Abstract
A small genetic region near the telomere of ovine chromosome 18 was previously shown to carry the mutation causing the callipyge muscle hypertrophy phenotype in sheep. Expression of this phenotype is the only known case in mammals of paternal polar overdominance gene action. A region surrounding two positional candidate genes was sequenced in animals of known genotype. Mutation detection focused on an inbred ram of callipyge phenotype postulated to have inherited chromosome segments identical-by-descent with exception of the mutated position. In support of this hypothesis, this inbred ram was homozygous over 210 Kb of sequence, except for a single heterozygous base position. This single polymorphism was genotyped in multiple families segregating the callipyge locus (CLPG), providing 100% concordance with animals of known CLPG genotype, and was unique to descendants of the founder animal. The mutation lies in a region of high homology among mouse, sheep, cattle, and humans, but not in any previously identified expressed transcript. A substantial open reading frame exists in the sheep sequence surrounding the mutation, although this frame is not conserved among species. Initial functional analysis indicates sequence encompassing the mutation is part of a novel transcript expressed in sheep fetal muscle we have named CLPG1.
[The sequence data described in this paper have been submitted to GenBank under the following accession numbers: G74891-G75331 for all STS generated; AF401294 for the amplicon identifying the specific callipyge mutation; AF533009 for the partial expressed transcript.]
Callipyge is a muscle hypertrophy phenotype in sheep (Jackson and Green 1993), resulting from an apparent single locus mutation in the telomeric region of ovine chromosome 18 (CLPG; Cockett et al. 1994). Dramatic effects on muscle development, carcass composition, shape, and meat quality are hallmarks of the callipyge syndrome (Koohmaraie et al. 1995; Freking et al. 1998b, 1999). The muscle hypertrophy phenotype is expressed in a unique parent of origin-dependent manner referred to as paternal polar overdominance (Cockett et al. 1996; Freking et al. 1998a). The only genotype that expresses muscle hypertrophy in this type of gene action is one in which the mutant callipyge allele (C) is inherited from the sire and a normal allele (N) from the dam (genotype CN). Interestingly, sheep with two copies of the mutant allele (genotype CC) did not express muscle hypertrophy even though other phenotypes such as increased longissimus muscle calpastatin enzyme activity and increased longissimus muscle shear force were observed relative to noncarrier (genotype NN) or maternal-derived heterozygous (genotype NC) genotypes (Freking et al. 1999). Understanding the mechanism by which the CLPG mutation alters these phenotypes would improve our basic knowledge of factors involved in muscle growth, carcass leanness, and meat quality, as well as delineate unique forms of genomic imprinting regulation.
Two independent efforts to identify the CLPG mutation in sheep have resulted in refinement of physical and comparative maps of the region on chromosome 18 (Fahrenkrug et al. 2000; Berghmans et al. 2001). Animals with genetic recombination events were used to reduce the candidate interval to an approximately 400-Kb region containing plausible candidate genes such as delta, Drosophila, homolog-like (DLK1; also known as PREF-1) and maternally expressed gene 3 (MEG3; also known as GTL2). The homologous regions on human chromosome 14 and mouse chromosome 12 have been intensively studied because DLK1 and MEG3 are reciprocally imprinted and expressed from the paternal and maternal alleles, respectively (Schmidt et al. 2000; Takada et al. 2000; Wylie et al. 2000). The human DLK1-MEG3 region also has a conserved spatial, structural, and epigenetic organization comparable to that of IGF2-H19 region (Wylie et al. 2000). These include common CTCF binding sites, differentially methylated CpG islands, and downstream enhancer sequences. Charlier et al. (2001b) reported that the homologous imprinted domain in sheep contains the imprinted transcripts DAT, PEG11, antiPEG11, and MEG8, in addition to DLK1 and MEG3. The imprint status of the first four of these genes in humans is unknown. The CLPG genotype status does not alter the imprinting polarity of these genes, but does affect the expression pattern of several transcripts from the CLPG interval during muscle development (Bidwell et al. 2001; Charlier et al. 2001a). However, the mutation responsible for different phenotypes associated with callipyge in sheep remains elusive. Our objective was to identify the specific DNA mutation responsible for the unique gene action producing the muscular hypertrophy phenotype.
RESULTS
Development of a DNA Panel for Mutation Detection
The region containing CLPG was small enough to make complete sequencing of the interval a realistic approach to discover the mutation. We chose a strategy of PCR amplification and direct sequencing of the products to detect nucleotide sequence differences among animals of known CLPG genotypes. Principal difficulties in applying this approach were the potential to overlook large-scale inversions and the presence of nucleotide sequence variation occurring in sheep independent of the causative mutation. To ensure detection of inverted chromosomal segments on mutated versus normal chromosomes, overlapping amplicons were used that spanned the entire interval, and DNAs from both homozygous classes were used as template. At the boundaries of segments inverted by a putative mutation, primer pairs would successfully amplify normal chromosomes but fail on templates homozygous for the mutation.
A more difficult problem was presented by the relatively high frequency of polymorphisms among sheep in our panel (see below). Initial sequencing utilized four affected (CN; hypertrophy phenotype), two normal (NN; normal phenotype), and two homozygous mutant (CC; normal phenotype) animals. It was postulated that single nucleotide polymorphisms (SNPs) heterozygous in affected and homozygous for alternative alleles in NN and CC animals were candidates for the causative mutation. Areas containing coding regions of previously identified candidate genes (DLK1, MEG3; Fahrenkrug et al. 2000) were first sequenced in these animals. Although polymorphisms were detected, none uniquely differentiated the CLPG alleles.
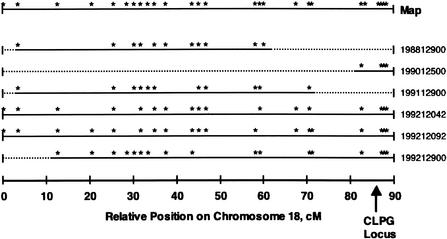
We sought a more efficient approach to identify all types of potential mutations within the broader region containing CLPG. An experiment at the U.S. Meat Animal Research Center (MARC) used nine Dorset rams expressing the muscle hypertrophy phenotype with pedigree information tracing back to Solid Gold, the first animal known to express the phenotype. These rams were extensively progeny-tested as part of the grandparent generation of the MARC resource population. All rams were proven to be heterozygous at CLPG (Freking et al. 1998a); however, there were significant differences in marker heterozygosity among this group of rams. Chromosome-wide marker heterozygosity for six rams chosen to be part of the mutation detection panel is shown in Figure 1. The CLPG locus has been mapped by breakpoint mapping to the interval between MULGE5 and OY3 microsatellite markers (Berghmans et al. 2001), at approximately position 86–87 cM in Figure 1. This figure illustrates that two of these rams (198812900 and 199112900) exhibited no marker informativeness in the critical region of chromosome 18. Both rams were born into the flock that produced Solid Gold.
Figure 1.
Marker informativeness among heterozygous (by progeny testing) CLPG Dorset rams used in the mutation discovery panel. Asterisks on the top row (Map) indicate relative positions of markers used to construct the chromosome 18 linkage group. The arrow (position 86–87 cM) indicates relative position of the CLPG locus. Markers heterozygous are presented as asterisks for each ram. All rams were genotyped for all markers on the chromosome 18 map. Regions of the linkage group for each ram not flanked by an informative marker are indicated by dotted lines. Of particular interest, rams 198812900 and 199112900 exhibit marker homozygosity in the critical region containing CLPG.
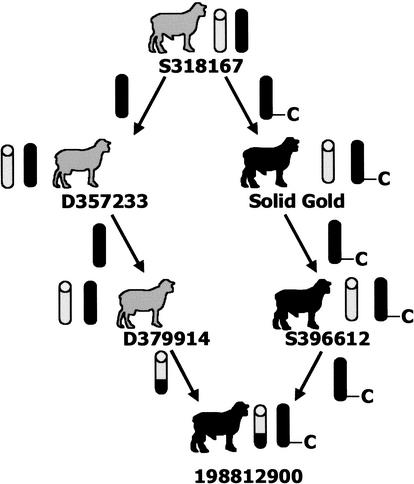
The lack of marker informativeness in the two rams, proven to be heterozygous at CLPG, suggested they would be useful for detecting the mutation responsible for the muscle hypertrophy phenotype. The pedigree for 198812900 was of particular interest, as the most telomeric heterozygous marker was microsatellite locus HH47, over 25 cM centromeric from CLPG (Freking et al. 1998a). Based on pedigree and marker data, it was hypothesized that this ram was identical-by-descent for the telomeric one-third of the chromosome, except for the CLPG mutation, which would be predicted to reside only on the paternally derived chromosome. One of the inbreeding paths identified for 198812900 fits this hypothesis (common ancestor S318167; Fig. 2). Ram S318167, the sire of Solid Gold, was normal in appearance according to his owner, indicating that the ram was not CN. Moreover, the owner stated that S318167 was used extensively in the flock but produced only a single offspring (Solid Gold) displaying characteristic muscular hypertrophy, suggesting that S318167 was NN, and that the CLPG mutation occurred in the germ cell that produced Solid Gold. Because 198812900 was only two generations removed from Solid Gold, the likelihood of additional mutations not involved in CLPG having accrued in the vicinity of the locus is small. On this basis, 198812900 was chosen for complete sequencing of the entire region containing DLK1 and MEG3, as detection of a polymorphism in this region should reveal the causative mutation.
Figure 2.
Ram 198812900 exhibits an inbreeding path with the sire (S318167) of Solid Gold contributing on both maternal and paternal sides of the pedigree. We hypothesized that this inbreeding path has allowed the region to be identical-by-descent, except for the CLPG mutation, which we propose occurred in the gamete that produced Solid Gold. The C identifies the mutated allele, and individuals with the muscle hypertrophy phenotype are identified as solid black sheep; those with normal phenotype are identified as gray sheep. Wild-type chromosomes are represented as gray chromatids. A recombination event is depicted to have occurred on the maternal side of the pedigree; however, the event could have occurred on either or both sides to generate marker informativeness in the centromeric region of chromosome 18.
A second ram, 199112900, also appeared to be identical-by-descent in the telomeric region with the exception of the mutation, as markers telomeric to ILSTS054 (position 71 cM) were not informative. However, the extent of the uninformative region was smaller than that of 198812900, and the dam of 199112900 was not recorded, so an inbreeding path such as the one defined for 198812900 could not be identified. Nevertheless, detection of a polymorphism in this ram could also uncover the CLPG mutation, providing confirmation of results obtained for 198812900. Therefore, these two rams were both used to detect the mutation, but completion of sequencing focused on 198812900 due to his known inbreeding.
These key rams permitted establishment of the most efficient screen available for the causative polymorphism. Genomic DNAs from the parents of Solid Gold were not available for testing the hypothesis for the origin of the causative mutation. A panel of animals for SNP discovery and validation was selected that included the six progeny-tested heterozygous Dorset rams (CN genotype, Fig. 1), two CN Dorset × Romanov F1 rams, two NN Romanov ewes, and two CC rams from an introgression flock at MARC. The two inbred CN rams were used for detection of the causative polymorphism, while NN and CC animals established normal and mutant alleles, respectively. Only products from primer pairs that successfully amplified all genotypes were used for sequencing. This prevented allele amplification bias that could potentially obscure the causative mutation.
Coverage of Candidate Interval by PCR Amplification and Sequencing
A total of 388 unique primer pairs successfully amplified sheep genomic DNA and produced sequence information targeting the region containing CLPG. Individual amplicons were not considered successfully amplified and finished as a sequence until at least one of the two homozygous CC templates, in addition to the inbred 198812900 ram, produced a quality sequence read. This approach allowed us to exclude allele amplification bias as the cause of homozygosity in 198812900. The resulting 441 contigs generated 400,989 overlapping bases of quality sequence (Phred score >20) in 198812900 for a total of 215-Kb coverage of the region. Individuals comprising the remainder of the panel contributed between 244,020 and 311,866 bp each of similar quality sequence. Comparison to the previously published sheep genomic sequence of the region (AF354168) suggested that over 97.5% of this candidate interval has been examined for variation in the inbred animal. A total of 5466 bases of the reference sequence was not covered specifically with 198812900, although only 3,335 of these bases are within the new telomeric exclusion boundary (see section below on Further Genetic Recombination Exclusion). Three small regions (total 1693 bp) generated amplicons that differed from the reference sequence in the middle of the amplicon, while matching one or both sides.
Mutation Discovery
In total, 616 polymorphisms were discovered, 1 SNP for every 340-bp unique sequence. Over two-thirds (67.7%) of the identified polymorphisms were purine-purine (A/G) or pyrimidine-pyrimidine (C/T) transition polymorphisms. Purine-pyrimidine transversions (A/T 3.7%; C/G 8.0%; A/C or G/T 12.8%) accounted for 24.5%, and small insertion-deletion events accounted for 7.6% of the total polymorphisms identified. No inversions or major deletions were observed from animals in our discovery panel.
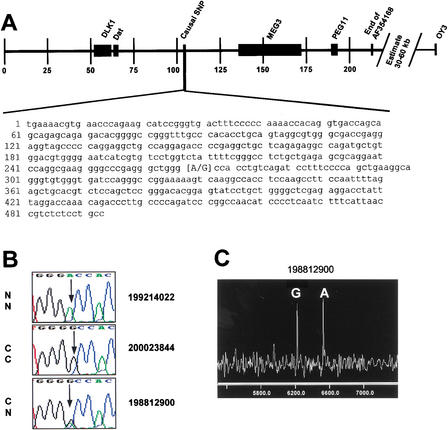
Data presented in Table 1 summarize the SNP information by animal. Heterozygosity for individuals varied widely over this region, as expected. The overall rate of heterozygosity per bp sequenced ranged from 0.0 to 0.0011738. Four of the Dorset rams (not inbred in the region), the two Romanov ewes, and the two F1 rams produced all of the heterozygous positions discovered. The remaining four individuals allowed us to exclude polymorphisms that were not causative for CLPG. As anticipated given the recency of the mutation, the two CC animals did not exhibit polymorphism over the entire sequenced interval. A single common haplotype was observed in phase with the C allele for the two CC rams and the two inbred CN rams. In this region, all eight chromosomes from these four animals were identical-by-descent to a gamete that resulted in the sire of Solid Gold, except at CLPG. Two haplotypes were observed for the entire region in the inbred heterozygous rams, differing only by a single A/G polymorphism located at position 103,894 of AF354168 and position 267 of the GenBank STS AF401294 (Fig. 3A,B). This polymorphism was the only position heterozygous for all CN rams in the discovery panel and homozygous at alternative alleles for the Romanov ewes (NN) and the composite rams (CC). It therefore met our established criterion for the polymorphism screen. Homozygous genotypic data from the remaining 615 polymorphic positions give direct evidence that 198812900 and 19912900, in addition to the two CC animals, are homozygous by descent for this entire interval.
Table 1.
SNP Information Obtained From Our Discovery Panel
| Breed | Callipyge genotypeb | Animal | Total number SNP genotypesc | Unique positions | Heterozygous positions | Hetd |
|---|---|---|---|---|---|---|
| Dorset(D) | CN | 198812900 | 1045 | 600 | 1 | 0.0000048 |
| Dorset | CN | 199112900 | 1020 | 606 | 1 | 0.0000048 |
| CIIIa | CC | 200023844 | 926 | 575 | 0 | 0.0 |
| CIII | CC | 200023886 | 898 | 564 | 0 | 0.0 |
| Romanov(R) | NN | 199214022 | 815 | 517 | 86 | 0.0004103 |
| Romanov | NN | 199214305 | 899 | 574 | 246 | 0.0011738 |
| Dorset | CN | 199012500 | 1006 | 596 | 145 | 0.0006919 |
| Dorset | CN | 199212042 | 886 | 561 | 236 | 0.0011261 |
| Dorset | CN | 199212092 | 850 | 540 | 232 | 0.0011070 |
| Dorset | CN | 199212900 | 951 | 573 | 81 | 0.0003865 |
| F1DxR | CN | 199360105 | 887 | 559 | 231 | 0.0011022 |
| F1DxR | CN | 199360365 | 822 | 521 | 184 | 0.0008779 |
CIII population contains introgressed CLPG allele from Dorset into terminal sire composite population (1/2 Columbia; 1/4 Suffolk; 1/4 Hampshire). The new composite population is on average 1/16 Dorset germplasm.
Genotype at CLPG locus with C representing the mutant allele, N the normal allele; the paternally-derived allele is listed first.
Multiple sequence reads contributed data for each animal-SNP combination.
Heterozygosity per base pair over the covered region of 209,580.
Figure 3.
Identification of the causal base change (SNP) for CLPG. (A) Position of the identified SNP relative to previously identified candidate genes (DLK1, DAT, MEG3, and PEG11). Microsatellite marker OY3 was the previously defined telomeric boundary. Scale is set to 1 = 1000 bp of the sheep genomic sequence in GenBank accession AF354168. The SNP is at position 267 of the sheep STS sequence in accession AF401294. (B) Sequence traces representing the different genotypes present at the noted SNP. A Romanov ewe (NN at CLPG) displays the homozygous adenine residue, while the progeny-tested composite ram (CC at CLPG) displays the homozygous guanine residue. The inbred ram 198812900 (CN at CLPG) exhibits both residues at this position. (C) Results from a MALDI-TOF mass spectrometry genotyping assay for this SNP. The two alleles are represented as different mass extension products from a probe primer with the 3′ end adjacent to the SNP (Table 2). Probe extension in the presence of a specific di-deoxy termination mix creates the different analyte masses for each allele.
Further Interval Exclusion Using Genetic Recombination
Several polymorphisms were developed into MALDI-TOF mass spectrometry assays (Table 2) and genotyped on a set of animals from a study designed to evaluate all 16 mating combinations of CLPG genotypes (K. Leymaster, unpubl.). Two individual animals had definitive phenotypic carcass data and recombinant marker genotypes within the region. These two individuals had evidence of recombination on an informative CLPG chromosome between genetic markers CSSM18 (position 84.9 cM) and haplotype for OY3 / OY15 / OY5 (position 88.6 cM), and were genotyped along with parental and grandparental DNAs for the new SNP markers.
Table 2.
Oligonucleotides for Amplification of Informative SNP Markers and Analytes Produced by MALDI-TOF MS Genotyping
| SNP Marker name | SNp position on AF354168 | Amplification primersa | PROBE primerb | Probe mass (m/z) | Amplicon length (bp) | Termination mixture | Analyte products | Analyte masses (m/z) |
|---|---|---|---|---|---|---|---|---|
| DLK1_406.1 | 47113 | cacctgcttgtcctctctcc | gaagccagagagcccgcg | 5543.7 | 328 | ddA/ddC | gaagccagagagcccgcga | 5840.9 |
| acttggtgcctcattcat | gaagccagagagcccgcggc | 6146.1 | ||||||
| DLK1_INTR1.1 | 54626 | cacagcacactgcctagctc | acaacaaacgtcaggccc | 5446.6 | 270 | ddC/ddG | acaacaaacgtcaggcccg | 5759.8 |
| cgccttacctatgagccttc | acaacaaacgtcaggccctc | 6024.0 | ||||||
| 9571-268.2 | 103894 | tggcaggagagacggttaat | gaaaggatctgacaggtgg | 5941.9 | 515 | ddA/ddC | gaaaggatctgacaggtggc | 6215.1 |
| gaaaacgtgaacccagaagc | gaaaggatctgacaggtggtc | 6519.3 | ||||||
| Meg3.1 | 139107 | aggagaacaaaggccacctc | cacgtagcaacccgctcg | 5429.6 | 229 | ddA/ddG | cacgtagcaacccgctcga | 5726.8 |
| ttcctttgtccctgtcaagc | cacgtagcaacccgctcgcg | 6032.0 | ||||||
| Meg3_enh2 | 151581 | gtgtagaagccgccattg | atgggatccccatgttga | 5514.6 | 355 | ddA/ddC | atgggatccccatgttgaa | 5811.9 |
| ggtgccccagagtgaaa | atgggatccccatgttgaga | 6141.1 | ||||||
| Meg3.13 | 157968 | ttgctaagcgtcccagtc | tgctcacccatccctccccac | 6183.1 | 539 | ddC/ddG | tgctcacccatccctccccacg | 6496.3 |
| acatctgaggctggtcactg | tgctcacccatccctccccacaag | 7122.7 | ||||||
| Meg3.12 | 158139 | ttgctaagcgtcccagtc | gaaagaggaggaattctgtg | 6270.1 | 539 | ddC/ddG | gaaagaggaggaattctgtgc | 6543.3 |
| acatctgaggctggtcactg | gaaagaggaggaattctgtgtg | 6887.6 | ||||||
| Meg3.10 | 158394 | ttgctaagcgtcccagtc | caatgaaaagcttgaag | 5251.5 | 539 | ddC/ddG | caatgaaaagcttgaagc | 5524.7 |
| acatctgaggctggtcactg | caatgaaaagcttgaagag | 5877.9 | ||||||
| Meg3.9 | 158520 | acacgtaagcatggcagatg | ggacacagaattcatctg | 5507.7 | 337 | ddA/ddT | ggacacagaattcatctga | 5804.9 |
| gcctcgaaaggcgaaaata | ggacacagaattcatctgca | 6094.1 | ||||||
| Meg3.8 | 158616 | acacgtaagcatggcagatg | ccctctgaatggtcatcc | 5410.6 | 337 | ddA/ddT | ccctctgaatggtcatcct | 5698.8 |
| gcctcgaaaggcgaaaata | ccctctgaatggtcatccct | 5988.0 | ||||||
| Meg3.7 | 158663 | acacgtaagcatggcagatg | gggtcagactccccaggccc | 6064.0 | 337 | ddA/ddT | gggtcagactccccaggccca | 6361.2 |
| gcctcgaaaggcgaaaata | gggtcagactccccaggcccgct | 6970.6 | ||||||
| Meg3.5 | 158998 | atggacaagcgataggtgct | atctcctatgtatcca | 4791.2 | 382 | ddA/ddC | atctcctatgtatccaa | 5088.4 |
| aaggactgtgggttttgtgc | atctcctatgtatccagc | 5393.6 |
Universal sequence (agcggataacaatttcacacagg) placed at 5′ end of the second locus specific primer listed.
PROBE primer is in sense direction relative to the first amplification primer.
Individual 199860459, produced by mating a CC ram to a CN dam, did not provide definitive phase information on the maternal gamete for the new markers. These markers generated only like-heterozygote genotypes or were noninformative. Individual 199860287 exhibited the extreme muscle hypertrophy phenotype and was produced by mating two NC parents. A recombination event on the paternally derived gamete showed CSSM18 to be in phase with the inherited C allele, while OY3 was not. Phase information on the paternal gamete at the MEG3.9 SNP locus was definitive and also not in phase with the C allele. This genotypic and phenotypic information dictates that CLPG is centromeric from the MEG3.9 position in the genome (base 158520 on AF354168) and the observed recombination event on this gamete is between markers CSSM18 and MEG3.9. Using a defined recombinant break point, we definitively identified the new telomeric boundary for the callipyge locus to be MEG3.9. The causative SNP remains within this newly defined genetic boundary.
Frequency of Causative SNP Allele in Diverse Populations
The causative SNP would be expected only in descendants of Solid Gold, whereas a closely linked polymorphism would likely exist among unrelated sheep. To provide further evidence that the SNP identified by the sequencing screen represents the causative mutation, a genetically diverse panel of breeds widely used in commercial sheep production was constructed (see MARC SheepDP v1.1 in Methods below). The objective was to estimate the frequency of alleles for polymorphisms that passed the initial screens in the discovery panel. A mass spectrometry assay for the causal SNP (9571–268.2 in Table 2) generated genotypes for 90 individuals across the nine breeds. Frequency of the normal allele (nucleotide A at position 267 on AF401294) was 100% for 180 alleles in this panel. Of particular interest was a sample of ten Dorset rams with normal muscle phenotype, as this was the breed of the progenitor animal. None of the 20 chromosomes in this sample had the causative G for A mutation. This information adds to the preponderance of evidence that this mutation represents CLPG.
Preliminary Functional Evaluation of Mutated Region
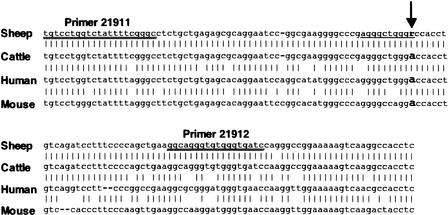
The mutation is not within the boundary of any previously identified transcript (Fig. 3). Therefore the mechanism by which this mutation causes the muscle hypertrophy phenotype is not obvious. Using a 144-bp sequence from the ovine genome centered on the identified SNP, corresponding regions of the cattle, human, and mouse genomes were identified between the DLK1 and MEG3 genes. Alignment of sheep, cattle, human, and mouse genomic sequences indicated a high degree of conservation in the region (Fig. 4). Complete conservation of sequence across these four species was observed for >74% of nucleotides. This indicates that this region has biological significance.
Figure 4.
Alignment of 144 bp of sheep genomic sequence flanking the identified SNP with homologous cattle, human, and mouse genomic sequence. The SNP is identified with the arrow. Percentage homology with the sheep sequence ranged from 99.3% identical (cattle) to 78.4% identical (mouse). The 10-bp motif for a MyoD recognition site is underlined. Primers used for RT-PCR experiments are double underlined.
Various GenBank databases were searched via BLASTN analysis to identify corresponding transcripts, including dbEST for human, mouse, and other species, using the genomic region sequence for each species as reference. No significant similarity to EST sequence in the database was identified that corresponded to the genomic sequence. Using the Mapviewer tool at the National Center for Biological Information (NCBI; www.ncbi.nlm.nih.gov/cgi-bin/Entrez/hum_srch), a predicted gene was identified in humans (LocusLink ID 123090; gene prediction method GenomeScan) that spans the area homologous to the CLPG mutation. The sequence containing the SNP lies within the 23,416-bp intron 6 of the predicted transcript, suggesting that it does not form a part of this putative human gene. Moreover, no significant open reading frame (ORF) containing the mutation is conserved among the genomic segments of the four species.
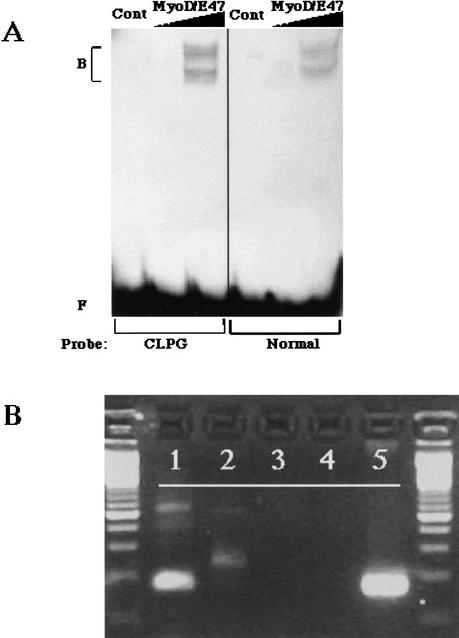
To determine whether the region containing this mutation might be involved in gene regulation, it was examined using the Transcription Element Search System (TESS; www.cbil.upenn.edu). A number of motifs were identified near the SNP that were consistent with binding of muscle-related transcription control factors (data not shown). Specifically, the SNP alters a sequence motif with homology to a muscle regulatory factor (MRF) binding site (Fig. 4). To determine whether this site can be recognized by MRFs, and whether the mutation affects this putative binding, oligonucleotides corresponding to the C and N alleles were synthesized and used in electrophoretic mobility shift assays (EMSAs) with MyoD protein in the presence of the E47 partner protein (Fig. 5A). Results demonstrated that both sequences bind the MyoD complex, with similar affinities. Thus, the mutation does not act through altered affinity for this muscle transcription factor complex. However, it is possible that binding affinity may be affected by epigenetic processes, as this region of the genome contains imprinted genes and CpG-rich imprinting regulatory elements that are methylated in a parent of origin-dependent manner (Wylie et al. 2000; Charlier et al. 2001b).
Figure 5.
Functional analysis of the region that encompasses the CLPG mutation. (A) An electrophoretic mobility shift assay using MyoD/E47 proteins and radiolabeled oligonucleotide probes representing the CLPG region. Probes were incubated either with no protein (Cont), 1 μL in vitro translated MyoD/E47, or 5 μL MyoD/E47, and the resulting complexes (B) were separated from free probe (F) by electrophoresis. MyoD/E47 forms two complexes with target DNA elements: the upper complex contains full-length proteins, and the lower complex contains a smaller translation product of E47 (Lemercier et al. 1998). (B) Expression of a transcript containing the CLPG mutation. RNA from fetal longissimus muscle was reverse transcribed with primer 21911 (primer sequence in Fig. 4) (lane 1) or 22051 5′- GCAAGGGTCTGTTTGGTCCTAA - 3′ (lane 2). Resulting cDNA was amplified with primer 21911 and a nested reverse primer, 22052 5′- GCTGGAGACGTGCAGCTCTAA - 3′. Control PCR with these primers utilized RNA from a mock reverse transcription to rule out DNA contamination (lane 3), no template negative control (lane 4), or ovine genomic DNA (lane 5).
Differential methylation of CpG regions is a key component of regulation of imprinting. One of the potential models for polar overdominance would involve a reversal of the imprint from one parental origin to the other. Although the mutation does not affect a site for methylation, it could potentially have cis-acting effects on local methylation patterns or efficiency. Epigenetic modifications to the immediate region surrounding the SNP were therefore evaluated using bisulphite sequencing. Eleven CpG sites near the mutation were evaluated for methylation status in all four CLPG genotypes from fetal (n = 8 animals) and adult stages (n = 8 animals). Fetal-stage DNA samples exhibited a consistent methylation pattern that did not differ between the genotypes. In adults, overall methylation levels were increased relative to the fetal samples. Furthermore, NN genotypes exhibited the highest degree of methylation, CN and NC genotypes an intermediate level, and CC genotypes the lowest level of methylation. However, the methylation exhibited in this region is not parent of origin-dependent, and phenotypic status does not correlate with the degree of methylation. This indicated that altered methylation in the vicinity of the SNP is not the mechanism by which this mutation affects the muscle hypertrophy phenotype.
To investigate the possibility that the sequence is part of a previously unrecognized expressed transcript, fetal sheep longissimus muscle RNA was reverse transcribed with random primers, and the resulting cDNA was amplified with primers (21911–21912) designed to amplify a 115-bp segment containing the mutation. This primer pair successfully produced a reverse transcriptase-dependent product with the correct sequence (data not shown), indicating that the sequence carrying the mutation represents a portion of an expressed transcript (Fig. 5B). Two additional reverse primers (22051 and 22052) were synthesized and also produced the appropriate products via RT-PCR in combination with primer 21911. To determine the direction of the transcript, cDNA synthesis was primed with either 21911 or 22051 prior to PCR with primers 21911–22052. Specific amplification was observed only from the cDNA produced with the 22051 primer, indicating that the transcript is produced in the direction heading from MEG3 to DLK1 (Fig. 5B). To obtain further sequence, cDNA produced using the 21911 primer was used for 5′ rapid amplification of cDNA ends (RACE) using a procedure dependent on the cap structure to identify the 5′ extent of the RNA (see Methods). The RACE product (accession number AF533009) included 547 bp of sequence identical to the sheep genomic sequence surrounding the mutation, and presumably defines the 5′ end of the transcript and a portion of the first exon. We will refer to the transcript in this direction as CLPG1.
The use of a modified poly(T) primer to perform 3′ RACE to obtain downstream sequence was unsuccessful despite multiple attempts on various RNA preparations from fetal longissimus muscle of various CLPG genotypes, and using various combinations of amplification primers. This suggests the possibility that the 3′ end of the transcript is very distant from the 5′ end, or that the sequence between the partial first exon and the polyadenylation site is recalcitrant to reverse transcription or PCR amplification. We attempted to establish the molecular weight of the RNA transcript by Northern blotting to address these possibilities. A blot was generated using 10–30 μg of RNA from several sheep tissues including fetal muscle, with the RT-PCR product as probe. An ovine GAPDH probe served as a positive control for RNA loading and exhibited hybridization signal for all samples. We failed to detect specific hybridization signals for the probe containing the CLPG mutation, indicating that the transcript is a very-high-molecular-weight transcript and/or present in low copy number (data not shown).
DISCUSSION
Discovery of the CLPG mutation offers potential for new insights into basic biology of imprinting regulation in this region of the genome. In addition, a better understanding of mammalian protein and adipose accretion as well as postmortem tenderization of muscle tissue would evolve. We previously refined the CLPG region to a small (3-cM interval) genetic interval containing a conserved orthologous comparative segment with bovine and human genomes (Fahrenkrug et al. 2000). Others completed a further refinement of physical and genetic maps (Berghmans et al. 2001) and generated a contig of the sheep genomic sequence (Charlier et al. 2001b) encompassing the interval. We describe the discovery of an SNP whose genome location and allelic concordance are consistent with it being the causative CLPG mutation that generates muscle hypertrophy and is expressed as a novel polar overdominance form of imprinting regulation. Although this finding ends a nearly ten-year effort to identify the CLPG mutation, it marks the starting point for determining the mechanism by which it leads to these marked phenotypic alterations.
A key element in our discovery effort was recognizing the value of two inbred animals that were heterozygous at CLPG. An efficient screening process was developed that allowed us to exclude over 600 SNP positions as candidates for the causal mutation. This panel subjected to comparative sequencing included two progeny-tested rams heterozygous for CLPG that were homozygous for all markers developed over the telomeric one-third of chromosome 18. Detection of a single common polymorphism in these two animals revealed the CLPG mutation. Phase of the mutation with respect to corresponding SNP alleles was determined in animals of the alternative homozygous genotypes. We developed a genotyping assay to validate this polymorphism and observed complete concordance in segregating populations of this SNP allele in phase with the mutant CLPG allele. Testing additional animals within and outside of the original breed confirmed the specificity of the SNP and supported the conclusion that it represents the causative CLPG mutation.
Except for the inbred CN rams and the two CC animals of the SNP discovery panel, nucleotide sequence variation was discovered at an expected rate in this region of the sheep genome. For example, base differences were observed on average every 184 bp in divergent crossbred pigs (Fahrenkrug et al. 2002), every 96 bp in a cattle diversity panel (Heaton et al. 2002), and every 90 bp in other regions of the sheep genome in a Sheep Diversity Panel (MARC SheepDP v1.1). Thus, there is no evidence that this is a hypermutable area of the sheep genome. Indeed, the existence of over 600 additional SNP-based genetic markers with observed homozygosity in the two inbred rams lends strong support to the contention that these chromosomes were identical-by-descent. The evidence presented here supports the hypothesis that the SNP identified occurred in the gamete that created Solid Gold. The possibility that an undiscovered mutation simultaneously occurred on the same gamete in this small region is highly unlikely.
Discovery of the CLPG mutation in a region of the genome absent of previously known expressed genes led us to question how this solitary SNP produces the unique genotype-phenotype interactions of this syndrome. Several transcripts, including several noncoding RNAs, in this chromosomal region exhibit preferential expression in skeletal muscle (Charlier et al. 2001b), indicating potential common regulatory mechanisms in the region. An initial hypothesis to explain the polar overdominance gene action of the muscle hypertrophy phenotype was that a mutation alters the polarity of imprinting for the entire region. All of the previously reported transcripts in this region have been observed to be imprinted; however, no polarity shift has been documented among the transcripts evaluated in sheep for different CLPG genotypes (Charlier et al. 2001a).
The degree of conservation of the sequence adjacent to the CLPG mutation across species indicates that this region has an important biological function. Sequence motifs associated with muscle regulatory factor binding were identified in this specific region; however, the mutation did not alter in vitro binding of MyoD. Epigenetic modifications observed by differentially methylated CpG sites are present in this region of the genome, but once again do not appear to be altered by CLPG genotypic status. Evidence has been generated, however, that demonstrated the region is expressed as an RNA transcript (CLPG1) in sheep muscle tissue. The 547-bp transcript identified in sheep contains an ORF predicting 123 amino acids in which the CLPG mutation would alter a serine codon to a proline, but this ORF is not conserved in human and mouse genomic sequence, and the likelihood that it produces the corresponding peptide is unknown. Moreover, the conservation of sequence between species, outside of the 144-bp region shown in Figure 4, is substantially lower. Current work is aimed at determining the full length of this novel CLPG1 transcript and establishing its functional role.
Retarded growth development and accelerated adiposity were recently observed in a knockout mouse for the DLK1 gene (Moon et al. 2002). This evidence would be consistent with a lean muscular phenotype in response to the constitutive overexpression of DLK1 for sheep with the CLPG muscle hypertrophy phenotype as observed by Charlier et al. (2001a). It is possible that the identified mutation within this new transcript could alter its function as an RNA effector molecule to regulate gene expression of DLK1 differently in animals heterozygous for the mutation on the paternal allele.
Associations of phenotypes with specific genes, and the subsequent identification of the causal variation, have previously relied upon the presence of known gene(s) in the region linked with nearby genetic markers. This facilitates interrogation of the sequence of coding regions of positional candidate genes to identify causative mutations (e.g., Kambadur et al. 1997; Cockett et al. 1999; Galloway et al. 2000; and Mulsant et al. 2001). To our knowledge, this is the first time that a novel transcript has been identified in livestock subsequent to discovery of a causal mutation associated with a phenotype. The discovery of this mutation and a new transcript encompassing the mutation will focus new investigations on both genetic and epigenetic aspects of this important genomic region.
METHODS
Bovine BAC Clones
Genomic sequence for this area of the sheep genome was unavailable at the start of this project, and sheep BAC clones containing the region had not been identified.
Two bovine BAC clones clones (486B7 and 540H9) were isolated from the RPCI-42 library (Warren et al. 2000) and were positive for both DLK1 and MEG3 genes. To generate initial genomic sequence for primer design, a total of 5 μg of DNA from a pool of these two BAC clones was partially digested (18 min at 37°C) with 0.5 U of the enzyme CviJ1. The nearly random distribution of fragments was separated on a 1% agarose gel, and the fraction between 1000 and 1200 bp was isolated using a commercial kit (Novagen). Size-selected fractions were cloned into dephosphorylated pBLUESCRIPT vector (Stratagene) prepared by EcoRV digestion. Eight 384-well plates of genomic subclones were picked and sequenced as described (Smith et al. 2000). Chromatograms were exported into the MARC relational database, bases called with Phred (Ewing and Green 1998; Ewing et al. 1998), and sequences assembled into contigs with Phrap (P. Green, unpubl.). Bovine contig consensus sequences were tentatively ordered relative to matching human genomic sequence obtained from two BAC clones AL132711 and AL117190 using pairwise BLAST (Tatusova and Madden 1999). Public release of the sheep genomic sequence (AF354168) of this region during the project redirected our primer design efforts to the ovine-specific sequence data.
Primer Design
Amplification primers were designed using Primer3 (Rozen and Skaletsky 2000) from either the available bovine or ovine genomic sequence data. Amplicons were designed as overlapping genomic segments of approximately 1000 bp each spanning 220,000 bp of the sheep genomic region from the published sequence (AF354168). Primers were ordered from a commercial vendor (Integrated DNA Technologies). Primer sequences that generated data during this project are available from the GenBank dbSTS accessions (see below).
SNP Detection
Sequencing of PCR products from sheep genomic DNA was conducted and analyzed as described (Fahrenkrug et al. 2001) using Phred, Phrap, Polyphred, and Consed software (Ewing et al. 1998; Ewing and Green 1998; Nickerson et al. 1997; Gordon et al. 1998; P. Green, unpubl.). Position and composition of each accepted polymorphism, animal genotypes, and contig sequences were parsed to the MARC database. Consensus sequences with denoted SNP positions contained within as IUB codes were submitted to dbSTS in GenBank (Accession numbers: G74891 to G75331).
SNP Discovery Panel
A panel of twelve sheep was utilized to identify SNPs by sequencing. This panel contained six Dorset rams (198812900, 199012500, 199112900, 199212042, 199212092, and 199212900). All of these animals are heterozygous for CLPG, tracing back to the presumed progenitor animal (Solid Gold) within three generations. Two Romanov ewes (199214022, 199214305) which do not contain the mutated allele, two heterozygous F1 Dorset × Romanov rams (199360105, 199360365), and two rams homozygous for the mutated allele comprised the rest of the panel. All individuals in the panel were progeny-tested for CLPG genotypic status. The two rams homozygous for CLPG (200023844, 200023886) are members of a separate composite population created by introgressing and fixing the mutated allele from Dorset rams by traditional backcrossing into a different genetic background. The key individual in our panel was a Dorset ram (198812900) chosen as the primary screen for the causative CLPG polymorphism. He was chosen because of the high degree of homozygosity for genetic markers from the telomeric one-third of the linkage group, and the pedigree information which indicated an inbreeding path to the sire (S318167) of Solid Gold (S354432) (Fig. 2). S318167 is both the paternal and maternal great grandsire of 198812900. This inbred individual (198812900) is identical-by-descent for the telomeric one-third of ovine chromosome 18 with the exception of the specific CLPG mutation that likely occurred on the sperm cell that produced Solid Gold. These characteristics make 198812900 the ideal screen for the causative polymorphism. Alternatively, sequencing of Solid Gold and his parents would also have revealed the mutation. Genomic DNA from the parents of Solid Gold is not available.
Sheep Diversity Panel (MARC SheepDP v1.1)
To evaluate the frequency of the candidate polymorphism, a panel of sheep breeds was developed. Ninety DNA samples were collected from nine genetically diverse breeds of sheep. Ten rams each, with no rams produced by a common sire, were sampled from the following breeds: Composite III (Leymaster 1991), Dorper, Dorset, Finnsheep, Katahdin, Suffolk, Texel, Rambouillet, and Romanov. These breeds represent wide ranges of performance for numerous economically important traits and all functions in crossbreeding systems. They represent a wide segment of the commercial sheep populations used in the U.S. The objective of using this panel was to evaluate the frequency of alleles for the polymorphism that passed the initial screen in the discovery panel.
SNP Genotyping
Assays for automated genotype scoring by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) were developed based on the Sequenom (Sequenom) genotyping technology. The MALDI-TOF MS system uses primer oligonucleotide base extension, nanoliter dispensing of extension products onto silicon chips (Little et al. 1997), and fully automated mass spectrometric analysis. Individual genotyping assays constructed are presented in Table 2. All assays were designed as a three-primer PCR amplicon with one of the gene-specific primers containing a universal tail primer sequence at the 5′ end to allow incorporation of a biotin-labeled universal primer as described in Stone et al. (2002).
Genotypes were captured at SNP marker loci for two groups of animals. A set of animals from a study designed to evaluate all 16 mating combinations at CLPG (K. Leymaster, unpubl.) was genotyped to evaluate two additional animals with definitive phenotypic carcass data and recombinant marker genotypes within the candidate interval. The second set of animals was the diverse breed panel described above (MARC SheepDP v1.1).
Linkage Analysis
Marker genotypes from the MALDI-TOF assays were put into a relational database (Keele et al. 1994). Genotypic data from the resource population were used to construct a linkage map of the region as described by Kappes et al. (1997) with Cri-Map version 2.4 (Green et al. 1990). The CHROMPIC option was used to evaluate location of phase changes of the recombinant animals in our panel given the known physical marker orders.
Methylation Analysis
Bisulphite treatment of genomic DNA was performed based on an adapted protocol from Grunau et al. (2001). Briefly, one μg of genomic DNA was denatured with 3M NaOH for 20 min at 42°C, followed by deamination in saturated sodium bisulphite/10mM hydroquinone solution, pH 5.0 for 4 h at 55°C. The DNA was desalted using the Wizard DNA Clean-up System (Promega), then desulfonated in 3M NaOH (20 min at 37°C) and ethanol precipitated. The samples were resuspended in 20 μL Tris-Cl, pH 8.0 and stored at 4°C. One μL of the bisulphite-treated DNA was used as template for PCR amplification. Amplification products were purified from agarose gels using GenElute spin columns (Sigma) and cloned into a pGEM T-Easy vector (Promega); individual clones were sequenced using radiolabeled terminator cycle sequencing (USB).
MyoD Binding Assay
Mouse MyoD and E47 proteins were synthesized in a single reaction in vitro using TNT Quick coupled rabbit reticulocyte lysate reagents (Promega). Substrate DNAs were 1 μg of pcDNA3-E47 and pcDNA3-E47 cDNA plasmids (Lemercier et al. 1998; generous gifts from Dr. S. Konieczny, Purdue University). Parallel reactions containing 35S-methionine (Amersham Pharmacia) were performed, and the radiolabeled proteins were analyzed by fluorography as previously described (Sloop et al. 2000; data not shown).
Electrophoretic mobility shift assays (EMSA) were performed as described (Sloop et al. 2000) using equivalent amounts of in vitro translated MyoD/E47. Negative control reactions contained either unprogrammed lysate or no protein. 32P-labeled DNAs representing the SNP region were generated from the following oligonucleotides: CLPG site, 5′ GGGAAAGGATCTGACAGGTGGCCCCAGCCCTCGG-3′, and normal site, 5′ GGGAAAGGATCTGACAGGTGGTCCCAGCCCTCGG-3′. The appropriate complementary sequences were used to generate the double-stranded targets.
RT-PCR and Northern Analysis
The highly conserved portion of the sheep genomic sequence surrounding the causative mutation was used to design primers 21911 and 21912 (Fig. 4) to amplify a 115-bp product from genomic DNA. Random hexamers, primer 21911, or primer 22051 (Fig. 5B) were used to prime cDNA synthesis from 1 μg of total RNA purified from sheep fetal longissimus muscle of all four CLPG genotypes (NN, CC, NC, and CN). Reverse transcription was performed with Avian Moloney Virus reverse transcriptase as recommended by the manufacturer (Invitrogen), in 25 μL total reaction volume. The cDNA product (1 or 5 μL ) was used as template for PCR using the 21911–21912 or 21911–22051 primer pairs, in 10 μL total reaction volume. The 5′RACE and 3′RACE reactions were performed using a GeneRacer™ kit (Invitrogen) as recommended by the manufacturer. Primer 21911 was used to prime cDNA synthesis for 5′RACE, followed by amplification employing 21911 – “5′RACE” primers in 10 μL reaction volume. One μL of this PCR product was used as template for a second round of amplification with a nested primer 22055 (5′-GGCTGGGGCCACCTGTCAGAT-3′) and the ”5′RACE nested” primer. The amplification product from this reaction was separated on agarose gel, eluted from the gel, and cloned using a TOPO TA™ kit (Invitrogen) prior to sequencing.
Standard Northern blot analysis was performed using 10 or 30 μg of total RNA separated on 1% agarose containing formaldehyde as described (Sambrook et al. 1989). RNA was transferred to Zetaprobe membrane (BioRad) via capillary transfer. Probe was prepared using a commercial kit (Superscript kit; Invitrogen) from either full-length bovine GAPDH or a clone containing the RT-PCR product from the 21911–22051 primer pair. Blots were exposed to a phosphorimaging screen for 24–72 h before images were collected.
WEB SITE REFERENCES
www.ncbi.nlm.nih.gov/cgi-bin/Entrez/hum_srch; Mapviewer tool at the National Center for Biological Information
www.cbil.upenn.edu; Transcription Element Search System
Acknowledgments
We acknowledge the excellent technical support of R. Godtel for primer design, sequencing, and SNP identification. We also thank L. Flathman, T. Happold, B. Lee, S. Simcox, and K. Tennill for sequence and genotype data acquisition. We thank Dr. S. Konieczny (Purdue University) for reagents. Supported in part by a grant to S.J.R. from the USDA/NRICGP/CSREES. This study was partly supported by the NIH grants CA25951 and ES08823 to R.L.J. Further information on genomic imprinting is available at http://www.geneimprint.com.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL freking@emailmarc.usda.gov; FAX (402) 762-4173.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.571002. Article published online before print in September 2002.
REFERENCES
- Berghmans S, Segers K, Shay T, Georges M, Cockett N, Charlier C. Breakpoint mapping positions the callipyge gene within a 450-kilobase chromosome segment containing the DLK1 and GTL2 genes. Mamm Genome. 2001;12:183–185. doi: 10.1007/s003350010246. [DOI] [PubMed] [Google Scholar]
- Bidwell CA, Shay TL, Georges M, Beever JE, Berghmans S, Cockett NE. Differential expression of the GTL2 gene within the callipyge region of ovine chromosome 18. Anim Genet. 2001;32:248–256. doi: 10.1046/j.1365-2052.2001.00776.x. [DOI] [PubMed] [Google Scholar]
- Charlier C, Segers K, Karim L, Shay T, Gyapay G, Cockett N, Georges M. The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat Genet. 2001a;27:367–369. doi: 10.1038/86856. [DOI] [PubMed] [Google Scholar]
- Charlier C, Segers K, Wagenaar D, Karim L, Berghmans S, Jaillon O, Shay T, Weissenbach J, Cockett N, Gyapay G, et al. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the Callipyge (CLPG) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 2001b;11:850–862. doi: 10.1101/gr.172701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockett NE, Jackson SP, Shay TL, Nielsen DM, Moore SS, Steele MR, Barendse W, Green RD, Georges M. Chromosomal localization of the callipyge gene in sheep (Ovis aries) using bovine DNA markers. Proc Natl Acad Sci. 1994;91:3019–3023. doi: 10.1073/pnas.91.8.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockett NE, Jackson SP, Shay TL, Farnir F, Berghmans S, Snowder GD, Nielsen DM, Georges M. Polar overdominance at the ovine callipyge locus. Science. 1996;273:236–238. doi: 10.1126/science.273.5272.236. [DOI] [PubMed] [Google Scholar]
- Cockett NE, Shay TL, Beever JE, Nielsen D, Albretsen J, Georges M, Peterson K, Stephens A, Vernon W, Timofeevskaia O, et al. Localization of the locus causing Spider Lamb Syndrome to the distal end of ovine Chromosome 6. Mamm Genome. 1999;10:35–38. doi: 10.1007/s003359900938. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fahrenkrug SC, Freking BA, Rexroad CE, III, Leymaster KA, Kappes SM, Smith TPL. Comparative mapping of the ovine CLPG locus. Mamm Genome. 2000;11:871–876. doi: 10.1007/s003350010150. [DOI] [PubMed] [Google Scholar]
- Fahrenkrug SC, Freking BA, Smith TPL, Rohrer GA, Keele JW. Single nucleotide polymorphism (SNP) discovery in porcine expressed genes. Anim Genet. 2002;33:186–195. doi: 10.1046/j.1365-2052.2002.00846.x. [DOI] [PubMed] [Google Scholar]
- Freking BA, Keele JW, Beattie CW, Kappes SM, Smith TPL, Sonstegard TS, Nielsen MK, Leymaster KA. Evaluation of the ovine callipyge locus: I. Relative chromosomal position and gene action. J Anim Sci. 1998a;76:2062–2071. doi: 10.2527/1998.7682062x. [DOI] [PubMed] [Google Scholar]
- Freking BA, Keele JW, Nielsen MK, Leymaster KA. Evaluation of the ovine callipyge locus: II. Genotypic effects on growth, slaughter, and carcass traits. J Anim Sci. 1998b;76:2549–2559. doi: 10.2527/1998.76102549x. [DOI] [PubMed] [Google Scholar]
- Freking BA, Keele JW, Shackelford SD, Wheeler TL, Koohmaraie M, Nielsen MK, Leymaster KA. Evaluation of the ovine callipyge locus: III. Genotypic effects on meat quality. J Anim Sci. 1999;77:2336–2344. doi: 10.2527/1999.7792336x. [DOI] [PubMed] [Google Scholar]
- Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Juengel JL, Jokiranta TS, McLaren RJ, Luiro K, Dodds KG, Montgomery GW, et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet. 2000;25:279–283. doi: 10.1038/77033. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. CONSED: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Green PK, Falls K, Crooks S. Documentation for CRI-MAP, version 2.4. St. Louis, MO: Washington University School of Medicine; 1990. [Google Scholar]
- Grunau, C., Clark, S.J., and Rosenthal, A. 2001. Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 29:e65. [DOI] [PMC free article] [PubMed]
- Heaton MP, Harhay GP, Bennett GL, Stone RT, Grosse WM, Casas E, Keele JW, Smith TPL, Chitko-McKown CG, Laegreid WW. Selection and use of SNP markers for animal identification and paternity analysis in U.S. beef cattle. Mamm Genome. 2002;13:272–281. doi: 10.1007/s00335-001-2146-3. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Green RD. Muscle trait inheritance, growth performance and feed efficiency of sheep exhibiting a muscle hypertrophy phenotype. J Anim Sci. 1993;71:241. (Abstr). [Google Scholar]
- Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in double muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7:910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- Kappes SM, Keele JW, Stone RT, McGraw RA, Sonstegard TS, Smith TPL, Lopez-Corrales, Beattie CW. A second generation linkage map of the bovine genome. Genome Res. 1997;7:235–249. doi: 10.1101/gr.7.3.235. [DOI] [PubMed] [Google Scholar]
- Keele JW, Wray JE, Behrens DW, Rohrer GA, Sunden SL, Kappes SM, Bishop MD, Stone RT, Alexander LJ, et al. A conceptual database model for genomic research. J Comput Biol. 1994;1:65–76. doi: 10.1089/cmb.1994.1.65. [DOI] [PubMed] [Google Scholar]
- Koohmaraie MK, Shackelford SD, Wheeler TL, Lonergan SM, Doumit ME. A muscle hypertrophy condition in lamb (callipyge): Characterization of effects on muscle growth and meat quality traits. J Anim Sci. 1995;73:3596–3607. doi: 10.2527/1995.73123596x. [DOI] [PubMed] [Google Scholar]
- Lemercier C, To RQ, Carrasco RA, Konieczny SF. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of myoD. EMBO J. 1998;17:1412–1422. doi: 10.1093/emboj/17.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leymaster KA. Straightbred comparison of a composite population and the Suffolk breed for performance traits of sheep. J Anim Sci. 1991;69:993–999. doi: 10.2527/1991.693993x. [DOI] [PubMed] [Google Scholar]
- Little DP, Cornish TJ, O'Donnell MJ, Braun A, Cotter RJ. MALDI on a chip: Analysis of arrays of low femtomole to subfemtomole quantities of synthetic oligonucleotides and DNA diagnostic products dispensed by a piezoelectric pipette. Anal Chem. 1997;69:4540–4546. [Google Scholar]
- Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, Sul HS. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002;22:5585–5592. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, Pisselet C, Riquet J, Monniaux D, Callebaut I, et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc Natl Acad Sci. 2001;98:5104–5109. doi: 10.1073/pnas.091577598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL. POLYPHRED: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Meth Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. pp. 7.43–7.45. [Google Scholar]
- Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The DLK1 and Gtl2 genes are linked and reciprocally imprinted. Genes & Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- Sloop KW, McCutchan-Schiller A, Blanton JR, Jr, Meier BC, Rohrer G, Smith TPL, Rhodes SJ. Biochemical and genetic characterization of the porcine Prophet of Pit-1 pituitary transcription factor. Mol Cell Endocrinol. 2000;168:77–87. doi: 10.1016/s0303-7207(00)00318-x. [DOI] [PubMed] [Google Scholar]
- Smith TPL, Godtel RA, Lee RT. PCR-based setup for high-throughput cDNA library sequencing on the ABI 3700 automated DNA sequencer. Biotechniques. 2000;29:698–700. doi: 10.2144/00294bm05. [DOI] [PubMed] [Google Scholar]
- Stone RT, Gross MW, Casas E, Smith TPL, Keele JW, Bennett GL. Use of bovine EST data and human genomic sequences to map 100 gene-specific bovine markers. Mamm Genome. 2002;13:211–215. doi: 10.1007/s00335-001-2124-9. [DOI] [PubMed] [Google Scholar]
- Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, Johnson MH, Paulsen M, Ferguson-Smith AC. Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol. 2000;10:1135–1138. doi: 10.1016/s0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. Blast 2 sequences—A new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Warren W, Smith TPL, Rexroad CE, III, Fahrenkrug SC, Allison T, Shu CL, Catanese J, de Jong PJ. Construction and characterization of a new bovine bacterial artificial chromosome library with 10 genome-equivalent coverage. Mamm Genome. 2000;11:662–663. doi: 10.1007/s003350010126. [DOI] [PubMed] [Google Scholar]
- Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000;10:1711–1718. doi: 10.1101/gr.161600. [DOI] [PMC free article] [PubMed] [Google Scholar]