appear to lie within low isochore regions (PLAGL1, IGF2R, PEG1/MEST, SNRPN, PEG3, GNAS, unpubl.).
Finally, the identification of these methylated CpG islands will facilitate comparison of their sequences to each other, as well as computational analysis of sequence motifs. For example, in preliminary experiments, we have identified several CTCF binding sites within at least 10 methylated CpG islands (R.S. Lee et al., unpubl.). We are currently performing biochemical experiments to determine whether CTCF binding is a common feature of these sequences.
METHODS
Isolation of Methylated CpG Islands from Genomic DNA
A two-step cloning procedure was used. In the first step, 200 μg of genomic DNA were digested overnight with 1000 units of Hpa II (LTI) followed by a 5-h digest with 600 units of Mse I (NEB), according to the manufacturer's conditions, and the volume was reduced using a SpeedVac concentrator (Savant). Fragments ≥1 kb were size selected using a Chromaspin+TE, 400 column (Clontech), and fragments between 1–9 kb were purified from a 0.8% gel by electroelution and an Elutip-D column (S&S). The eluate was ethanol precipitated, cloned into the compatible Nde I site of pGEM-4Z, which was first modified to abolish the Sma I site, transformed into the competent cells of the restriction-deficient strain XL2-Blue MRF‘ (Stratagene), and plated onto LB-ampicillin agar plates. Library DNA was prepared directly from plates using a plasmid Maxi kit (Qiagen). In the second step, 100 μg of the Mse I library DNA were digested with 1,000 U of Eag I (NEB) according to the manufacturer’s conditions. The digest was ethanol precipitated, and 100- –1500-bp fragments were size-selected by purification from a 1.5% agarose gel, cloned into the Eag I site of pBC (Stratagene), and transformed into XL1-Blue MRF‘ (Stratagene). To eliminate methylated CpG islands that corresponded to dispersed repetitive sequences, we derived the Mse I library adding restriction enzymes designed to cleave those sequences and render them unclonable. For 28S and ribosomal DNA, we used Asc I. For SVA, we used Dra III + Sal I, followed by either Acc I or TthIII1.
DNA Sequencing
DNA sequencing was performed using an ABI 377 automated sequencer following protocols recommended by the manufacturer (Perkin-Elmer). The sequences were analyzed by BLAST search (Altschul et al. 1990) of the GenBank and Celera databases.
Southern Hybridization
Genomic DNA was digested with Mse I alone or Mse I together with a methylcytosine-sensitive (Hpa II, LTI, or Sma I, NEB) or methyl-insensitive (Msp I or Xma I, NEB) restriction endonuclease according to the manufacturer’s conditions. Southern hybridization was performed as described (Dyson 1991).
Imprinting Analysis of Elongin A3 Gene
Fetal tissues and matched maternal decidua were obtained from the University of Washington Fetal Tissue Bank. We identified polymorphisms by sequencing fetal and maternal PCR amplified genomic DNA. The following conditions were used for PCR amplifications: 95°C, 2 min; then 40 cycles of 95°C 1 min, 60°C 30 sec, 72°C 1 min; then 72°C for 9 min. Total RNA was isolated from fetal tissues using RNeasy mini kit (Qiagen). To eliminate DNA contamination from RNA preparations, samples were treated with preamplification-grade DNase I (Invitrogen) according to supplied protocols. RT-PCR was carried out using the Superscript II preamplification system (Invitrogen) and was performed for each sample in the presence and absence (negative controls) of RT. cDNA samples were sequenced only when no bands were obtained with the negative controls. The primers used for the imprinting analysis were EL2AL-1093–1112F: 5′-TCT GCT GTC CGC TTT TGA GG -3′ and EL2AL-1526–1550R: 5′-ATC GGA TTT TCG TGG TCA CTA CTC G-3'. DNA and cDNA sequencing was run on an ABI-377 automated sequencer following protocols recommended by the manufacturer (Perkin-Elmer).
Acknowledgments
We thank J. Boeke, M. Boguski, S. Kern, B. Migeon, R. Ohlsson, and members of the Feinberg laboratory for helpful discussions. We thank Tracy Litzi for technical assistance. This work was supported by NIH grant CA65145 (A.P.F.). L. S.-A. was a student in the graduate program in human genetics, and R. S. L. is a student in the graduate program in biochemistry, cellular, and molecular biology.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL afeinberg@jhu.edu; FAX (410) 614–9819.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.224102. Article published online before print in March 2002.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird AP. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima T, Drewell RA, Arney KL, Inoue J, Makita Y, Hata A, Oshimura M, Wake N, Surani MA. A conserved imprinting control region at the HYMAI/ZAC domain is implicated in transient neonatal diabetes mellitus. Hum Mol Genet. 2001;10:1475–1483. doi: 10.1093/hmg/10.14.1475. [DOI] [PubMed] [Google Scholar]
- Arima T, Drewell RA, Oshimura M, Wake N, Surani MA. A novel imprinted gene, HYMAI, is located within an imprinted domain on human chromosome 6 containing ZAC. Genomics. 2000;67:248–255. doi: 10.1006/geno.2000.6266. [DOI] [PubMed] [Google Scholar]
- Aso T, Yamazaki K, Amimoto K, Kuroiwa A, Higashi H, Matsuda Y, Kitajima S, Hatakeyama M. Identification and characterization of Elongin A2, a new member of the Elongin family of transcription elongation factors, specifically expressed in the testis. J Biol Chem. 2000;275:6546–6552. doi: 10.1074/jbc.275.9.6546. [DOI] [PubMed] [Google Scholar]
- Barlow DP. Gametic imprinting in mammals. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The human genome: Organization and evolutionary history. Ann Rev Genet. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird AP, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Brock GJ, Charlton J, Bird AP. Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene. 1999;240:269–277. doi: 10.1016/s0378-1119(99)00442-4. [DOI] [PubMed] [Google Scholar]
- Brock GJR, Bird AP. Mosaic methylation of the repeat unit of the human ribosomal RNA genes. Hum Mol Genet. 1997;6:451–456. doi: 10.1093/hmg/6.3.451. [DOI] [PubMed] [Google Scholar]
- Choi Y-C, Gu W, Hecht NB, Feinberg AP, Chae C-B. Molecular cloning of mouse somatic and testis-specific H2B histone genes containing a methylated CpG island. DNA Cell Biol. 1996;15:495–504. doi: 10.1089/dna.1996.15.495. [DOI] [PubMed] [Google Scholar]
- Cockell M, Gasser SM. Nuclear compartments and gene regulation. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci. 1996;93:7149–7153. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson NJ. In: Essential molecular biology: A practical approach. Brown TA, editor. Vol. 2. Oxford: IRL Press; 1991. pp. 111–156. [Google Scholar]
- Feinberg AP. Cancer epigenetics takes center stage. Proc Natl Acad Sci. 2001;98:392–394. doi: 10.1073/pnas.98.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Methylation meets genomics. Nat Genet. 2001;27:9–10. doi: 10.1038/83825. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- Ferguson-Smith AC, Sasaki H, Cattanach BM, Surani MA. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Hayashizaki Y, Shibata H, Hirotsune S, Sugino H, Okazaki Y, Sasaki N, Hirose K, Imoto H, Okuizumi H, Muramatsu M, et al. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat Genet. 1994;6:33–40. doi: 10.1038/ng0194-33. [DOI] [PubMed] [Google Scholar]
- Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DR, Gnarra GR, Linehan WM, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- Kawajiri K, Watanabe J, Gotoh O, Tagashira Y, Sogawa K, Fujii-Kuriyama Y. Structure and drug inducibility of the human cytochrome P-450c gene. Eur J Biochem. 1986;159:219–225. doi: 10.1111/j.1432-1033.1986.tb09857.x. [DOI] [PubMed] [Google Scholar]
- Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG., Jr Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- Kim J, Lu X, Stubbs L. Zim1, a maternally expressed mouse Kruppel-type zinc-finger gene located in proximal chromosome 7. Hum Mol Genet. 1999;8:847–854. doi: 10.1093/hmg/8.5.847. [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Hausmann S, Stojmenovic G, Dixkens C, Bink K, Schulz-Schaeffer W, Altmann M, Engel W. SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics. 1999;62:216–222. doi: 10.1006/geno.1999.6005. [DOI] [PubMed] [Google Scholar]
- Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Lee MP, Hu R-J, Johnson LA, Feinberg AP. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- McMahon FJ, Hopkins PJ, Xu J, McInnis MG, Shaw S, Cardon L, Simpson SG, MacKinnon DF, Stine OC, Sherrington R, et al. Linkage of bipolar affective disorder to chromosome 18 markers in a new pedigree series. Am J Hum Genet. 1997;61:1397–1404. doi: 10.1086/301630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee D, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, de la Casa-Esperon E, Sapienza C. Natural selection and the function of genome imprinting: Beyond the silenced minority. Trends Genet. 2000;16:573–579. doi: 10.1016/s0168-9525(00)02134-x. [DOI] [PubMed] [Google Scholar]
- Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevtseva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, et al. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation and genomic imprinting. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- Serrano A, Garcia A, Abril E, Garrido F, Ruiz-Cabello F. Methylated CpG points identified within MAGE-1 promoter are involved in gene repression. Int J Cancer. 1996;68:464–470. doi: 10.1002/(SICI)1097-0215(19961115)68:4<464::AID-IJC11>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem. 1994;269:8466–8476. [PubMed] [Google Scholar]
- Shiraishi M, Chuu YH, Sekiya T. Isolation of DNA fragments associated with methylated CpG islands in human adenocarcinomas of the lung using a methylated DNA binding column and denaturing gradient gel electrophoresis. Proc Natl Acad Sci. 1999;96:2913–2918. doi: 10.1073/pnas.96.6.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine OC, Xu J, Koskela R, McMahon FJ, Gschwend M, Friddle C, Clark CD, McInnis MG, Simpson SG, Breschel TS. Evidence for linkage of bipolar disorder to chromosome 18 with a parent-of-origin effect. Am J Hum Genet. 1995;57:1384–1394. [PMC free article] [PubMed] [Google Scholar]
- Strom TM, Hortnagel K, Hofmann S, Gekeler F, Scharfe C, Rabl W, Gerbitz KD, Meitinger T. Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum Mol Genet. 1998;7:2021–2028. doi: 10.1093/hmg/7.13.2021. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ho C, Ahuja N, Jair K-W, Li Q, Ohe-Toyota M, Baylin SB, Issa J-PJ. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- Wang AH, Kruhlak MJ, Wu J, Bertos NR, Vezmar M, Posner BI, Bazett-Jones DP, Yang XJ. Regulation of histone deacetylase 4 by binding of 14–3–3 proteins. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen PH, Patel P, Chinault AC, Mohandas T, Shapiro L. Differential methylation of hypoxanthine phosphoribosyltransferase genes on active and inactive human X chromosomes. Proc Natl Acad Sci. 1984;81:1759–1763. doi: 10.1073/pnas.81.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZB, Hsieh S, Bently DR, Campbell DR, Volanakis JE. A variable number of tandem repeats locus within the human complement C2 gene is associated with a retroposon derived from a human endogenous retrovirus. J Exp Med. 1992;175:1783–1787. doi: 10.1084/jem.175.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]