Abstract
The Conserved Domain Architecture Retrieval Tool (CDART) performs similarity searches of the NCBI Entrez Protein Database based on domain architecture, defined as the sequential order of conserved domains in proteins. The algorithm finds protein similarities across significant evolutionary distances using sensitive protein domain profiles rather than by direct sequence similarity. Proteins similar to a query protein are grouped and scored by architecture. Relying on domain profiles allows CDART to be fast, and, because it relies on annotated functional domains, informative. Domain profiles are derived from several collections of domain definitions that include functional annotation. Searches can be further refined by taxonomy and by selecting domains of interest. CDART is available at http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi.
The public release of multiple genomes has led to a large amount of sequence data that requires increasing expertise to query and understand. As of this writing, the NCBI Entrez Protein Database (Wheeler et al. 2002) contains more than 800,000 nonredundant protein sequences. This data growth is further complicated by the fact that experimental evidence about proteins has lagged the rapid growth of sequence data, leading to incorrect or insufficiently precise annotation. Because many of these new sequences are predicted, they are often labeled solely by sequence similarity. This can lead to an incorrect inference if the annotator does not take into account factors such as the extent of the sequence similarity and its relationship to functional domains and residues (Ponting and Dickens 2001) or if the similar protein is incorrectly annotated itself. Additionally, sequence similarity search algorithms, such as BLAST and PSI-BLAST (Altschul et al. 1997), implicitly deal with functional domains, whereas explicit domain annotation can be of great use in understanding homology, especially when searching iteratively. One potential solution to these problems is to create new search algorithms that allow scientists to efficiently and accurately comprehend similarity based on functional domains.
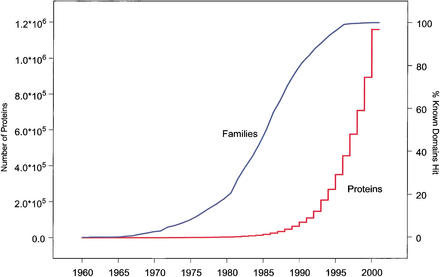
Most proteins are composed from a finite lexicon of evolutionarily conserved functional domains. Several efforts are under way to create comprehensive databases of protein domains, including Pfam (Bateman et al. 2002), SMART (Letunic et al. 2002), and CDD (Marchler-Bauer et al. 2002). Figure 1 displays the number of proteins discovered each year and the number of presently known nonredundant domains found in these proteins. Although the number of proteins discovered grows at increasingly higher rates, the number of domains found appears to be asymptotically reaching a limit. Even with a finite number of domains, however, efficiently searching for domains in a large number of sequences can be computationally expensive. Fortunately, a fast algorithm to find domains in sequences has been developed, RPS-BLAST (reverse-position-specific BLAST; Marchler-Bauer et al. 2002).
Figure 1.
The growth over time of the number of proteins known versus the growth in the number of unique domains. The left axis and red line are the cumulative sum of proteins in the NCBI Entrez Protein Database discovered in that year and in all previous years. The right axis and blue line are the total number of unique domains from the Conserved Domain Database that can be found in the cumulative set of proteins for each year, found by running RPS-BLAST on the set of proteins. Note that the zero slope in recent years may be caused by the need to accumulate multiple sequences to create a domain profile. However, this does not explain the inflection point in the curve beginning in 1990.
CDART is a Web-based tool that uses the domain definitions and annotations from the CDD database (which imports alignments from SMART and Pfam) and RPS-BLAST to allow users to rapidly query the Entrez Protein database by domain. To use the tool, a user enters a protein of interest, and RPS-BLAST is run on the protein to deduce its domain architecture. This domain architecture is then used to query CDART's database to find proteins with similar domain architectures, and the resulting proteins are displayed in a compact list. To show how distant homologies can be easily found, we use the tumor suppressor BRCA1 as an example.
RESULTS AND DISCUSSION
An Example
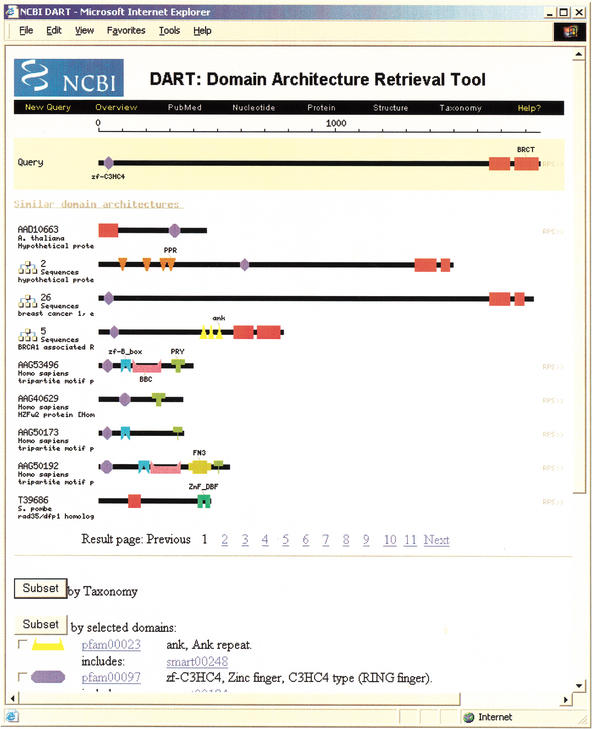
The CDART query page can be found on the Internet at http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi. On this page, one may enter a sequence accession or FASTA formatted sequence. For this example we enter the accession for human BRCA1, NP_009225. Pressing the search button runs RPS-BLAST on the sequence, comparing the sequence to the domain definitions in the CDD database. The search completes in a few seconds, and the results are displayed in Figure 2.
Figure 2.
CDART results page for the tumor suppressor protein BRCA1 (accession NP_009225). Domains found in BRCA1 are shown in beads-on-a-string style at the top of the page and include zinc fingers and BRCT protein–protein interaction domains. Similar domain architectures are listed below using the same style. If an architecture contains more than one protein, it is preceded by a graphical icon, and clicking on the icon gives the full list of proteins with that architecture. At the bottom of the page are controls to subset the list of architectures by taxonomy and by domain.
The top section of the results Web page displayed in Figure 2 shows the domains found in BRCA1 using a beads-on-a-string style. The domains are an N-terminal zinc finger and two C-terminal BRCT domains. This domain annotation, which is taken from CDD, immediately indicates that this protein binds to DNA and that it interacts with other proteins via the BRCT domain, information particularly useful if the function of BRCA1 had not been known.
The middle section of the Web page lists domain architectures of proteins found in Entrez that contain at least one domain found in the query sequence. These architectures are defined by the sequence of unique domains, where sequentially repeated domains are collapsed into a single occurrence of the domain. This culling of repeated domains is done for several reasons: repeats may be duplicated more easily than other types of domain insertions; the choice of the beginning residue of a repeat can be arbitrary and affects the number of repeats found; and the number of repeats included in the definition of a domain can cause variation in the number of repeats found in a hit. The domain architectures are ranked by the total number of domain clusters in common with the query. A better ranking would take into account the evolution of domain architectures. Unfortunately, the evolution of domain architectures remains a subject of research at present, and, to our knowledge, no reconstruction has yet been encoded in a way that CDART could use for ranking.
If there is more than one protein with a given architecture, the group is represented by an example sequence and description. Clicking on the description results in a Web page that lists all proteins in the group. The “more>” link next to each protein runs RPS-BLAST on the protein to give a detailed sequence alignment to the domain definitions.
The list of similar proteins extends over several pages and can be examined by clicking on the page numbers at the bottom of the page. In this case, >800 proteins are found homologous to the query protein. Running BLASTP using the same query sequence returns 340 results. The increase in neighbors is due to the double comparison done in CDART (protein to domain then domain to protein) and the high sensitivity of the RPS-BLAST algorithm and domain profiles.
Subsetting by Domain
At the bottom of the results page shown in Figure 2 is a form to subset the results by domain. Similar or redundant domains are grouped together and are represented by the same symbol in the display. The grouping together of domains is accomplished by examining overlapping hits of the domains to proteins in nr, using the algorithm described in the Methods section.
For example, one can select the BRCT domain and click the subset button to retrieve all proteins that contain the BRCT domain. Using PSI-BLAST, this type of subsetting requires advance knowledge of where domains exist on the query protein and restricting the search to the part of the protein that contains the domain of interest. During each iteration of PSI-BLAST, the user has to manually select sequences that construct a Position Specific Scoring Matrix (the statistical profile of a protein motif) with the desired sensitivity, a task made more difficult if the sequences have been incorrectly annotated. In domain databases like Pfam, SMART, and CDD, the sequences chosen and the extent of the domain have been screened by a knowledgeable curator who edited the multiple sequence alignments used to create the corresponding domain profile. To illustrate this difference, querying PSI-BLAST with BRCA1 returns a large number of similar proteins, but the results are largely dictated by a large, uncharacterized domain in the center of the protein. To concentrate on a smaller domain like BRCT, the user would have to know to manually limit the query to the BRCT domain. For example, using PSI-BLAST without limiting the query fails to find PARP, an NAD+ ADP-ribosyltransferase involved in a variety of biological processes such as DNA repair, cell, cycle, transformation, carcinogenesis, and apoptosis (Hanai et al. 1998). Because it contains zinc-finger and BRCT domains and participates in similar cellular functions, PARP contains similarities to BRCA1 that may be of interest to an investigator. The CDART query for BRCA1 lists PARP proteins in the first few pages of results.
It is important to note, however, that CDART is limited to known domain definitions and that these domain definitions may not span the phylogenetic clade of interest. This coverage problem will be significantly reduced as more domains are discovered or defined and existing domain definitions are expanded. Presently 67% of the proteins in Entrez have one or more domain hits annotated by CDART.
Taxonomic Restriction
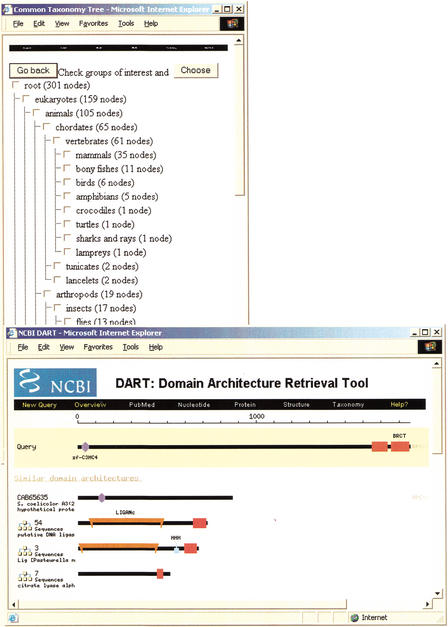
The list of similar proteins in DART can be restricted taxonomically by clicking on the “Subset by Taxonomy” button at the bottom of the results page. Figure 3 displays the form used to select the parts of the taxonomic tree that are of interest. The number of proteins found under each taxonomic node is displayed next to the taxonomic common name, and a checkbox allows selection of the node, which also selects all underlying taxonomic nodes. The user has two choices at this point, either selecting one or more of the general taxonomic nodes and clicking on the “Go Back” button to select all organisms under the selected nodes, or clicking on the “Choose” button, which allows the user to prune the taxonomic tree at the species level.
Figure 3.
The top left window allows users to select taxonomic groups of interest to subset CDART results. The lower right window is the results page for the search using BRCA1 subset by taxonomy to include only bacterial sequences. Note the large family of DNA ligases that contain the BRCT domain.
Using the example of BRCA1, the user can select the taxonomic node “Eubacteria,” and CDART will return a list of bacterial proteins containing BRCA1 domains (see Fig. 3). Interestingly, there is a large family of DNA ligases that contain a single copy of the BRCT domain. Many of these ligases are involved in cell cycle checkpoint functions responsive to DNA damage (Bork et al. 1997).
Querying by Domain
The incorporation of CDD into NCBI's Entrez database allows the user to retrieve proteins that contain a particular domain. To do this, the user can go to the Entrez Domains database at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=cdd and search for a domain of interest. Once this domain is found, clicking on the “Proteins” link in the domain record launches CDART, which lists all of the proteins in nr that contain that domain.
Comparison With Other Resources
Because several groups have made domain-based resources available on the Internet, it is instructive to show how CDART compares with and improves on these resources.
The Karolinska Institutet version of Pfam at http://www.cgr.ki.se/Pfam/ provides a search for proteins with similar domain architectures appended to the standard Pfam protein search. This tool is unique in that it includes domain definitions from Pfam B, which is a computationally generated, uncurated set of domain definitions, and also classifies proteins by specific variations from the domain architecture of the query protein. This search is limited to the SWISS-PROT protein database (Bairoch and Apweiler 2000) and does not explicitly search SMART. In comparison with CDART, this search does not group proteins by domain architecture, and the number of domain architectures returned is limited. For example, the PARP protein found in the CDART search detailed above does not appear in the results of this search tool. Subsetting by taxonomy or domain is not supported, although the user can manually query Pfam for a particular domain combination.
The SMART database at http://smart.embl-heidelberg.de/ is a resource that allows the identification and annotation of genetically mobile domains and the analysis of domain architectures. SMART includes a sequence analysis tool that, given a query sequence, displays SMART and Pfam domains, signal peptides, and protein homologs. This tool allows the user to search for proteins with identical domain architectures and organizations and show their taxonomy. Unlike CDART, the tool does not search for similar domain architectures, nor does it allow subsetting by taxonomy or by domain, although, like Pfam, the user can manually query for a particular domain combination.
The clustering algorithm used in CDART is necessary to reduce the redundancy of domain definitions in CDD. The Interpro database (Apweiler et al. 2001) at http://www.ebi.ac.uk/interpro/ clusters together and annotates a variety of protein signature databases, including the relationships between domains in Pfam and SMART. The relationships between these protein signature databases are complex, for example, between a motif and a domain, thus in Interpro they are manually curated, although the curation is based in part on automatically generated data. In contrast, because CDART only requires similarity between the domain definitions in CDD, it is able to create the relationships algorithmically.
METHODS
Creation of the CDART Database
An essential step to finding similar proteins is to calculate the domain architectures of all available proteins. Proteins are extracted from the NCBI nonredundant protein database (nr). RPS-BLAST is used to apply the domain definitions from the Conserved Domain Database (CDD; Marchler-Bauer et al. 2002) to the nr protein set, and hits with an e-value <0.01 are recorded. To filter out low-complexity sequence, the seg filter is applied. To reduce false positives, each hit must be at least 40% of the length of the domain definition. Hits to sequences are then sorted by domain.
The CDD database contains redundant domain definitions and domain definitions that are closely related, and it simplifies the CDART results to reduce this redundancy. To do this, we perform an all-against-all comparison of each domain's sequence hits to the sequence hits of all other domains. The comparison looks for overlapping sequence hits, defined as a >50% overlap of the length of either domain's hit to the sequence. If >15% of the total number of hits to nr for either domain being compared is exceeded, then both domains are recorded as being similar. At the end of this comparison, all similar domains are clustered together using single linkage clustering.
Using the sequence hits and the clusters of similar domains, domain architectures are calculated for each protein in nr. These architectures are an N-terminus to C-terminus listing of the domain clusters found in each protein with consecutive repeats of a domain cluster collapsed to a single repeat. The architectures are recorded in the CDART database along with taxonomic information taken from the NCBI Entrez Taxonomy Database (Wheeler et al. 2002).
Querying CDART
When given a query sequence by the user, CDART runs RPS-BLAST to find domain hits. Each of these domain hits is assigned to a domain cluster. CDART then retrieves all domain architectures that contain any of the domain clusters in the query. These domain architectures are then ranked and listed by the total number of domain clusters in common with the query.
For display purposes, overlapping hits in both the query and similar sequences are eliminated using the following algorithm: The highest scoring hit in a sequence is selected. Any overlapping hits are discarded, where overlapping is defined as >50% overlap of either hit. Then the next highest scoring hit is selected and the process is repeated until there are no remaining overlaps.
WEB SITE REFERENCES
http://smart.embl-heidelberg.de/; SMART program.
http://www.cgr.ki.se/Pfam/; Pfam program.
http://www.ebi.ac.uk/interpro/; Interpro program.
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=cdd; Entrez CDD.
http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi; CDART.
Acknowledgments
We greatly appreciate the programming assistance of the NCBI Information Engineering Branch. In particular, we thank Jim Ostell for technical help in creating Figure 1, the BLAST group for providing RPS-BLAST, and the Taxonomy group for a variety of useful resources, including the common tree program used to do the taxonomic subsetting. The NCBI Structure group provided many helpful comments in the creation of CDART. We are also grateful to the NIH intramural research program for support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lewisg@mail.nih.gov; FAX (301) 435-7794.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.278202.
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MDR, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. The Pfam Protein Families Database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- Hanai S, Uchida M, Kobayashi S, Miwa M, Uchida K. Genomic organization of Drosophila poly(ADP-ribose) polymerase and distribution of its mRNA during development. J Biol Chem. 1998;273:11881–11886. doi: 10.1074/jbc.273.19.11881. [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: A database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C P, Dickens NJ. Genome cartography through domain annotation. Genome Biol. 2001;2:comment2006.1–2006.6. doi: 10.1186/gb-2001-2-7-comment2006. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Lash AE, Leipe DD, Madden TL, Pontius JU, Schuler GD, Schriml LM, Tatusova TA, Wagner L, et al. Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res. 2002;30:13–16. doi: 10.1093/nar/30.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]