Abstract
Remnants of more than 3 million transposable elements, primarily retroelements, comprise nearly half of the human genome and have generated much speculation concerning their evolutionary significance. We have exploited the draft human genome sequence to examine the distributions of retroelements on a genome-wide scale. Here we show that genomic densities of 10 major classes of human retroelements are distributed differently with respect to surrounding GC content and also show that the oldest elements are preferentially found in regions of lower GC compared with their younger relatives. In addition, we determined whether retroelement densities with respect to genes could be accurately predicted based on surrounding GC content or if genes exert independent effects on the density distributions. This analysis revealed that all classes of long terminal repeat (LTR) retroelements and L1 elements, particularly those in the same orientation as the nearest gene, are significantly underrepresented within genes and older LTR elements are also underrepresented in regions within 5 kb of genes. Thus, LTR elements have been excluded from gene regions, likely because of their potential to affect gene transcription. In contrast, the density of Alu sequences in the proximity of genes is significantly greater than that predicted based on the surrounding GC content. Furthermore, we show that the previously described density shift of Alu repeats with age to domains of higher GC was markedly delayed on the Y chromosome, suggesting that recombination between chromosome pairs greatly facilitates genomic redistributions of retroelements. These findings suggest that retroelements can be removed from the genome, possibly through recombination resulting in re-creation of insert-free alleles. Such a process may provide an explanation for the shifting distributions of retroelements with time.
Since Barbara McClintock discovered transposable elements (TEs) in maize (McClintock 1956), it has become well established that such elements are universal. Although there are examples of both loss and increase of host fitness because of the activity of transposable elements, their population dynamics are far from being understood, and the forces underlying their genomic distributions and maintenance in populations are a matter of debate (Biemont et al. 1997; Charlesworth et al. 1997). The prevailing view is that TEs are essentially selfish DNA parasites with little functional relevance for their hosts (Doolittle and Sapienza 1980; Orgel and Crick 1980; Yoder et al. 1997). According to this hypothesis, the interaction of TEs with the host is primarily neutral or detrimental and their abundance is a direct result of the ability to replicate autonomously. It is generally accepted that selection is the major mechanism controlling the spread and distribution of TEs in natural populations of model organisms (Charlesworth and Langley 1991). Although the exact mechanisms through which selection acts are controversial, the processes controlling transposition involve selection against the deleterious effects of TE insertions close to genes (Charlesworth and Charlesworth 1983; Kaplan and Brookfield 1983) and selection against rearrangements caused by unequal recombination (ectopic exchange) in meiosis (Langley et al. 1988). More recently, the ubiquitous nature of TEs has gained increasing attention and it is now becoming accepted that TEs give rise to selectively advantageous adaptive variability that contributes to evolution of their hosts (McDonald 1995; Brosius 1999). However, the mechanisms responsible for maintenance, dispersion, fixation, and genomic clearance of TEs remain largely unknown.
Although most work on TEs has focused on model organisms, sequencing of the human genome has revealed that nearly half of our DNA is derived from ancient TEs, mainly retroelements (Smit 1999; International Human Genome Sequencing Consortium 2001). The wealth of human genomic information now allows comprehensive explorations into the evolutionary history and genomic distribution patterns of transposable elements with a view to increasing our understanding of the forces that have shaped our genome and its mobile inhabitants. The retroelements present in the human genome are divided in two major types, the non-LTR and LTR retroelements (International Human Genome cConsortium 2001). The non-LTR retroelements are represented by the autonomous L1 and L2 elements (LINE repeats) and the non-autonomous Alu and MIR (SINE) repeats and have been extensively studied (Smit 1999; International Human Genome Sequencing Consortium 2001; Ostertag and Kazazian 2001; Batzer and Deininger 2002), but appreciation of the heterogeneous collection of LTR retroelements is more limited. These sequences make up 8% of the human genome (International Human Genome Sequencing Consortium 2001) and include defective endogenous retroviruses (ERVs) (Wilkinson et al. 1994; Sverdlov 2000; Tristem 2000), related solitary LTRs, and sequences with LTR-like features for which no homologous proviral structure has been found. More than 200 families of LTR retroelements are defined in Repbase (Jurka 2000), but they can be grouped into six broad superfamilies (see Methods). Although some of the LTR retroelement families, particularly members of class I and II ERVs, presumably entered the primate germ line as infectious retroviruses and then amplified via retrotransposition (Wilkinson et al. 1994; Sverdlov 2000; Tristem 2000), other LTR families likely represent ancient retrotransposons that amplified at different stages during mammalian evolution (Smit 1993).
The vast majority of human retroelements were actively transposing at various stages prior to and during the radiation of mammals and are now deeply fixed in the primate lineage. Essentially only the youngest subtypes of Alu (Batzer and Deininger 2002) and L1 elements (Ostertag and Kazazian 2001) are still actively retrotransposing in humans. Some ERVs belonging to the Class II HERV-K family are human specific (Medstrand and Mager 1998) and a few are polymorphic (Turner et al. 2001), but no current activity of human ERVs has been documented. Here we show that genomic densities of human retroelements vary with distance from genes and that their distributions with respect to surrounding GC content also shift as a function of their age.
RESULTS AND DISCUSSION
Distributions of Retroelements in Different GC Domains
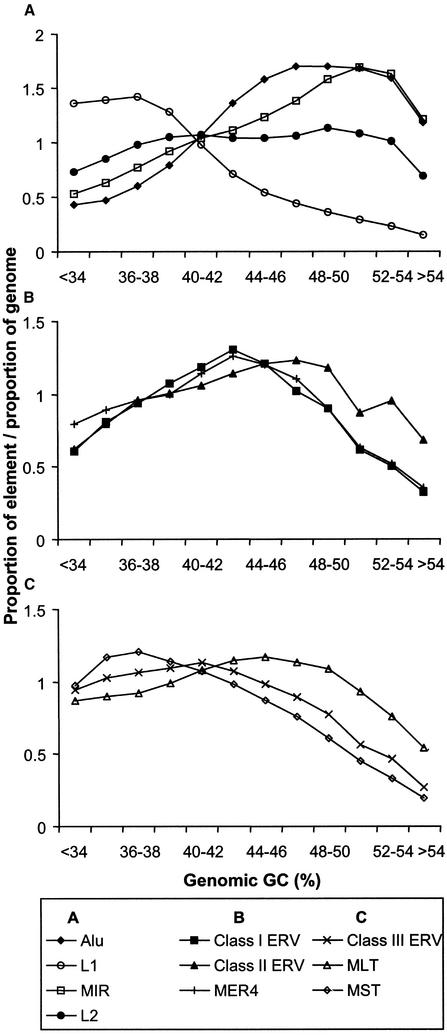
To begin our analysis, we measured the density of various retroelements with respect to GC content in 20-kb windows across the human genome sequence. As reported previously (Smit 1999; International Human Genome Sequencing Consortium 2001), L1 elements are predominantly found in the AT-rich regions, L2 elements are more uniformly distributed whereas Alu and MIR repeats reside in the higher GC fractions of the genome (Fig. 1A) in comparison to the entire genome which has an average GC content of 40% (International Human Genome Sequencing Consortium 2001). For the different LTR superfamilies, an uneven distribution in GC occupancy is also observed. The relatively young Class I ERVs and the nonautonomous MER4 sequences, which may have been propagated by Class I elements, have very similar broad distributions that peak in regions of “medium” GC. Class II ERVs, which include the youngest known HERVs (Medstrand and Mager 1998; Turner et al. 2001), have a distribution more skewed toward higher GC regions (Fig. 1B). Distributions of the older Class III ERVs and their distantly related MLT and MST elements are generally biased toward low GC regions, except for MLT elements, which are spread more uniformly (Fig. 1C).
Figure 1.
Density of retroelements in different GC fractions in the human genome, calculated over 20-kb windows across the genome sequence. (A–C) The density of various retroelement classes. Those represented in each panel are indicated in the box below the graphs. The bins from left to right correspond to an increasing 2% GC fraction.
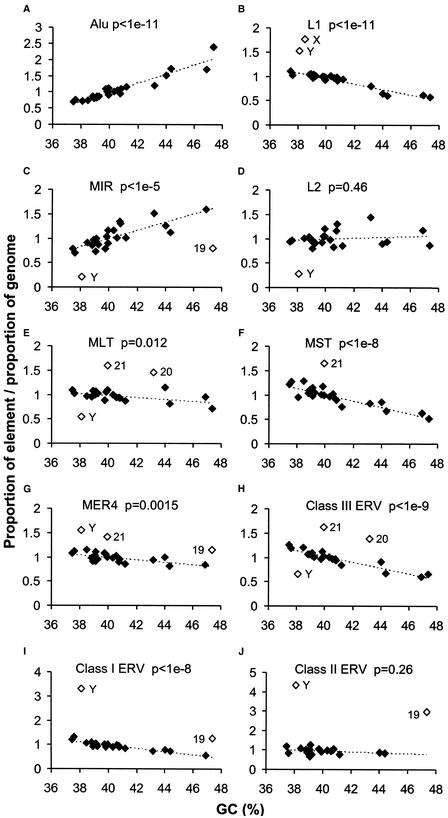
To determine whether retroelement densities on each chromosome agree with overall densities shown in Figure 1, we plotted densities against estimated gene (data not shown) or average GC content of each chromosome (Fig. 2). As expected, the two distribution profiles are almost identical because of the strong correlation between GC content and gene density (International Human Genome Sequencing Consortium 2001). The density of Alu elements increases as a strict function of increasing GC content and MIR elements also generally follow this trend (Fig. 2A,C). In contrast, there is generally a negative or no correlation between the density of L1, L2, or LTR elements and gene density or GC content (Fig. 2). The Class II ERVs and the MLT elements show little, if any, bias for GC-poor chromosomes, whereas the L1, Class I, III, and MST groups are overrepresented on these chromosomes. Class I–II elements are dramatically overrepresented on chromosome Y, as noted before (Kjellman et al. 1995; Smit 1999; International Human Genome Sequencing Consortium 2001), and also somewhat on 19. Abundance of the youngest ERVs on chromosome Y may be due to recombination isolation and absence of major recent rearrangements on much of this chromosome (Graves 1995; Lahn et al. 2001), and because chromosome 19 is much more gene dense than the other chromosomes (International Human Genome Sequencing Consortium 2001), one possible explanation for the overrepresentation of the same ERVs on this autosome is that these elements had an initial integration preference for regions near genes or gene-related features such as CpG islands. We also noted an underrepresentation on Y of the old L2, MIR, and MLT retroelements, which is consistent with major rearrangements and deletions of Y during mammalian evolution (Lahn et al. 2001). Similar trends are observed for MER4 distributions and their autonomous class I counterparts (overrepresentation on Y and 19), and for the nonautonomous MaLR (MLT and MST) elements and their apparent autonomous class III ERVs (overrepresentation on 21). Alu, L1, MER4, and class I and II ERV sequences represent the “young” elements that have actively amplified during the last 40 MYR of primate evolution, whereas other element types were already inactivated for transposition by this time (International Human Genome Sequencing Consortium 2001). All “young” retroelements except Alu sequences are overrepresented on Y. Even though some of the LTR superfamilies show a stronger negative correlation than others, the distribution profiles demonstrate that various retroelement families cluster preferentially in different genomic landscapes and are in agreement with the general trends observed in Figure 1.
Figure 2.
Density of retroelements as a function of average GC content of each human chromosome. The line connecting solid diamonds indicates the general correlation trend between retroelement and GC content of individual chromosomes. The level of significance (P values) of the correlation for each data set is indicated. Open diamonds were excluded from the correlation analysis and indicate over- or underrepresentation of retroelement density on a particular chromosome. Chromosomes 20, 21, and 22 were excluded from the Class II graph (J) because they had <100 supporting elements.
Arrangements of Retroelements With Respect to Genes
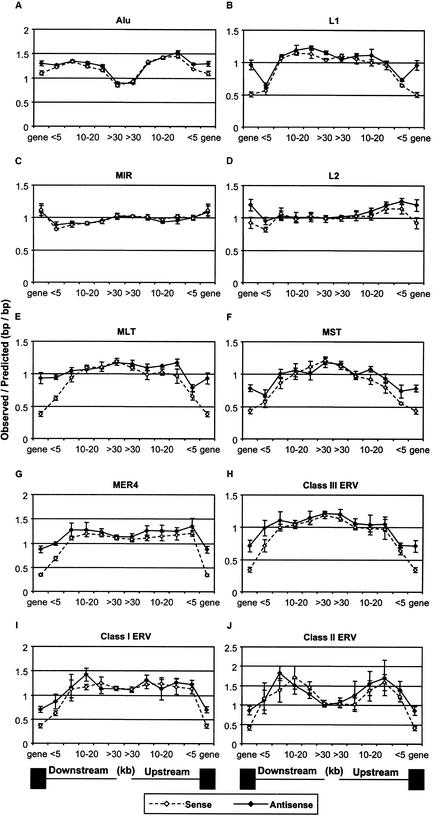
Given the results in Figures 1 and 2, we looked in more detail at the distribution of retroelements by locating all elements in the human genome relative to annotated genes. Although it is reasonable to assume that locations with respect to genes affect retroelement dispersal and fixation patterns, the aim of this analysis was to attempt to obtain a measure of this effect. Our strategy was to determine how closely retroelement densities with respect to genes could be predicted based on the surrounding GC content. DNA regions located upstream of each gene's transcriptional start site and downstream of the polyadenylation site were divided into segments of various size fractions (see Methods) and the density of each retroelement class in either transcriptional orientation with respect to the gene was determined. Regions within the boundaries of a gene, including the introns, were assigned a single segment. The local GC content of each segment was also calculated and used to determine an expected retroelement density based on the whole genome distributions indicated in Figure 1 (see Methods) and the results shown in Figure 3. To obtain estimates of the variation associated with this type of analysis, we divided the genome into four “subgenomes” as detailed in Methods and performed the analysis independently for each. The points in the graphs represent the mean and standard deviation derived from values obtained for each subgenome.
Figure 3.
Ratios of observed to predicted retroelement densities with respect to genes in the human genome. The points above “gene” and “<5” of each graph indicate the density in gene regions, and in the first 5 kb either 5′ or 3′ of genes. The other bins are 5–10, 10–20, 20–30, and >30 kb either upstream or downstream of genes. Open symbols and broken lines indicate elements in the same or sense orientation with respect to the nearest gene and solid symbols and lines indicate elements in the reverse direction. Standard deviation error bars, which are too small to see in some cases, were determined as described in Methods. Solid boxes below the graphs represent gene regions and the lines indicate the distance bins of the intergenic regions. It should be noted that the vast majority of retroelements within genes are located in introns.
Dividing the genome based on proximity to genes revealed several intriguing patterns. First, densities of the relatively old MIR and L2 elements in intergenic regions generally conform to that predicted from the GC content of each region. That is, the ratio of observed-to-expected density is close to one (Fig. 3C,D). Second, for the SINE (Alu and MIR) elements, densities within genes are close to that predicted or are overrepresented based on average GC content of gene regions (Fig. 3A,B,D). In contrast, L1 elements and all six LTR classes, particularly those in the same transcriptional direction, are underrepresented within genes (Fig. 3B,E–J). L1 sequences and the older MLT, MST, and Class III elements are also underrepresented in the 0–5-kb regions both upstream and downstream of genes, whereas the younger class I and MER4 elements are underrepresented in the downstream region only. The higher tendency for LTR elements and L1s within genes to be oriented in the antisense direction has been noted previously (Smit 1999) and likely reflects less fixation because of interference by retroelement regulatory motifs, such as polyadenylation signals, when genes and elements are located in the same transcriptional direction. However, this is the first study to demonstrate lower densities of LTR and L1 elements within genes relative to that predicted based on the surrounding GC content. In addition, the fact that an orientation bias for some elements extends to significant distances away from genes has not been reported previously. Moreover, our analysis indicates that the densities of most LTR elements and L1s are highest in regions furthest from genes. These patterns suggest that L1 and LTR elements are excluded from genes and nearby regions by selection. Interestingly, the density distribution of Alu elements with respect to genes is opposite to that observed for L1 and most LTR elements in that the density is lowest in regions most distant from genes and they are overrepresented (as predicted by GC content) in regions within and near genes. It is also noteworthy that densities of the relatively young LTR class II elements peak in the region 5–20 kb 5′ or 3′ of genes and, indeed, are overrepresented in these areas compared to the expected densities based on regional GC content (Fig. 3J). Such a pattern may reflect a preference for this class of elements to integrate near genes.
The statistical significance of these results is shown in Table 1, which lists the resulting P values for three sets of comparisons. The top of the table compares the sense versus antisense distributions and confirms the significance of the orientation biases discussed above. MIR elements are the only group to show no significant orientation bias. In contrast, an orientation bias extends up to 20 kb 5′ of genes for MLT and MST elements. The bottom two panels in Table 1 compare densities of retroelements in each orientation at each intergenic location to the densities of retroelements in regions most distant (>30 kb) from genes. These latter comparisons illustrate that the retroelement density differences plotted relative to gene location are highly significant. For example, the densities of Alu sequences at all locations are highly significantly different from their density in regions >30 kb from genes.
Table 1.
Significance (P Values) of Retroelement Locations With Respect to Genes
| Sense vs. Antisense | ||||||||||
| Alu | L1 | MIR | L2 | MLT | MST | MER4 | Class III | Class I | Class II | |
|---|---|---|---|---|---|---|---|---|---|---|
| ina | 0.001 | 4.9E-05 | 0.34 | 0.005 | 2.9E-05 | 9.7E-05 | 9.2E-06 | 3.6E-04 | 1.6E-04 | 3.7E-04 |
| 0-5 dnstb | 0.051 | 0.041 | 0.042 | 0.007 | 9.1E-06 | 0.069 | 3.9E-05 | 0.011 | 0.02 | 0.44 |
| 5-10 dnst | 0.48 | 0.22 | 0.17 | 0.32 | 0.03 | 0.034 | 0.054 | 0.13 | 0.44 | 0.037 |
| 10-20 dnst | 0.014 | 0.19 | 0.43 | 0.32 | 0.24 | 0.26 | 0.2 | 0.35 | 0.012 | 0.18 |
| 20-30 dnst | 0.002 | 0.034 | 0.29 | 0.37 | 0.37 | 0.092 | 0.089 | 0.26 | 0.14 | 0.078 |
| >30 dnst | 0.019 | 0.003 | 0.4 | 0.39 | 0.35 | 0.29 | 0.3 | 0.21 | 0.41 | 0.2 |
| >30 upstc | 0.035 | 0.047 | 0.41 | 0.24 | 0.057 | 0.29 | 0.21 | 0.14 | 0.38 | 0.34 |
| 20-30 upst | 0.25 | 0.16 | 0.23 | 0.45 | 0.037 | 0.27 | 0.018 | 0.14 | 0.13 | 0.17 |
| 10-20 upst | 0.42 | 0.053 | 0.067 | 0.054 | 0.008 | 0.012 | 0.11 | 0.28 | 0.25 | 0.23 |
| 5-10 upst | 0.043 | 0.17 | 0.066 | 0.23 | 0.014 | 0.042 | 0.15 | 0.25 | 0.23 | 0.35 |
| 0-5 upst | 6.0E-05 | 0.001 | 0.29 | 0.024 | 0.015 | 0.009 | 0.096 | 0.03 | 0.15 | 0.15 |
| in | 0.001 | 4.9E-05 | 0.34 | 0.005 | 2.9E-05 | 9.7E-05 | 9.2E-06 | 3.6E-04 | 1.6E-04 | 3.7E-04 |
| Antisense vs. >30 kb from genes | ||||||||||
| Alu | L1 | MIR | L2 | MLT | MST | MER4 | Class III | Class I | Class II | |
| in | 8.9E-06 | 0.015 | 0.13 | 0.007 | 0.003 | 1.2E-04 | 0.001 | 6.9E-05 | 4.3E-05 | 0.02 |
| 0-5 dnst | 9.9E-07 | 2.3E-06 | 0.006 | 0.1 | 1.3E-04 | 1.0E-04 | 0.005 | 0.012 | 0.015 | 0.27 |
| 5-10 dnst | 7.1E-07 | 0.45 | 0.009 | 0.29 | 0.006 | 0.018 | 0.08 | 0.12 | 0.42 | 1.8E-04 |
| 10-20 dnst | 4.9E-07 | 0.058 | 0.003 | 0.49 | 0.026 | 0.097 | 0.07 | 0.002 | 0.005 | 0.006 |
| 20-30 dnst | 1.1E-06 | 0.002 | 0.074 | 0.5 | 0.011 | 0.03 | 0.032 | 0.18 | 0.41 | 0.005 |
| 20-30 upst | 4.2E-07 | 0.38 | 0.2 | 0.27 | 0.06 | 0.007 | 0.033 | 0.01 | 0.013 | 0.17 |
| 10-20 upst | 1.5E-07 | 0.38 | 0.011 | 0.012 | 0.045 | 0.064 | 0.061 | 0.054 | 0.46 | 0.014 |
| 5-10 upst | 2.1E-07 | 0.004 | 0.083 | 0.001 | 0.38 | 0.004 | 0.022 | 0.029 | 0.028 | 0.022 |
| 0-5 upst | 3.0E-07 | 3.0E-06 | 0.34 | 7.7E-05 | 2.4E-05 | 4.8E-04 | 0.033 | 1.2E-06 | 0.06 | 0.016 |
| in | 8.9E-06 | 0.015 | 0.13 | 0.007 | 0.003 | 1.2E-04 | 0.001 | 6.9E-05 | 4.3E-05 | 0.02 |
| Sense vs. >30 kb from genes | ||||||||||
| Alu | L1 | MIR | L2 | MLT | MST | MER4 | Class III | Class I | Class II | |
| in | 1.7E-04 | 8.6E-07 | 0.069 | 0.14 | 4.4E-07 | 2.2E-06 | 1.3E-07 | 1.6E-07 | 1.5E-07 | 1.3E-05 |
| 0-5 dnst | 1.0E-06 | 6.9E-06 | 9.3E-05 | 0.002 | 2.5E-06 | 2.3E-05 | 1.0E-05 | 1.9E-04 | 2.0E-06 | 0.3 |
| 5-10 dnst | 9.0E-07 | 0.45 | 0.003 | 0.12 | 0.003 | 0.001 | 0.39 | 0.003 | 0.48 | 0.055 |
| 10-20 dnst | 2.9E-06 | 0.007 | 0.003 | 0.39 | 0.15 | 0.019 | 0.034 | 0.02 | 0.18 | 0.005 |
| 20-30 dnst | 3.9E-06 | 0.096 | 0.011 | 0.24 | 0.28 | 0.22 | 0.03 | 0.068 | 0.078 | 0.002 |
| 20-30 upst | 1.8E-07 | 0.4 | 0.36 | 0.24 | 0.012 | 0.002 | 0.44 | 0.002 | 0.073 | 0.4 |
| 10-20 upst | 1.4E-07 | 0.053 | 0.066 | 0.24 | 0.015 | 0.003 | 0.23 | 0.002 | 0.04 | 0.041 |
| 5-10 upst | 4.7E-07 | 0.02 | 0.41 | 0.012 | 0.026 | 4.6E-04 | 0.14 | 0.045 | 0.33 | 0.002 |
| 0-5 upst | 6.8E-07 | 6.8E-07 | 0.13 | 0.012 | 2.1E-05 | 1.3E-06 | 0.017 | 8.3E-06 | 0.48 | 0.043 |
| in | 1.7E-04 | 8.6E-07 | 0.069 | 0.14 | 4.4E-07 | 2.2E-06 | 1.3E-07 | 1.6E-07 | 1.5E-07 | 1.3E-05 |
Shaded regions are significant (P < 0.05). aWithin a gene. bdnst: kb downstream of the nearest gene. cupst: kb upstream of the nearest gene.
Shifting Retroelement Distributions With Age
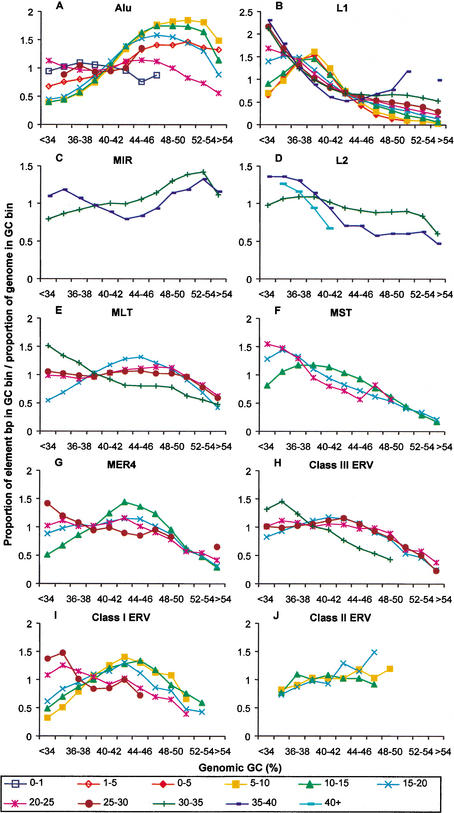
It is apparent that the retroelement distributions in genes and intergenic regions (Fig. 3) do not fully conform to the genome-wide distribution patterns of elements observed in Figures 1 and 2. Furthermore, for Alu repeats, it has been reported previously that young elements (<1 myr) have a preference for AT-rich regions whereas older Alus show an increasing density in GC-rich DNA (Smit 1999; International Human Genome Sequencing Consortium 2001) (see Fig. 4A) and hypotheses to explain this phenomenon have been proposed (Schmid 1998; Brookfield 2001; International Human Genome Sequencing Consortium 2001; Pavlicek et al. 2001). Transposition into AT-rich regions might be expected to lead to accumulation of TEs in this gene-poor part of the genome (e.g., the heterochromatin) where recombination is strongly reduced and element interference with genes is less pronounced. However, the observed density differences of the youngest Alu elements (present in AT-rich regions) as opposed to older elements (in GC-rich regions) do not follow this expectation. A possible explanation for the age-related Alu density differences is that these retroelements are removed preferentially from their initial integration sites in the AT-rich regions of the genome prior to fixation. However, because there is a gradual density increase of Alu elements by age in the GC-rich fraction, it is possible that already fixed elements are gradually lost from the AT-rich region while they are maintained in GC-rich regions.
Figure 4.
Retroelement densities of different divergence classes in various GC fractions of the human genome. The density distribution of each retroelement divergence cohort was plotted in GC bins as indicated in the legend to Figure 1. The divergence classes are indicated in % divergence from the consensus sequence below the graphs. Data points missing in traces are due to GC bins containing <100 elements. Standard deviations were calculated (see Methods) but are not shown in the interest of clarity.
To investigate whether other retroelements also change their genomic distribution with age, we determined the distribution patterns of LTR elements, SINEs, and LINEs of different ages as a function of GC content (Fig. 4). As discussed above, it is apparent that the youngest Alu elements (0–1% divergent), many of which are polymorphic insertions (Carroll et al. 2001; Batzer and Deininger 2002), are distributed differently than the next youngest (fixed) Alus of the 1–5% divergence group and that the densities of the next two Alu age cohorts (5–15% divergent) are skewed even further to GC-rich regions (Fig. 4A). Notably, this figure also reveals that the oldest Alu repeats are less prevalent in GC-rich domains and, indeed, have a density distribution closer to that of the youngest age class. This density pattern of the oldest Alu elements was not evident in a similar analysis reported previously (International Human Genome Sequencing Consortium 2001). In that study, Alu elements were divided by subfamily instead of divergence and the density of the oldest subfamily, AluJ, was still highly skewed to GC-rich regions. However, the AluJ subfamily was considered as a single large cohort, the members of which have divergences ranging from <10% to >25%. When the more divergent AluJ members of 15%–20% and 20%–25% divergence are separated into their own groups, their densities are essentially identical to the patterns presented in Figure 4A (data not shown). Thus, the different methods for separating Alu elements accounts for the differences between our analysis and that in the genome consortium study.
Results of similar analyses conducted for the other retroelements reveal some provocative trends. As noted before (Smit 1999) and as shown in Figure 4B, young L1 elements are preferentially found in the AT-rich fraction in the genome and older elements tend to be found in the most AT-dense part of the genome. Analysis of the ancient L2 and MIR repeats was hampered by the short average length of most elements, which prevented an accurate determination of their divergence from a consensus sequence (age) (see Methods for details). However, for the two divergence classes that could be reliably determined, the oldest L2 and MIR sequences also show an increased density in the less GC-rich sections of the genome compared with their younger counterparts (Fig. 4C,D).
For most of the LTR elements, we observe a trend similar to that seen for the L2 and MIR sequences. For elements belonging to the MLT, MST, MER4, and Class I and III ERV groups, densities of the youngest members of these superfamilies peak in regions of higher GC compared with their older relatives (Fig. 4E–I). That is, the highest concentrations of these elements appear to gradually shift to regions of lower GC with increasing age. This tendency is not evident for the Class II ERVs (Fig. 4J). Potential explanations for this trend will be discussed below.
To determine whether the shifting patterns observed in Figure 4 are statistically significant, we again divided the genome into four subgenomes and redid the analysis for each of these. Each point in the graphs could then be assigned a mean and standard deviation based on values obtained for each subgenome. The t-test was used to determine whether the density distribution of a particular age cohort was significantly different when compared with the next oldest cohort. Table 2 lists the P values resulting from this analysis. For all retroelements except the Class II ERVs, the majority of the density points are significantly different (P < 0.05) for at least one comparison between adjoining age cohorts. Indeed, for the most numerous elements, Alu and L1, almost all comparisons are statistically significant. If the youngest and oldest age cohort of each superfamily are compared, all except the Class II ERVs are highly significant (data not shown).
Table 2.
Significance (P-Values) of Distributional Differences Between Divergence Cohorts
| Alu | L1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0-1: | 1-5: | 5-10: | 10-15: | 15-20: | 20-25: | 0.5: | 5-10: | 10-15: | 15-20: | 20-25: | 25-30: | 30-35: | |
| 1-5a | 5-10 | 10-15 | 15-20 | 20-25 | 25-30 | 5-10 | 10-15 | 15-20 | 20-25 | 25-30 | 30-35 | 35-40 | |
| <34b | 0.20 | 0.10 | 0.50 | 0.36 | 0.038 | 0.49 | 0.26 | 0.18 | 0.34 | 0.30 | 0.48 | 0.43 | |
| 34-36 | 0.002 | 2.5E-06 | 0.43 | 7.9E-05 | 1.6E-07 | 0.07 | 0.06 | 0.002 | 2.0E-07 | 0.08 | 0.021 | 0.007 | 0.006 |
| 36-38 | 0.001 | 1.8E-04 | 0.25 | 1.3E-04 | 5.9E-07 | 0.31 | 0.004 | 0.27 | 0.031 | 0.002 | 0.001 | 0.016 | 0.47 |
| 38-40 | 4.4E-04 | 2.1E-04 | 0.002 | 9.2E-05 | 0.003 | 0.39 | 0.41 | 0.006 | 1.5E-04 | 9.2E-05 | 2.7E-05 | 0.13 | 0.020 |
| 40-42 | 0.007 | 0.17 | 0.013 | 0.002 | 0.006 | 0.24 | 0.029 | 0.010 | 8.0E-05 | 0.005 | 0.011 | 0.28 | 0.001 |
| 42-44 | 0.06 | 4.2E-05 | 0.009 | 0.46 | 0.001 | 0.14 | 0.37 | 0.36 | 0.003 | 0.042 | 0.18 | 0.12 | 0.05 |
| 44-46 | 4.1E-04 | 4.0E-05 | 0.025 | 0.002 | 1.4E-05 | 0.016 | 0.08 | 0.002 | 0.043 | 0.023 | 0.001 | 0.032 | 0.019 |
| 46-48 | 2.3E-04 | 6.1E-05 | 0.29 | 4.9E-04 | 9.1E-07 | 0.023 | 6.2E-05 | 0.005 | 0.002 | 0.09 | 0.003 | 0.25 | |
| 48-50 | 4.0E-04 | 0.022 | 2.9E-04 | 3.5E-05 | 0.020 | 3.9E-05 | 0.010 | 0.021 | 0.009 | 3.9E-05 | 0.048 | ||
| 50-52 | 4.1E-04 | 0.040 | 1.2E-04 | 1.4E-06 | 0.10 | 0.001 | 0.001 | 0.008 | 0.017 | 0.013 | 0.001 | ||
| 52-54 | 6.8E-05 | 4.0E-04 | 7.7E-05 | 4.2E-06 | 0.002 | 0.002 | 0.001 | 0.001 | 3.9E-04 | ||||
| >54 | 0.017 | 3.2E-07 | 3.1E-07 | 0.001 | 2.6E-04 | 6.4E-06 | 1.4E-05 | 1.1E-04 | 1.8E-06 | 0.001 | |||
| MIR | L2 | MLT | MST | MER4 | |||||||||
| 30-35: | 30-35: | 35-40: | 15-20: | 20-25: | 25-30: | 10-15: | 15-20: | 10-15: | 15-20: | 20-25: | |||
| 35-40 | 35-40 | 40+ | 20-25 | 25-30 | 30-35 | 15-20 | 20-25 | 15-20 | 20-25 | 25-30 | |||
| <34 | 0.23 | 0.24 | 0.11 | 0.45 | 0.22 | 0.16 | 0.30 | 0.12 | 0.36 | 0.22 | |||
| 34-36 | 5.7E-05 | 3.8E-06 | 0.05 | 7.6E-05 | 0.12 | 0.001 | 1.5E-05 | 0.10 | 1.5E-04 | 0.029 | 0.10 | ||
| 36-38 | 9.2E-05 | 0.001 | 0.20 | 0.010 | 0.020 | 8.9E-05 | 3.7E-04 | 0.24 | 0.005 | 0.30 | 0.08 | ||
| 38-40 | 0.50 | 0.026 | 0.48 | 0.001 | 0.34 | 0.006 | 0.015 | 0.13 | 0.35 | 0.27 | 0.07 | ||
| 40-42 | 0.001 | 0.019 | 0.07 | 3.6E-04 | 0.23 | 0.014 | 1.2E-04 | 0.024 | 0.012 | 0.31 | 0.044 | ||
| 42-44 | 0.001 | 3.2E-05 | 8.4E-06 | 0.08 | 1.7E-04 | 0.001 | 0.14 | 2.8E-04 | 0.45 | 0.004 | |||
| 44-46 | 0.001 | 2.3E-04 | 9.1E-05 | 0.16 | 0.001 | 0.001 | 0.014 | 0.010 | 0.023 | 0.06 | |||
| 46-48 | 0.036 | 0.001 | 0.043 | 0.023 | 0.002 | 0.001 | 0.026 | 0.016 | 0.11 | 0.38 | |||
| 48-50 | 0.029 | 3.4E-04 | 0.25 | 0.039 | 0.004 | 0.031 | 0.38 | 0.21 | 0.17 | 0.29 | |||
| 50-52 | 0.06 | 1.6E-04 | 0.045 | 0.46 | 1.1E-04 | 0.011 | 0.40 | 0.38 | |||||
| 52-54 | 0.43 | 0.009 | 0.032 | 0.30 | 0.019 | 0.14 | 0.45 | 0.23 | |||||
| >54 | 0.016 | 0.029 | 6.9E-05 | 0.22 | 0.004 | 0.16 | 0.15 | 0.002 | 0.014 | ||||
| Class III | Class I | Class II | |||||||||||
| 15-20: | 20-25: | 25-30: | 5-10: | 10-15: | 15-20: | 20-25: | 5-10: | 10-15: | |||||
| 20-25 | 25-30 | 30-35 | 10-15 | 15-20 | 20-25 | 25-30 | 10-15 | 15-20 | |||||
| <34 | 0.33 | 0.48 | 0.26 | 0.12 | 0.29 | 0.09 | 0.33 | ||||||
| 34-36 | 0.005 | 0.029 | 0.003 | 0.022 | 0.05 | 0.001 | 0.06 | 0.30 | 0.30 | ||||
| 36-38 | 0.21 | 0.08 | 4.1E-05 | 0.020 | 0.18 | 0.041 | 0.06 | 0.050 | 0.028 | ||||
| 38-40 | 0.008 | 0.13 | 0.39 | 0.07 | 0.038 | 0.39 | 0.13 | 0.32 | 0.20 | ||||
| 40-42 | 0.016 | 0.16 | 0.026 | 0.13 | 0.07 | 0.010 | 0.29 | 0.21 | 0.07 | ||||
| 42-44 | 0.023 | 0.020 | 2.8E-04 | 0.035 | 0.21 | 0.011 | 0.28 | 0.46 | 0.08 | ||||
| 44-46 | 0.036 | 0.08 | 0.001 | 0.48 | 0.06 | 0.030 | 0.25 | 0.001 | 0.18 | ||||
| 46-48 | 0.10 | 0.36 | 1.4E-06 | 0.38 | 0.006 | 0.003 | 0.18 | 0.001 | |||||
| 48-50 | 0.06 | 0.21 | 0.011 | 0.004 | 0.08 | 0.031 | |||||||
| 50-52 | 0.033 | 0.24 | 0.30 | 0.031 | 0.15 | ||||||||
| 52-54 | 0.041 | 0.19 | 0.05 | ||||||||||
| >54 | 0.003 | 0.001 | |||||||||||
Shaded regions are significant (P < 0.05).
Divergence cohorts compared.
GC content (%).
One qualification regarding this data concerns the method used to identify retroelements of different ages. Elements were classified as belonging to divergence cohorts based on percent substitution from their consensus sequence (Jurka 2000). The consensus sequence corresponds to the approximate sequence at the time of integration in the genome, where retroelements in higher divergence cohorts indicate an older time of integration relative to the retroelements of lower divergence values (International Human Genome Sequencing Consortium 2001; Li and Graur 1991; Shen et al. 1991; Smit et al. 1995). Therefore, the validity of this method is highly dependent on having accurate consensus sequences for all subfamilies. It is quite possible, and even likely, that some elements have been assigned an incorrect age because of extreme heterogeneity of some of the retroelement classes, particularly among the LTR groups. However, if this was a major problem, one would not expect to observe a consistent shift in density in one direction – namely toward lower GC regions with increasing divergence.
Length Differences Do Not Account for the Shifting Patterns
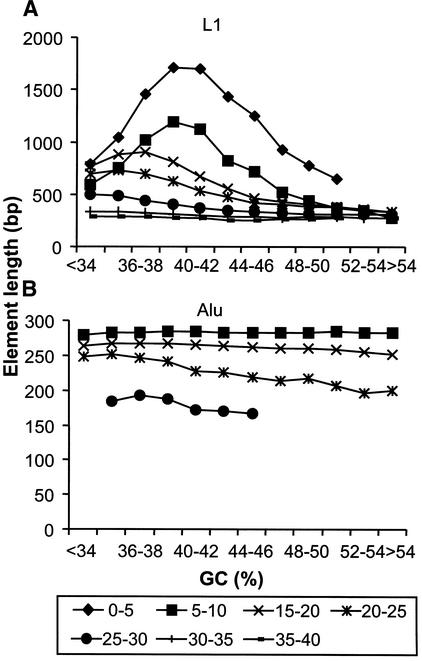
To investigate potential mechanisms that may underlie the age-related distribution differences, we used two different methods to try to determine whether differential rates of retroelement deletions in different genomic GC regions account for the shifting patterns observed in Figure 4. First, we examined the relative length of elements in different GC fractions. The results of this analysis indicated that retroelements gradually become shorter as they age, presumably because of small deletions or loss of recognition of diverged segments by RepeatMasker, but the shortening is largely independent of the surrounding GC content (data not shown). The two exceptions to this general observation are represented by L1 elements and older Alu sequences (Fig. 5). The average length of younger L1 elements (<10% divergence) peaks in the 38%–42% GC fractions, which might explain the abundance of L1 base pairs in this region (Fig. 4B). In the case of Alu elements in the 20%–30% divergence cohorts, there is a slight decrease in apparent length with increasing GC content (Fig. 5B), but this is not enough to account for the density pattern of this age group (Fig. 4A). In addition, the small degree of shortening as measured here does not explain the rapid enrichment of younger Alu elements in higher GC fractions.
Figure 5.
Length distribution of retroelements with respect to surrounding GC content. Retroelements of each group were classified as belonging to divergence cohorts as described in the text. The average length in base pairs (bp) of each retroelement divergence cohort contained within each GC bin (see legend to Fig. 1) is shown for L1 (A) and Alu (B) elements. GC bins containing <100 elements were excluded from the graphs.
Delay of Alu Density Changes on the Y Chromosome
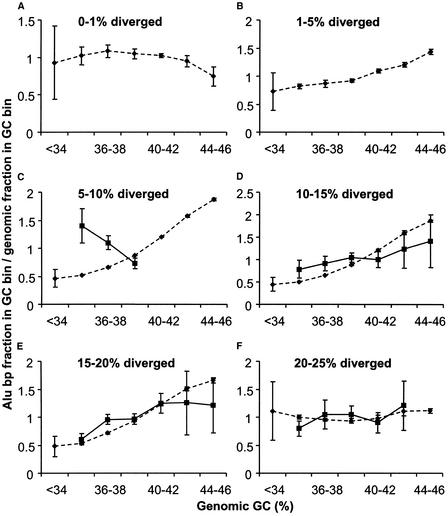
As another way of investigating the change in distribution of younger Alus toward GC-rich regions, we analyzed Alu density patterns on the Y chromosome, much of which does not recombine (Graves 1995), and detected a major difference on this chromosome compared with the whole genome (Fig. 6). Alu elements on chromosome Y <5% divergent are not numerous enough to include in this analysis. However, the density pattern of Alus in the 5%–10% divergence class is strikingly opposite to that observed in the whole genome in that they are much more prevalent in AT-rich regions compared with GC-rich regions (Fig. 6C). The distributions of older Alu elements (<10% divergent from the consensus) with respect to GC content are consistent with the patterns seen in the entire genome (Fig. 6D–F). Table 3 shows the P values resulting from this analysis. This finding suggests that the density shift of Alus from AT-rich to GC-rich regions during evolution was significantly delayed on the Y chromosome and, therefore, that the ability to recombine with a homologous chromosome greatly facilitated this shift.
Figure 6.
Density of Alu divergence cohorts in different GC fractions on chromosome Y compared with the whole genome. Solid lines indicate Alu elements on chromosome Y; broken lines represent the Alu density in the whole genome. (A–F) The density of specific divergence classes, which are indicated on the top of each panel. There were insufficient numbers of Alu elements on the Y chromosome in the first two divergence cohorts to be plotted in A and B. The density distribution of each Alu divergence class is plotted against the local 20-kb genome GC content. Standard deviations were calculated as described in Methods.
Table 3.
Significance (P-Values) of Distributional Difference Between Alus on the Y Chromosome vs. the Whole Genome
| 5-10a | 10-15 | 15-20 | 20-25 | |
|---|---|---|---|---|
| 34-36b | 0.0012 | 0.023 | 0.13 | 0.022 |
| 36-38 | 4.5E-04 | 0.014 | 0.0022 | 0.28 |
| 38-40 | 0.021 | 0.022 | 0.28 | 0.14 |
| 40-42 | 0.039 | 0.41 | 0.27 | |
| 42-44 | 0.10 | 0.24 | 0.34 | |
| 44-46 | 0.11 | 0.08 |
Shaded regions are significant (P < 0.05).
Divergence cohorts compared.
GC content (%).
Potential Explanations for Alu Distribution Patterns
The density patterns of Alu elements do not conform to trends observed for other retroelements. These elements integrate into the AT-rich part but accumulate in GC-rich DNA (International Human Genome Sequencing Consortium 2001) (Fig. 4A) and at least three hypotheses have been proposed to account for this phenomenon. One proposed explanation is that the GC-rich Alu elements are more stable in regions where the surrounding GC content is similar (Pavlicek et al. 2001). However, we have observed that partial deletions or apparent shortening of various Alu age groups are uniformly distributed irrelevant of GC occupancy (Fig. 5B). This finding does not seem to support such a hypothesis, although it is possible that the tendency of retroelements to remain in regions of matching GC content does play some role. A second hypothesis proposes that Alu elements are selectively retained in GC-rich regions because having these elements close to genes is of functional benefit (Britten 1997; Kidwell and Lisch 1997; Schmid 1998). Figure 3A shows that the Alu density near genes is higher than predicted based on GC content. That is, the tendency of Alu elements to be located near genes is not fully explained by the general GC-richness associated with coding regions and such a pattern may therefore reflect a functional role for these elements. However, other observations appear discordant with this view. For example, it is known that the developmentally critical HoxD gene cluster is almost devoid of retroelements (International Human Genome Sequencing Consortium 2001). A recent study has also found that SINEs (Alu and MIR elements) are less frequently associated with imprinted than nonimprinted genomic regions (Greally 2002). Certain classes of genes may therefore need to exclude such sequences from their environment to ensure proper function or regulation. A third hypothesis proposes that the maintenance of Alus in GC-rich regions may be due to the adverse effects that deletions and unequal recombinations could have in gene-rich regions (Brookfield 2001). Indeed, because of the vast numbers of Alu elements, it is likely that specific recombinational mechanisms have been a major force in shaping the distribution of Alus in the genome. It has recently been demonstrated that the efficiency of Alu–Alu recombination in yeast increases as a pair of elements are placed closer together (Lobachev et al. 2000). Such closely spaced Alu pairs are found only occasionally in the human genome (Lobachev et al 2000; Stenger et al. 2001), possibly because of clearance of these elements through the mechanism of inverted repeat (IR)-mediated recombination (Leach 1994). Alu elements seem quite promiscuous for recombination because two elements up to 20% divergent are still able to recombine efficiently (Lobachev et al. 2000). Furthermore, there are many examples of Alu-mediated recombination resulting in mutations in humans (Batzer and Deininger 2002). These findings suggest a possible explanation for the changing Alu distribution profiles shown in Figure 4A and their enrichment near genes. Considering the high number of genomic Alu elements and the fact that they preferentially target AT-rich regions, these domains must have suffered a massive build-up of Alu integrations. Such accumulation likely resulted in increased recombination as the occurrence of closely spaced, highly related Alus increased, which could have led to loss of both newly integrated and fixed Alu elements in the AT-rich fraction of the genome. In regions close to genes, it is possible that Alu–Alu recombination events are less likely to be allowed or become fixed because of an increased chance of simultaneously removing gene regulatory domains (Brookfield 2001). This could help explain the overrepresentation of Alu elements near genes without invoking a functional role. The fact that we observe no increased density in GC- or gene-rich regions for the oldest Alus could be explained by the fact that Alus in these age cohorts are much less numerous and therefore would have been less subject to loss via recombination in AT-rich regions. Alu elements of 20%–30% divergence are present in only ∼25,000 copies whereas younger Alus in the 5%–10%, 10%–15%, and 15%–20% divergence classes are present in ∼300,000, ∼480,000, and ∼210,000 copies, respectively. Furthermore, because of their higher divergence values, the oldest Alus would also have been less able to recombine with their younger, more numerous relatives when the latter populated the genome.
Differences in recombination are likely also responsible for the fact that Alu elements are not over represented on chromosome Y as are other “young” retroelements such as Class I and II ERVs (International Human Genome Sequencing Consortium 2001) (Fig. 2). This finding suggests that Alus are lost more readily than the LTR elements. However, loss of Alu elements on the Y appears delayed compared with on the autosomes (Fig. 6), likely because only intrachromosomal/IR recombination can operate on most of the Y. IR recombinations seem to work more efficiently when two elements are closely located (Lobachev et al. 2000) and it is likely that this is true also for intrachromosomal recombinations in general. Thus, we postulate that LTR elements are removed less efficiently than Alu elements because of their much lower copy number and, therefore, larger average interelement distance.
Concluding Remarks
One view of transposable elements considers them to be selfish DNA of no use to the host (Doolittle and Sapienza 1980; Orgel and Crick 1980; Yoder et al. 1997), whereas others hypothesize that their fixation reflects functional interactions with the host (McDonald 1995; Brosius 1999). Our data support the idea that retroelements have a general negative impact on the host because of a gradual accumulation of most retroelement superfamilies in the AT-rich fraction and on the Y chromosome (which is predicted to occur according to the selfish DNA hypothesis) (Charlesworth et al. 1997). However, these findings also support a concept in which retroelements gradually are cleared (or maintained) from the host genome, a relationship that seems dependant on the age of their association. (Di Franco et al. 1997; Junakovic et al. 1998; Torti et al. 2000; Kidwell and Lisch 2001). The fact that densities of old MIR and L2 retroelements near genes are close to that predicted by average GC content suggests a relatively benign relationship between these retroements and genes. In contrast, retroviral elements may have interfered more often with gene function because of initial integration site preference into gene-rich regions. The density pattern of the relatively young class II ERVs (Fig. 3J) supports this suggestion. Of those LTR elements that have been fixed in the population (i.e., almost all of those in humans), our analyses have revealed that the highest densities of the older elements gradually shift with age to AT-rich or gene-poor DNA. Furthermore, we have shown that all types of LTR retroelements are significantly underrepresented within genes. Because LTRs carry transcriptional regulatory signals very similar to those in cellular genes (Majors 1990), it seems reasonable that insertion of an LTR close to or within a gene would frequently be disadvantageous unless it is efficiently silenced by methylation or other mechanisms (Yoder et al. 1997; Whitelaw and Martin 2001). Such insertions with a marked negative impact will be selected against with no chance to spread to fixation. However, it is known that a mutation with a selective disadvantage can still be fixed through genetic drift, especially if the effective population size is small (Li and Graur 1991). It is possible that some LTR elements, despite being fixed in the species, had a slight negative impact and were gradually eliminated with time. Alternatively, mechanisms unrelated to selection, such as differential rates of recombination in different GC domains, may also explain the shifting density patterns of LTR retroelements. The fact that the youngest Class II ERVs do not show the same density pattern shifts as seen for most of the LTR superfamilies could be because there has not been sufficient evolutionary time for their distribution to be shaped by selective forces and/or recombination.
Once fixed in the population, it is not possible for an insertion to be eliminated unless insert-free alleles are re-created. Although unequal crossing-over between homologous chromosomes may be the main mechanism responsible for elimination of retroelements in GC-rich regions, which have higher rates of recombination (Fullerton et al. 2001), intrachromosomal deletions and IR-mediated recombination might enhance this effect, especially in regions of high retroelement density. Such processes could regenerate insert-free alleles and again provide an opportunity for the original insertion to be lost from the population through natural selection or drift.
Although these studies have attempted to address some of the potential mechanisms or forces that have shaped the genomic distributions of human retroelements, further studies are warranted to elucidate the complex evolutionary and functional relationships between these sequences and their host genome.
METHODS
Description of Retroelements
Human retroelements are classified into two major classes: non-LTR and LTR retroelements. The former category contains the LINEs, represented by the L1 and L2 elements, whereas the Alu and MIR elements belong to SINEs. For this analysis, LTR retroelements were divided into the following 6 groups (Smit 1999; Jurka 2000; International Human Genome Sequencing Consortium 2001; Medstrand and Mager 2002): class I ERVs, which are similar to type C or γ retroviruses such as murine leukemia virus; class II ERVs, which are similar to type B or β retroviruses like mouse mammary tumor virus; class III ERVs (also called ERV-L), which have limited similarity to spuma retroviruses; MER4 elements, which are nonautonomous class I-related ERVs; and MST (named for a common restriction enzyme site MstII) and MLT (mammalian LTR transposon) elements, which are both part of the large nonautonomous mammalian apparent LTR retrotransposon (MaLR) superfamily. Solitary LTRs outnumber LTR elements with internal sequences by approximately 10-fold.
Data Sources
Genomic sequence and annotated gene data for all figures were derived from the August 6, 2001, draft human genome assembly at http://genome.ucsc.edu. Retroelement locations derived from RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html), GC content calculated in nonoverlapping windows of 20-kb sequence gap data, and known gene data from the Reference Sequence database were all downloaded from this site. After compilation, data points were included in graphs only if supported by >100 retroelements. Element count was calculated to reflect as nearly as possible the number of individual integrations of the element. That is, nearby repeat segments (within 20 kb of each other) having the same family name and RepeatMasker alignment parameters (alignment score, substitution, and gap levels) were combined and treated as a single element. Subfamily assignments and divergence values were taken directly from RepeatMasker output files. Internal sequences of LTR elements were excluded from the analysis. Data was further conditionally discarded in figures where retroelement divergence is used as a measure of age. In some cases where element length was very short (<150 bp), it was noted that RepeatMasker assigned an artificially low divergence value because of the alignment method used in finding repeats. This was a particular problem for the old MIR and L2 sequences. An attempt was therefore made to ensure that relative divergence indeed represented age by plotting element length versus assigned divergence values. Because repeats in general grow shorter as they age (see, e.g., Fig. 5), retroelement divergence cohorts were considered anomalous and discarded if they did not follow this trend.
Density Analysis
The retroelement data were compiled by repeat superfamily, divergence from consensus, and surrounding genomic GC content. The density function in Figures 1, 4, and 6 was calculated as the fraction of the retroelement base pairs in a given GC bin divided by the fraction of the genome in that GC bin. Thus, it affords a measure of preference of a particular age class for different GC contents. When an age class of an element had a significant presence in only some of the GC bins, the effective genome size for that age class was calculated from the sizes of only those GC bins. Thus, for the Figure 6 genomic data, the “whole genome” is that fraction of the genome with GC content <46%. In Figure 2, the “bin” considered was an individual chromosome. With these considerations in mind, the calculations of density are identical.
For Figure 2 (retroelement density versus GC content on each chromosome), correlation coefficients (r) and level of significance (P values) were calculated for each data set. The graphs of chromosomal retroelement density as a function of gene density are not shown but are almost identical because of the highly significant correlation between GC content and gene density (International Human Genome Sequencing Consortium 2001).
For Figure 3, a script divided the chromosomes into eleven segment types or bins: within the transcript start and end positions of known (annotated) genes and 0–5, 5–10, 10–20, 20–30, and >30 kb upstream and downstream of genes. The majority of the genome was located either within genes (22% of the total) or at distances >30 kb from genes (63% of the total). In each segment, the script determined the base-pair contribution of each retroelement type and noted the orientation of the element with respect to the nearest gene. The GC content of each segment was calculated and then the density data from Figure 1 was used to predict the base pair contribution by each retroelement type in the segment. Predictions done within genes or at distances >30 kb from genes were compiled from predictions made from 10 kb subsegments. Half of the predicted retroelement base pairs were assumed to be in the sense orientation and half in antisense. Finally, the observed base pairs in each bin were divided by the cumulative predicted base pairs for each retroelement type.
P values shown in Tables 1, 2, and 3 and variability of the data in Figures 3, 4, and 6 were calculated as follows. The sequence segments comprising the whole genome were divided up into four “subgenomes” of equal composition. The retroelement distributions were calculated in each subgenome, and the means and standard deviations of retroelement distributions were calculated. After appropriate normalization, the significance (P value) of the difference between different retroelement distributions was tested by the one-tailed unpaired t-test.
WEB SITE REFERENCES
http://genome.ucsc.edu; UC Santa Cruz genome browser.
http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker.
Acknowledgments
We thank Christine Kelly for help with manuscript preparation. We also thank an anonymous reviewer for many helpful comments. This work was supported by a grant from the Canadian Institutes of Health Research to D.M. with core support provided by the British Columbia Cancer Agency. P.M. was supported by a fellowship from the Knut and Alice Wallenberg Foundation, Sweden and by grants from Magn. Bergvalls Foundation and Ake Wibergs Foundation, Sweden.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dixie@interchange.ubc.ca; FAX (604) 877–0712.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.388902.
REFERENCES
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Biemont C, Tsitrone A, Vieira C, Hoogland C. Transposable element distribution in Drosophila. Genetics. 1997;147:1997–1999. doi: 10.1093/genetics/147.4.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ. Mobile elements inserted in the distant past have taken on important functions. Gene. 1997;205:177–182. doi: 10.1016/s0378-1119(97)00399-5. [DOI] [PubMed] [Google Scholar]
- Brookfield JF. Selection on Alu sequences? Curr Biol. 2001;11:R900–R901. doi: 10.1016/s0960-9822(01)00547-4. [DOI] [PubMed] [Google Scholar]
- Brosius J. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica. 1999;107:209–238. [PubMed] [Google Scholar]
- Carroll ML, Roy-Engel AM, Nguyen SV, Salem AH, Vogel E, Vincent B, Myers J, Ahmad Z, Nguyen L, Sammarco M, et al. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J Mol Biol. 2001;311:17–40. doi: 10.1006/jmbi.2001.4847. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The population dynamics of transposable elements. Genet Res. 1983;42:1–27. [Google Scholar]
- Charlesworth B, Langley CH. Population genetics of transposable elements in Drosophila. In: Selander RK, Clark AG, Whittam TS, editors. Evolution at the molecular level. Sunderland, MA: Sinauer Associates; 1991. pp. 150–176. [Google Scholar]
- Charlesworth B, Langley CH, Sniegowski PD. Transposable element distributions in Drosophila. Genetics. 1997;147:1993–1995. doi: 10.1093/genetics/147.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Franco C, Terrinoni A, Dimitri P, Junakovic N. Intragenomic distribution and stability of transposable elements in euchromatin and heterochromatin of Drosophila melanogaster: Elements with inverted repeats Bari 1, hobo, and pogo. J Mol Evol. 1997;45:247–252. doi: 10.1007/pl00006227. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Fullerton SM, Bernardo Carvalho A, Clark AG. Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol. 2001;18:1139–1142. doi: 10.1093/oxfordjournals.molbev.a003886. [DOI] [PubMed] [Google Scholar]
- Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes: An evolving understanding. BioEssays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Greally JM. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc Natl Acad Sci. 2002;99:327–332. doi: 10.1073/pnas.012539199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Junakovic N, Terrinoni A, Di Franco C, Vieira C, Loevenbruck C. Accumulation of transposable elements in the heterochromatin and on the Y chromosome of Drosophila simulans and Drosophila melanogaster. J Mol Evol. 1998;46:661–668. doi: 10.1007/pl00006346. [DOI] [PubMed] [Google Scholar]
- Jurka J. Repbase update: A database and electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Kaplan NL, Brookfield JFY. The effect on homozygosity of selective differences between sites of transposable elements. Theor Popul Biol. 1983;23:273–280. doi: 10.1016/0040-5809(83)90018-7. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci. 1997;97:7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Perspective: Transposable elements, parasitic DNA, and genome evolution. Evolution Int J Org Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- Kjellman C, Sjogren HO, Widegren B. The Y chromosome: A graveyard of endogenous retroviruses. Gene. 1995;161:163–170. doi: 10.1016/0378-1119(95)00248-5. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Pearson NM, Jegalian K. The human Y chromosome, in the light of evolution. Nat Rev Genet. 2001;2:207–216. doi: 10.1038/35056058. [DOI] [PubMed] [Google Scholar]
- Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 1988;52:223–235. doi: 10.1017/s0016672300027695. [DOI] [PubMed] [Google Scholar]
- Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- Li WH, Graur D. Fundamentals of molecular evolution. Sunderland, MA: Sinauer Associates; 1991. [Google Scholar]
- Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA, Resnick MA. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. The structure and function of retroviral long terminal repeats. Curr Top Microbiol Immunol. 1990;157:50–92. doi: 10.1007/978-3-642-75218-6_3. [DOI] [PubMed] [Google Scholar]
- McClintock B. Controlling elements and the gene. Cold Spring Harbor Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- McDonald JF. Transposable elements: possible catalysts of organismic evolution. Trends Ecol Evol. 1995;10:123–126. doi: 10.1016/s0169-5347(00)89012-6. [DOI] [PubMed] [Google Scholar]
- Medstrand P, Mager DL. Human specific integrations of the HERV-K endogenous retrovirus family. J Virol. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— . Encyclopedia of the human genome. London, UK: Nature Publishing Group; 2002. Retroviral repeat sequences. . (In press.) [Google Scholar]
- Orgel LE, Crick FHC. Selfish DNA: The ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH. Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Pavlicek A, Jabbari K, Paces J, Paces V, Hejnar JV, Bernardi G. Similar integration but different stability of Alus and LINEs in the human genome. Gene. 2001;276:39–45. doi: 10.1016/s0378-1119(01)00645-x. [DOI] [PubMed] [Google Scholar]
- Schmid CW. Does SINE evolution preclude Alu function? Nucleic Acids Res. 1998;26:4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol Evol. 1991;33:311–20. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- Smit AFA. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Smit AF, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Stenger JE, Lobachev KS, Gordenin D, Darden TA, Jurka J, Resnick MA. Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- Sverdlov ED. Retroviruses and primate evolution. BioEssays. 2000;22:161–171. doi: 10.1002/(SICI)1521-1878(200002)22:2<161::AID-BIES7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Torti C, Gomulski LM, Moralli D, Raimondi E, Robertson HM, Capy P, Gasperi G, Malacrida AR. Evolution of different subfamilies of mariner elements within the medfly genome inferred from abundance and chromosomal distribution. Chromosoma. 2000;108:523–532. doi: 10.1007/s004120050404. [DOI] [PubMed] [Google Scholar]
- Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- Whitelaw E, Martin DK. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nature Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- Wilkinson DA, Mager DL, Leong JC. Endogenous human retroviruses. In: Levy J, editor. The Retroviridae. New York: Plenum Press; 1994. pp. 465–535. [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]