Abstract
Draft sequencing is a rapid and efficient method for determining the near-complete sequence of microbial genomes. Here we report a comparative analysis of one complete and two draft genome sequences of the phytopathogenic bacterium, Xylella fastidiosa, which causes serious disease in plants, including citrus, almond, and oleander. We present highlights of an in silico analysis based on a comparison of reconstructions of core biological subsystems. Cellular pathway reconstructions have been used to identify a small number of genes, which are likely to reside within the draft genomes but are not captured in the draft assembly. These represented only a small fraction of all genes and were predominantly large and small ribosomal subunit protein components. By using this approach, some of the inherent limitations of draft sequence can be significantly reduced. Despite the incomplete nature of the draft genomes, it is possible to identify several phage-related genes, which appear to be absent from the draft genomes and not the result of insufficient sequence sampling. This region may therefore identify potential host-specific functions. Based on this first functional reconstruction of a phytopathogenic microbe, we spotlight an unusual respiration machinery as a potential target for biological control. We also predicted and developed a new defined growth medium for Xylella.
[The sequence data from this study have been submitted to GenBank under accession nos. NC_002723 (X. fastidiosa Almond [Dixon]) and NC_002722 (X. fastidiosa Oleander [Ann-1]).
Xylella fastidiosa is a Gram-negative bacterium belonging to the gamma subgroup of the proteobacteria (Wells et al. 1987). This microorganism is an important plant pathogen causing many economically important crop diseases, such as Pierce's disease (PD) of grapevine and citrus variegated chlorosis in citrus (Moller et al. 1974; Purcell 1997). Almond leaf scorch was first described in 1974 (Moller et al. 1974), and electron microscope studies indicated the presence in almond of the same bacterium previously associated with PD of grapevines (Mircetich et al. 1976). Cross-inoculation studies showed that PD and the almond leaf scorch strains were pathogenic to both almond and grape (Davis and Thompson 1980). Oleander leaf scorch was first noticed in 1994 in southern California. The glassy-winged sharpshooter, Homalodisca coagulata, was found to be the vector for this bacterium. It is possible that several other varieties of sharpshooters can transmit X. fastidiosa between oleander plants (Purcell et al. 1999). The oleander leaf scorch strain could not be reisolated from grapevine in a greenhouse after inoculation using needle puncture (Purcell et al. 1999). This result provides evidence for host specificity among strains of X. fastidiosa. Previous studies classified X. fastidiosa as a single species, but differentiated members of the species, depending on such criteria as host specificity and pathogenicity (Hendson et al. 2001 and references therein). We define each strain of X. fastidiosa to be a pathovar for a specific plant, as suggested previously (Hendson et al. 2001). We will therefore refer throughout to the citrus, almond, and oleander strains as Xf pv citrus (XFA), Xf pv almond (XFX), and Xf pv oleander (XFY).
The complete genome sequence of the X. fastidiosa 9a5c strain (XFA), which causes citrus variegated chlorosis , was recently published (Simpson et al. 2000). In this study, we report the gapped-genome sequences of Xf pv almond (Dixon strain) (XFX) and Xf pv oleander (Ann-1 strain) (XFY) and compare them to the complete genomic sequence of XFA. Here we present highlights of the first functional reconstruction of a phytopathogenic microbe based on the three sequenced genomes. This approach has led us to the identification of a putative drug target in the aerobic respiratory chain of this organism, as well as to a prediction of the specific growth requirements of this bacterium, which were used to develop a defined medium for optimal growth. Furthermore, based on a global alignment of the proteomes for these genomes, we identified a region in XFA that is absent in the other two genomes that potentially encodes host-specific functions. Comparisons of curated pathways and their components present in the complete XFA genome relative to the gapped XFY and XFX genomes allows the prediction of functions potentially missed during gapped-genome sequencing. Such an approach provides an added level of quality control to the analysis of sequenced genomes.
RESULTS AND DISCUSSION
Gapped-Genome Sequencing of New Xf Strains
The capacity to produce draft genomes based on a random shotgun approach (Fleischmann et al. 1995) far exceeds our capacity to produce completely finished genomes. The draft-sequencing process is faster (two to three weeks compared with months or years for finished sequence) and much less expensive. The gapped-genomic sequence of the two new Xylella strains (XFX and XFY) represents more than 95% of the complete sequence, with 9.4- and 8.1-fold sequence coverage, respectively (Table 1). As we hereby show, the microbial draft sequences (high-quality annotated assemblies from >8-fold coverage) offers an extremely useful form of information, leading to an effective sampling of >95% of the gene repertoire of a given microbial genome. The genome size of both almond (XFX) and oleander (XFY) strains is very similar to the completely sequenced XFA genome, with 2.4 Mb (XFX) and 2.6 Mb (XFY), compared with 2.7 Mb for the XFA genome. All three strains display a similar GC content. We identified 2681 (XFX) and 2870 (XFY) ORFs distributed over 121 and 93 contigs, respectively, compared with the 2985 ORFs of the XFA genome. Using the ERGO bioinformatics suite, we assigned functions to 62% of the genes in each of the two new Xylella genomes, compared with the 58% of genes in the XFA genome. Of the remaining 38% of the ORFs in the XFX and XFY genomes, most show sequence similarity to other proteins of unknown function (hypothetical proteins), and only 3%–4% of the total ORFs in the two genomes did not show any sequence similarity to other known proteins (unique ORFs).
Table 1.
Comparative Statistics of Sequenced Genomes of Xylella fastidiosa Strains
| Xf pv citrus (XFA)a | Xf pv oleander (XFY)b | Xf pv almond (XFX)c | ||||
|---|---|---|---|---|---|---|
| Affected plants | Citrus | Oleander | Almond | |||
| Disease caused | Citrus variegated chlorosis | Oleander leaf scorch | Almond leaf scorch | |||
| Sequencing source | ONSA | This study | This study | |||
| DNA, total sequenced bases | 2,731,748 | 2,625,581 | 2,434,849 | |||
| Contigs | 3 | 93 | 121 | |||
| Coverage | Complete | ∼8.1X | ∼9.4X | |||
| G+C content | 53% | 52% | 52% | |||
| Total ORFs | 2985 | 2870 | 2681 | |||
| Assigned functions | 58% (1684 ORFs) | 62% (1713 ORFs) | 62% (1593 ORFs) | |||
| Unassigned functions | 42% | 38% | 38% | |||
| Unique | Hypothetical | Unique | Hypothetical | Unique | Hypothetical | |
| 7% | 35% | 4% | 34% | 3% | 35% | |
| ORFs in ortholog clusters | 60% | 58% | 59% | |||
| ORFs in paralog clusters | 21% (639) | 23% (660) | 22% (585) | |||
| Paralog clusters | 252 | 230 | 220 | |||
| ORFs in chromosomal clusters | 32% | 29% | 26% | |||
Statistics derived from this study.
Xylella fastidiosa pv almond Dixon strain.
Xylella fastidiosa pv oleander Ann-I strain.
(ONSA) Organization for Nucleotide Sequencing and Analysis.
Nearly 59% of the ORFs in any of the three Xylella genomes can be grouped in ortholog clusters and 22% in paralog clusters. Interestingly, the number of paralog clusters in XFY is less than that in XFA, but there are more ORFs in the XFY paralog clusters compared with the XFA strain. This result indicates that there is greater functional redundancy in the oleander strain, XFY.
Identification of Xf Oleander (XFY) Episome
The sequencing and assembly of the XFY genome revealed that it possesses a putative 30-kb episome (Table 2). The plasmid bears 36 ORFs, of which three encode proteins involved in replication (RXFY0974, RXFY0979) and plasmid maintenance and stability (RXFY0987). A Type IV secretion pathway (chromosomal cluster) that includes VirB2 (RXFY02430), B4 (RXFY00976), B5 (RXFY00992), B6 (RXFY00989), B8 (RXFY00988), B9 (RXFY00984), B10 (RXFY00977), B11 (RXFY00991), and VirD4 (RXFY00973), as well as a putative transcriptional regulator (RXFY00983) are encoded on the 30-kb replicon. The VirB system is designed to mediate the plasmid transfer from a donor to a recipient bacterium. Two members of the DNA conjugative transfer pathway (TraM, RXFY02499 and TrbN, RXFY00996) have also been found along with a “nickase” (RXFY0985), which is responsible for initiating transposition by introducing single-stranded nicks in the circular plasmid DNA.
Table 2.
Comparative Statistics of the Xf pv oleander (XFY) and Xf pv citrus (XFA; p51) Eplsomes
| Plasmid statistics | XFY | XFAa |
|---|---|---|
| Length (bp) | 30,270 | 51,158 |
| G+C ratio | 49% | 49.6% |
| Protein-encoding regions | 26.9% | 86.9% |
| ORFs | 36 | 70 |
| ORFs with functional assignment | 25 (69%) | 34 (48%) |
| Hypothetical proteins | 8 | 31 |
| ORFs without significant database match | 3 | 5 |
Based on previously published data (Simpson et al. 2000).
Pathway Analysis
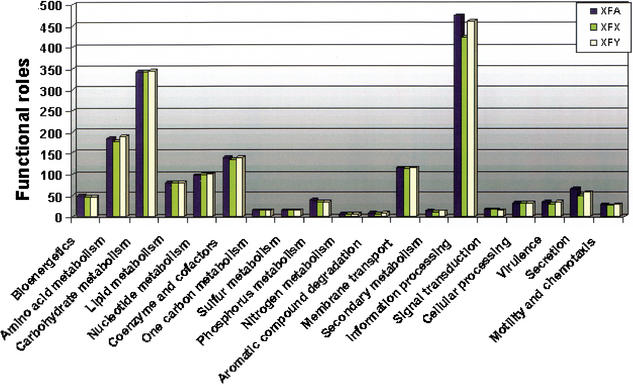
As part of the X. fastidiosa functional reconstruction (described later), ORFs were assigned to the appropriate metabolic and nonmetabolic pathways (Kyrpides et al. 2000; Selkov et al. 2000). Figure 1 shows the statistics of functional roles in all three organisms according to subsystem in more detail. Table 3 indicates that a similar number of pathways are present in all three Xf strains. Because the citrus (XFA) genome is complete, the percent of functions that appear missing from pathways (Table 3) is likely to represent instances of nonorthologous gene replacement, that is, the replacement of a particular protein function by unrelated protein(s) with the same cellular function (Forterre 1999; Galperin and Koonin 1999). Therefore, the number of functions missing from pathways for XFA represents the relative baseline of functions that appear as “absent”. Given that ERGO integrates a database of cellular pathways, it is possible to predict which of the functional roles that appear as missing from the XFX/XFY pathways are likely to reside in their nonsequenced regions. We define a functional role as a step in a pathway, and one functional role may have more than one biochemical function. For example, there are 40 and 97 functional roles missing from the genomes of XFY and XFX, respectively, but present in XFA (Table 3). A comparison of these genomes shows a similar number of functional roles associated with each subsystem. Most of the missing functions in almond and oleander strains are present in informational processing pathways (Table 3). Some of these missing functions represent components of the core functional machinery (for example, subunits of the large and small ribosomal complexes), and therefore we would expect them to be present in the nonsequenced gaps. More rigorous investigation of these cases indicated that although these functions are indeed present in these genomes, some are associated with ORFs residing on contigs with <20 reads, which have lower coverage and sequence quality (see Methods). We speculate that the functional absences present in the secretion subsystem in XFX (Table 3) are likely due to the fact that a plasmid-borne ‘pathogenicity island’ was not clearly identified in XFX, in contrast to the XFA and XFY. Thus, the pathway analysis approach introduces an additional quality control step in the reconstruction of a partial genome and allows a greater degree of functionality to be assigned to sequenced genomes.
Figure 1.
Comparative disposition of functional roles in cellular subsystems in Xf genomes. The number of functional roles present in the each of the described subsystems was plotted for each of the Xylella genomes.
Table 3.
Prediction of Functions Missing from Pathways in Gapped Xf Genomes
| XFA (citrus) | XFY (oleander) | XFX (almond) | |
|---|---|---|---|
| Pathways asserted | 958 | 965 | 938 |
| Functional roles missing from pathways | 352 | 359 | 379 |
| Examples of functions missing from pathways | |||
| Ribosomal SSU | — | 1 | 10 |
| Ribosomal LSU | — | 4 | 17 |
| Aminoacyl tRNA synthetases | — | 1 | 2 |
| Recombinational repair | — | — | 1 (recF) |
| Replication, elongation | — | — | 1 (DNA Pol IIIβ) |
| Total | — | 6 | 31 |
(LSU) large ribosomal subunit; (SSU) small ribosomal subunit.
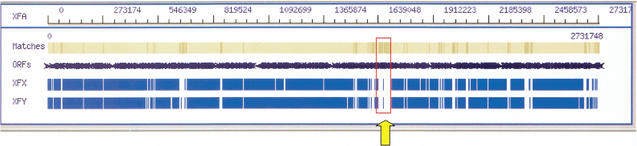
Phage Integration Region Identifies Potential Host-Specific Functions
During our comparative analysis of the three Xylella genomes, we noticed that the XFA genome contains two tRNA genes (encoding glycine and threonine, at 1,638,644 and 1,707,587 bp) that flank a region marked by the presence of prophage genes absent from the XFX and XFY genomes. This region was identified in the XFA genome using a comparative genome/proteome alignment tool called GenomeWalk, the visual rendering of which is shown in Figure 2. The putative phage encoded in this region is most similar to the Siphophage group of double-stranded DNA phage based on whole-genome level and taxonomic comparisons (Rohwer and Edwards 2002). The DNA sequence in this region has a mean GC content of 65.9%, significantly greater than that found in the XFA genome as a whole (52.6%), indicating that it has been inherited relatively recently in the evolution of the Xf genome. This region also contains genes encoding integrase enzymes and DNA replication machinery, as well as genes encoding structural proteins such as capsid and tail proteins.
Figure 2.
Graphical alignment of Xylella fastidiosa (Xf) proteomes using GenomeWalk. A unique 65-kb phage insertion region (red rectangle, brown shading) in the Xf citrus strain (XFA) is absent in the Xf almond (XFX) and oleander (XFY) strains (arrow).
Closer inspection of this prophage region reveals that it contains two unusual “operons”. These operons are striking by the presence of oxidoreductase, dehydrogenase, esterase, hydrolase, and isomerase enzymes, as well as outer membrane permease/transporter proteins, indicating that they are involved in carbon-utilization pathways. Both operons are completed by LysR-family transcriptional regulators that presumably control transcription of the genes in response to environmental signals. It is unclear which carbon-utilization pathways these enzymes are involved in, although the limited distribution (no close orthologs previously identified) indicates that these operons may provide host-specific functions to XFA that are not present in other Xf strains. The completeness of the proteins encoded in this region, the lack of similar prophage in related Xf genomes, and the similarity between this phage and prophage found in the enteric Salmonella typhi genome recently sequenced (Parkhill 2000) might indicate that this is still a viable phage and not a cryptic remnant of an invading parasite.
Functional Reconstruction
We embarked on a functional reconstruction of the X. fastidiosa (Xf) genomes based on the genomic DNA sequences. A more detailed description of how this is done has been published previously (Kyrpides et al. 2000; Selkov et al. 2000; DelVecchio et al. 2002; Kapatral et al. 2002). Here, we present selected highlights of this analysis specifically pertaining to respiratory mechanisms and some of the core metabolism that is relevant to the growth of the bacteria.
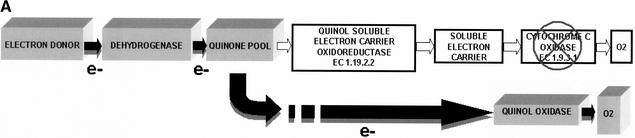
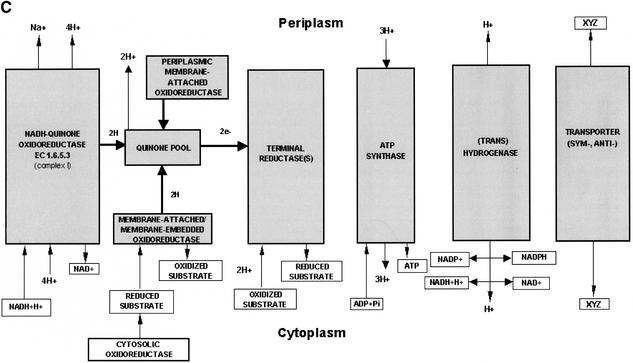
Reconstruction of electron transport in Xylella spp. was performed by analyzing the respiratory complexes, both anaerobic and aerobic, as well as proton transport and ATPase. In contrast to previous interpretations (Simpson et al. 2000), the resulting overview indicates that the Xf strains possess quite a simple and unusual aerobic respiratory complex (Fig. 3A,B) that does not have the adaptive capabilities of, for instance, Escherichia coli or Bacillus subtilis. Functional analysis in silico shows the presence of the least energy-efficient type of aerobic respiration of any known organism reported to date. The ORF numbering corresponds to the XFA genome, but orthologs in the XFX and XFY strains are present. All three Xf possess cytochrome o (bo) ubiquinol oxidase (EC 1.10.3.-) as the only terminal oxidase (RXFA01387–1390). Such a limited system of aerobic respiration indicates that this type of energy metabolism is not a prevalent pathway for this bacterial family. In addition, it indicates that Xf are capable of aerobic respiration at high aeration levels but not under oxygen-limiting conditions, because of the absence of cytochromes with high oxygen affinity. Surprisingly, there is no trace of a cytochrome c oxidase (or other type of quinol oxidase as the terminal oxidase) despite the availability of the entire operon encoding the ubiquinol-cytochrome c reductase complex (EC 1.10.2.2; complex III: Rieske iron-sulfur protein, cytochrome b and cytochrome c1 precursor [RXFA00908–910], respectively). Thus, we can suggest that, because the cytochrome o (bo) ubiquinone oxidase is the only available terminal oxidase, it provides a strong potential control point and drug target for these phytopathogens. Although soluble periplasmic cytochrome c has not been found in the Xf genomes, another protein, the copper-containing protein azurin (RXFA00507), could perform electron transport in the periplasm between ubiquinol-cytochrome c reductase and the missing cytochrome c oxidase.
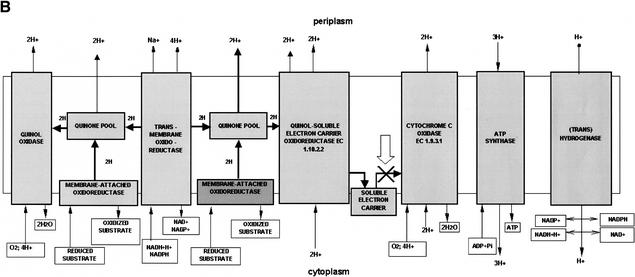
Figure 3.
Graphical visualization of the functional reconstruction of Xylella fastidiosa aerobic and anaerobic respiration. (A) Diagrammatic representation of aerobic respiration. Three-dimensional boxes show the presence of functional components and filled arrows indicate electron flow. (B) Detailed spatial disposition of membrane components of aerobic respiratory chain. The absence of cytochrome c oxidase is designated by the cross and arrow. (C) Overview of the anaerobic respiratory chain. Reconstructions show proton translocation and oxidative phosphorylation.
Anaerobic respiration in Xf is also quite limited (Fig. 3C). The entire system for nitrate utilization to dinitrogen is absent. Additionally, the enzymes converting nitrate into ammonia appear to be absent. Xf clearly prefers anaerobic respiration based on sulfur metabolism. The entire operon that encodes enzymes for the use and reduction of sulfate to sulfide is present (sulfate adenylyltransferase [EC 2.7.7.4; RXFA01500–1501], adenylylsulfate kinase [EC2.7.1.25; RXFA01501], phosphoadenosine phosphosulfate reductase [EC 1.8.99.4; RXFA01497], and sulfite reductase [EC 1.8.1.2; RXFA01498–1499]). Indication that another type of anaerobic respiration may be present is suggested by the presence of arsenate reductase (RXFA00115). The simplicity and the type of anaerobic respiratory complex indicate that the enzymes of the sulfate–sulfide reduction pathway could also serve as a potential drug target.
Xf has a well-defined operon encoding H+-transporting ATP synthase (EC 3.6.1.34; RXFA01142–1149). There is no indication of any other type of ATP synthase. Proton balance seems to be maintained mainly by NADH-quinone oxidoreductase (EC 1.6.5.3; RXFA00305–318), cytochrome o ubiquinol oxidase, quinone pool, and ubiquinol-cytochrome c reductase that pump protons out and the reverse catalyzed by the H+-transporting ATP synthase.
X. fastidiosa is able to use very few sugars (glucose, fructose, mannose, ribose, glycerol, N-acetylglucosamine) and cellulose. Paradoxically, despite its ability to use these sugars, it was not apparent from previous studies (Simpson et al. 2000) how these sugars are transported into the cell. We have identified candidates for some of the sugar transport systems, which have orthologs in Xf pv almond and Xf pv oleander. For example, based on the chromosomal neighborhood of the XFA ORF, RXFA01462, which is positioned at the beginning of a N-acetylglucosamine utilization operon, this ORF is likely to be the N-acetylglucosamine and glucosamine transporter. The Xylella genomes contain one family 1 ABC transporter, the subunits of which are dispersed throughout the genome. The sugar transport permease (RXFA02446, RXFA02467) and the periplasmic binding protein (RXFA02448) are found adjacent to an enzyme involved in glucose metabolism. The ATPase subunit (RXFA01067) is found in a distant region of the genome. We propose that this sugar transporter is likely to be involved in glucose uptake.
The range of amino acids that might be used by Xylella as a sole carbon source seems to be limited to d- and l-alanine (via alanine racemase and d-amino acid dehydrogenase), glycine (via glycine cleavage system) and l-glutamate (via glutamate dehydrogenase). Thus, some carbon and energy source, other than amino acids and an oligopeptide mixture, should be provided for efficient growth of Xylella. This carbon source might be glycerol (all genes for glycerol utilization were found, including glycerol uptake protein, glycerol kinase, and glycerol-3-phosphate dehydrogenases), malate or oxaloacetate (C4-dicarboxylate transporter and NADP-dependent malic enzyme are clustered together), glucose, fructose, or N-acetylglucosamine. Most likely, an optimal growth medium for Xf would be predicted to contain glycerol as a carbon and energy source and l-glutamate as both a nitrogen and carbon source. In addition, the clear preference of Xf for sulfate reduction-based anaerobic respiration could indicate that the best medium for the Xf family would contain an increased concentration of sulfur-containing compounds.
Growth Medium
Culture media for Xf were first developed for both grape and almond strains (Davis et al. 1978) and eventually for other X. fastidiosa strains such as those from peach and periwinkle (Davis et al. 1981a). Determination of the minimum metabolic requirements for this organism would thus allow improvement in medium composition for optimal growth and make this organism more tractable for study. Based on our functional reconstruction of the sequenced Xf genomes, we generated predictions of the nutritional profile of these phytopathogens, and we designed growth experiments to test and develop a defined medium. A commonly used growth medium was modified by adding one or more of the predicted compounds. The standard growth medium used was PW (Davis et al. 1981b). Medium components that were changed included those that are putative sources for carbon, nitrogen, and iron. The bacterial strain used for the experiments was Xf pv almond (Dixon). Numbers of colony-forming units (CFU) of Xf were determined after 7 days growth at 28°C.
We compared the growth of Xf pv almond on standard PW as the growth medium (1.13 × 109 CFU ml-1), and determined that the addition of fructose (8.40 × 108 CFU ml-1), oxaloacetate (5.80 × 108 CFU ml-1), N-acetylglucosamine (2.90 × 108 CFU ml-1), or α-keto-glutarate (7.00 × 108 CFU ml-1) could substitute for the Bovine serum albumin (BSA) constituent of the PW medium when added separately. In comparison, glycerol (2.93 × 107 CFU ml-1) and glucose (5.70 × 106 CFU ml-1) could not substitute as well for BSA in the PW-based medium. Sucrose did not substitute for BSA, as was mentioned in previous studies (Davis et al. 1981b). The different iron sources used were hemin chloride (1.13 × 109 CFU ml-1), ferric pyrophosphate (4.60 × 108 CFU ml-1), or iron sulfate (3.10 × 108 CFU ml-1). These iron sources affected the growth of Xf similarly. The addition of the amino acids glycine (4.40 × 108 and 2.9 × 108 CFU ml-1) or glycine plus l-alanine (9.50 × 108 and 6.83 × 108 CFU ml-1) to the PW base did not appreciably improve growth, although Xylella approached the growth properties with the PW-based medium. Indeed, when glycine was omitted from the modified medium, less growth was observed (data not shown). Xylella appear to grow better in the presence of glycine, although the exact reason for this is unclear. When l-cysteine was added to PW, no growth was observed. Our results indicate that when BSA is removed from the PW-based medium, the best growth conditions are provided by the addition of fructose, glutamine, glycine, and iron sulfate. Thus, Xf was found to grow well on media components predicted by the metabolic reconstruction when they were added as substitutes for BSA in the basal medium.
The aspects of the genome comparisons and functional reconstructions presented here represent the relative highlights of the biological analysis discerned from the genome analysis of the phytopathogenic X. fastidiosa (Xf) species. A more complete description of the genome analyses and reconstructions are presented elsewhere (Bhattacharyya et al., in press). However, there are two intriguing questions posed from this study. First, does the unique phage insertion sequence in the Xf citrus strain indeed bear potential host-specific functions? Second, can the strong potential control point identified in the aerobic respiratory chain provide a potential drug target in these phytopathogens? We envision that microarray, genetic, proteomic, and pharmacological approaches will provide the tools to address these new challenges identified from this in silico analysis of the genome sequences of these organisms.
METHODS
Sequencing and Assembly
Genomic DNA for strains of both Xf pv almond (Dixon) and oleander (Ann-1) were obtained from the collection of isolates from A.H.P, at the University of California, Berkeley. Using a whole genome shotgun approach, purified DNA was fragmented using a GeneMachines Hydroshear, end repaired and size fractionated on agarose gels. Fragments in the 3- to 4-kb size range were excised, eluted, and blunt-end ligated to SmaI-linearized pUC18 DNA. DNA from overnight cultures (180 μL) of these libraries was purified using an automated 96-well SPRI protocol (Hawkins et al. 1994). These templates were cycle-sequenced using both universal M13-forward and M13-reverse primers with dye-terminator chemistry (AP Biotech). The resulting products were sequenced on MegaBACE 1000 capillary sequencers. Raw traces were preprocessed using Cimarron software version 2.1905 (AP Biotech) and base-called using Phred (Ewing et al. 1998). Read lengths in libraries typically averaged 500–550 bases with Phred ≥20. Individual sequence reads were assembled using Phrap (http://bozeman.mbt.washington.edu/phrap.docs/phrap.html), and contig order/orientation was established by using technology developed at the Joint Genome Institute (JGI) (e.g., readsincontig, phrap2gapspan) in conjunction with the graphical sequence editor, Consed (Gordon et al. 1998). Libraries were sequenced to at least 8× depth of coverage. The DNA sequences of the two Xylella strains (almond and oleander strains) are available at the JGI Web server (http://www.jgi.doe.gov/JGI_microbial/html/).
ORF Prediction
ORFs were predicted with a proprietary ORF-calling software system developed at Integrated Genomics. The system automatically combines its own statistically predicted ORFs with ORFs derived from external sources (if available) and BLAST and FASTA similarities. ORFs were predicted using this approach for all three Xf genomes including the previously published Xf pv citrus (Simpson et al. 2000) whose DNA sequence was extracted from GenBank. The ORF prediction tools were not run on contigs with Phred <20 reads.
Genome Analysis: Annotations, Functional Reconstructions, and Comparative Genomics
We used the ERGO bioinformatics suite (http://www.integratedgenomics.com) (Kyrpides et al. 2000; Selkov et al. 2000; DelVecchio et al. 2002; Kapatral et al. 2002). ERGO contains an integration of over 450 genomes (including complete and partial, public, and proprietary from all kingdoms), with an extensive manually curated set of functional annotations and over 5000 manually curated cellular pathways, which is part of the Integrated Genomics proprietary Pathway Database. The analysis of the three Xf strains was performed as previously described (Kyrpides et al. 2000; Selkov et al. 2000; DelVecchio et al. 2002; Kapatral et al. 2002). The gene clustering based on sequence (i.e., ortholog and paralog clusters), as well as the one based on chromosomal context (i.e., chromosomal and fusion clusters), were computed on the basis of proprietary technology developed at Integrated Genomics. Comparative genomics technology such as GenomeWalk (available through ERGO suite) was also used. GenomeWalk provides a graphical whole genome comparison environment that facilitates the identification of unique chromosomal regions between closely related genomes. Workbench allows the identification of the common and unique clusters of genes between genomes, by using clustering algorithms to calculate protein clusters between a set of organisms.
WEB SITE REFERENCES
http://www.jgi.doe.gov/JGI_microbial/html/index.html; JGI Web server.
http://www.integratedgenomics.com; ERGO bioinformatics suite
Acknowledgments
We thank members of the bioinformatics and genome analysis group at Integrated Genomics, specifically, Gordon Pusch, Lynn Jablonski, Olga Vassieva, Allen Bartman, and Warren Gardner for assistance during preliminary stages of the genome analysis. We would also like to thank Ken Frankel (JGI) for his contribution in the genomic sequence analysis. Funding for this work was supported by the Integrated Genomics research and development program, and by the U.S. Department of Energy research grants, from the Office of Biological and Environmental Research, by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under contract No. DE-AC03 76SF00098, and Los Alamos National Laboratory under contract No. W-7405-ENG-36.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL anamitra@integratedgenomics.com; FAX (312) 226-9446.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.370702.
REFERENCES
- Bhattacharyya A, Stilwagen S, Ivanova N, D'Souza M, Bernal A, Lykidis A, Kapatral V, Anderson I, Larsen N, Los T, et al. Proc. Natl. Acad. Sci. 2002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MJ, Thompson SV. Etiological role of a xylem-limited bacterium causing Pierce's disease in almond leaf scorch. Phytopathology. 1980;70:472–475. [Google Scholar]
- Davis MJ, Purcell AH, Thompson SV. Pierce's disease of grapevines: Isolation of the causal bacterium. Science. 1978;199:75–77. doi: 10.1126/science.199.4324.75. [DOI] [PubMed] [Google Scholar]
- Davis MJ, Whitcomb RF, Gillespie AG., Jr . Fastidious bacteria of plant vascular tissue and invertebrates (including so called rickettsia-like bacteria) In: Starr MP, Stolp H, Truper HG, Balows A, Schlegel HG, editors. The prokaryotes: A handbook on habits, isolation, and identification of bacteria. Heidelberg: Springer-Verlag; 1981a. pp. 2172–2188. [Google Scholar]
- Davis MJ, French WJ, Schaad NW. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr Microbiol. 1981b;6:309–314. [Google Scholar]
- DelVecchio VG, Kapatral V, Redkar RJ, Patra G, Mujer C, Los T, Ivanova N, Anderson I, Bhattacharyya A, Lykidis A, et al. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc Natl Acad Sci. 2002;99:443–448. doi: 10.1073/pnas.221575398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Forterre P. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol Microbiol. 1999;33:457–465. doi: 10.1046/j.1365-2958.1999.01497.x. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Functional genomics and enzyme evolution. Homologous and analogous enzymes encoded in microbial genomes. Genetica. 1999;106:159–170. doi: 10.1023/a:1003705601428. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Hawkins TL, O'Connor-Morin T, Roy A, Santillan C. DNA purification and isolation using a solid-phase. Nucleic Acids Res. 1994;22:4543–4544. doi: 10.1093/nar/22.21.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendson M, Purcell AH, Chen D, Smart C, Guilhabert M, Kirkpatrick B. Genetic diversity of Pierce's disease strains and other pathotypes of Xylella fastidiosa. Appl Environ Microbiol. 2001;67:895–903. doi: 10.1128/AEM.67.2.895-903.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, Bhattacharyya A, Bartman A, Gardner W, Grechkin G, et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol. 2002;184:2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides NC, Ouzounis CA, Iliopoulos I, Vonstein V, Overbeek R. Analysis of the Thermotoga maritima genome combining a variety of sequence similarity and genome context tools. Nucleic Acids Res. 2000;28:4573–4576. doi: 10.1093/nar/28.22.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mircetich SM, Lowe SK, Moller JW, Nyland G. Etiology of almond leaf scorch disease and transmission of the causal agent. Phytopathology. 1976;66:17–24. [Google Scholar]
- Moller WJ, Sanborn RR, Mircetich SM, Williams HE, Beutel JA. A newly recognized leaf scorch disease of almond. Plant Dis Rep. 1974;58:99–101. [Google Scholar]
- Parkhill J. In defense of complete genomes. Nature. 2000;18:493–494. doi: 10.1038/75346. [DOI] [PubMed] [Google Scholar]
- Purcell AH. Xylella fastidiosa, a regional problem or global threat? Journal of Plant Pathology. 1997;79:99–105. [Google Scholar]
- Purcell AH, Saunders SR, Hendson M, Grebus ME, Henry MJ. Causal role of Xylella fastidiosa in oleander leaf scorch disease. Phytopathology. 1999;89:53–58. doi: 10.1094/PHYTO.1999.89.1.53. [DOI] [PubMed] [Google Scholar]
- Rohwer F, Edwards R. The phage proteomic tree: A genome-based taxonomy for phage. J Bacteriol. 2002;184:4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkov E, Overbeek R, Kogan Y, Chu L, Vonstein V, Holmes D, Silver S, Haselkorn R, Fonstein M. Functional analysis of gapped microbial genomes: Amino acid metabolism of Thiobacillus ferrooxidans. Proc Natl Acad Sci. 2000;97:3509–3514. doi: 10.1073/pnas.97.7.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Reinach FC, Arruda P, Abreu FA, Acencio M, Alvarenga R, Alves IM, Araya JE, Baia GS, Baptista CS, et al. The genome sequence of the plant pathogen Xylella fastidiosa. Nature. 2000;406:151–157. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- Wells JM, Raju BC, Hung HY, Weisberg WG, Mandelco-Paul L, Brenner DJ. Xylella fastidiosa new-genus new-species gram-negative xylem-limited fastidious plant bacteria related to Xanthomonas spp. Int J Syst Bacteriol. 1987;37:136–143. [Google Scholar]