Abstract
Wine yeast strains show a high level of chromosome length polymorphism. This polymorphism is mainly generated by illegitimate recombination mediated by Ty transposons or subtelomeric repeated sequences. We have found, however, that the SSU1-R allele, which confers sulfite resistance to yeast cells, is the product of a reciprocal translocation between chromosomes VIII and XVI due to unequal crossing-over mediated by microhomology between very short sequences on the 5′ upstream regions of the SSU1 and ECM34 genes. We also show that this translocation is only present in wine yeast strains, suggesting that the use for millennia of sulfite as a preservative in wine production could have favored its selection. This is the first time that a gross chromosomal rearrangement is shown to be involved in the adaptive evolution of Saccharomyces cerevisiae.
[The sequence data from this study have been submitted to EMBL under accession nos. AF239757, AF239758, and AJ458340–AJ458367. The following individual kindly provided reagents, samples, or unpublished information as indicated in the paper: N. Goto-Yamamoto.]
The unaware use of yeast for winemaking by the first agricultural civilizations has been reported as far back as 7400 years ago. Until the middle of the last millennium, wines were mainly produced around the Mediterranean Sea and the Caucasus. Since then, winemaking has spread with the European colonizers throughout the temperate regions of the world (Pretorius 2000).
Although different genera and species of yeasts are found in musts, the species Saccharomyces cerevisiae is mainly responsible for the transformation of musts into wines. The origin of S. cerevisiae is controversial. Some authors propose that this species is a “natural” organism present in plant fruits (Mortimer and Polsinelli 1999). Others argue that S. cerevisiae is a domesticated species originated from its closest relative S. paradoxus, a wild species found all around the world (Vaughan-Martini and Martini 1995). This debate is important in postulating the original genome of S. cerevisiae and how the strong selective pressure applied since its first unconscious use in controlled fermentation processes has reshaped it. Useful phenotypic traits such as fast growth in sugar-rich media, high alcohol production and tolerance, and good flavor production selected for billions of generations have had strong influences on the S. cerevisiae genome.
In contrast to most S. cerevisiae strains used in the laboratory, which are either haploid or diploid and have a constant chromosome electrophoretic profile, wine yeast strains are mainly diploid, aneuploid, or polyploid, homothallic, and highly heterozygous (Bakalinsky and Snow 1990; Barre et al. 1993; Codón et al. 1995), and show a high level of chromosome length polymorphisms (Bidenne et al. 1992; Rachidi et al. 1999). Moreover, wine yeast strains seem not to remain genetically uniform (Pretorius 2000). Their exacerbated capacity to reorganize its genome by chromosome rearrangements such as Ty-promoted chromosomal translocations (Longo and Vézinhet 1993; Rachidi et al. 1999), mitotic crossing-over (Aguilera et al. 2000), and gene conversion (Puig et al. 2000) promotes a faster adaptation to environmental changes than spontaneous mutations, which occur at comparatively very low rates. The ploidy of the wine yeasts may confer advantages in adapting to variable external environments or increasing the dosage of some genes important for fermentation (Bakalinsky and Snow 1990; Salmon 1997).
In addition, the possibility of adaptive gross genomic changes occurring during laboratory growth conditions has been demonstrated with DNA chip technology by Hughes et al. (2000). Those authors showed in multiple cases that the deletion of a gene strongly favors the acquisition of a second copy of a whole chromosome or a chromosomal segment containing a compensatory copy of a close homolog of the deleted gene.
In a comparative study of transcriptomes, we found that SSU1, a gene that mediates sulfite efflux in S. cerevisiae and, hence, confers sulfite resistance (Park and Bakalinsky 2000), showed a significantly higher expression in the T73 wine yeast strain than in a laboratory strain (Hauser et al. 2001). In contrast to the allele present in the laboratory strains, a highly sulfite-resistant wine strain exhibited a translocation involving the promoter region of the gene (SSU1-R allele), which produces an increase in the sulfite resistance (Goto-Yamamoto et al. 1998). In the present study, we explored the organization of this gene at the molecular level in different wine yeast strains.
RESULTS
Genome Organization of the SSU1 Locus in T73 Wine Yeast Strain
Sulfite-generating compounds are widely used during wine making as bacterial inhibitors (Pretorius 2000). Therefore, sulfite resistance is a desired trait for wine yeast strains. Interestingly, our comprehensive study of gene expression in the T73 wine yeast strain grown under standard laboratory conditions (Hauser et al. 2001) revealed that the SSU1 gene, which mediates sulfite efflux in S. cerevisiae (Park and Bakalinsky 2000), is expressed more in the wine strain than in a reference laboratory strain (S288c background). High levels of SSU1 mRNA have also been observed in other wine yeast strains, such as Y-9, and this has been correlated with increased sulfite resistance (Goto-Yamamoto et al. 1998). Y-9 strain possesses an SSU1 allele (SSU1-R) with an upstream sequence completely replaced due to a putative translocation (Goto-Yamamoto et al. 1998).
To understand the mechanisms underlying the increased expression of SSU1 in wine yeast strains, we decided to investigate the promoter sequence of this gene. For this purpose, PCR primers for the selective amplification of the SSU1 region were designed according to the sequences of the laboratory S288c background (SSU1 allele) and Y-9 (SSU1-R allele) wine yeast strain (Fig. 1). Both primer pairs (SSU1MD/SSU1R for the SSU1 allele, and ECM34D/SSU1R for the SSU1-R allele) amplified in T73 fragments of the predicted lengths, 569 and 573 bp, respectively (data not shown). Their sequences were in both cases 100% identical to those described for S288c background (from MIPS database) and Y-9 strain (Goto-Yamamoto et al. 1998). Consequently, we can conclude that the T73 wine yeast strain is a heterozygote containing both SSU1 and SSU1-R alleles.
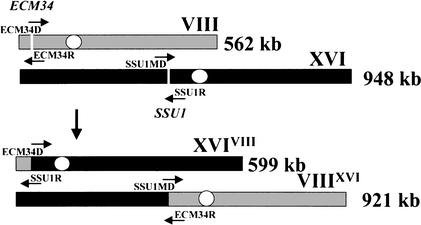
Figure 1.
Diagram representing the reciprocal translocation between chromosomes VIII and XVI observed in wine yeast strains. This translocation was mediated by crossing-over between microhomology regions of the promoters of the ECM34 and SSU1 genes, the locations of which, on chromosomes VIII and XVI, respectively, are indicated by white bars. Small arrows indicate the PCR primers used to amplify those regions involved in the recombination.
The promoter sequence of the SSU1-R allele presents a very high similarity with the promoter sequence of ECM34, a gene of unknown function from chromosome VIII (this study: accession no. AF239758, and Goto-Yamamoto et al. [1998]: accession no. AB002531). It is worth noting that the sequence of the SSU1-R allele contains four repeats of a 76-bp sequence, which is a single copy of 77 bp in ECM34 from S288c background (Fig. 2). These results strongly suggest that a reciprocal translocation between chromosomes VIII and XVI could have occurred in the wine yeasts (Fig. 1).
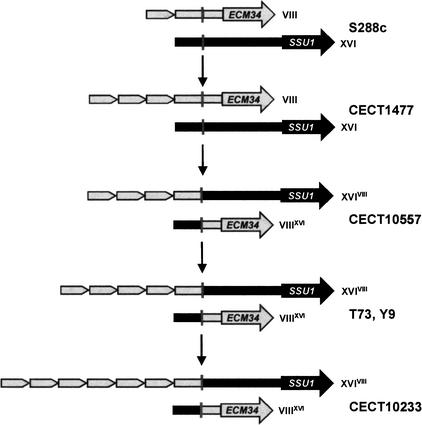
Figure 2.
Diagrams representing the organization of the ECM34 (in gray) and SSU1 (in black) nonrecombinant alleles and their corresponding recombinant variants obtained by an illegitimate crossing-over of a microhomology region (see Fig. 3), indicated by a vertical line, located in the promoters of both genes. This illegitimate crossing-over was involved in the generation of the reciprocal translocation between chromosomes VIII and XVI found in Saccharomyces strains (Fig. 1). Thick arrows represent the protein-coding regions of each gene. The pentagon block corresponds to a 76-bp sequence that has been found several times repeated in the promoters of both nonrecombinant ECM34 (strain CECT 1477) and recombinant SSU1-R (several strains). Strains bearing these sequences are indicated for each diagram.
By designing a new primer from the ECM34 coding sequence (ECM34R), two new PCR amplifications were performed with the ECM34D+ECM34R and SSU1MD+ECM34R primer pairs (Fig. 1). The first primer pair allowed us to amplify from T73 a band of 207 bp corresponding to the “standard” ECM34 locus, and the second pair, a band of ∼450 bp (accession number AF239757), which corresponded with a putative reciprocal translocation between chromosomes VIII and XVI at the 5′ upstream regions of the SSU1 and ECM34 genes (30 bp upstream from the ECM34 ATG start codon) (Fig. 2).
This putative translocation explains the contour-clamped homogeneous electrical field electrophoresis (CHEF) analysis we previously observed for the T73 strain (Puig et al. 2000). That analysis revealed the existence of anomalous-sized bands in the region of chromosomes VIII and XVI (Figs. 1 and 2 from Puig et al. 2000). A probe from chromosome VIII (CUP1 sequence) hybridized in a Southern blot with two chromosomal bands of ∼560 (chromosome VIII) and ∼920 Kb (chromosome VIIIXVI), and a probe from chromosome XVI (CAR1 sequence) hybridized with bands of ∼920 (chromosome VIIIXVI) and ∼950 Kb (chromosome XVI). This strain had a sporulation efficiency of 60% and a spore viability of 70%. However, only 13% of the dissected tetrads produced four viable spores (Puig et al. 2000). In all complete viable tetrads analyzed, the polymorphic bands observed after hybridization with probes from chromosomes VIII and XVI segregated at 2:2 (Puig et al. 2000), indicating that each pair of bands is allelic. These results confirm that a reciprocal translocation involving chromosomes VIII and XVI occurred (Fig. 1), and that the T73 strain is heterozygous for the reciprocal translocation, containing both the translocated and the “standard” chromosomal arrangements. Because translocated chromosomes contain all of the original genes from chromosomes VIII and XVI, spores are viable only if they contain either both “standard” chromosomes or both translocated ones.
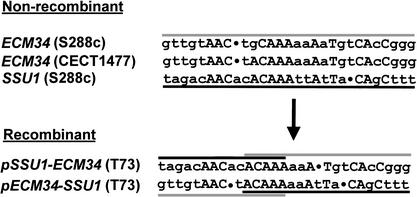
The reciprocal translocation between chromosomes VIII and XVI could be originated by different molecular mechanisms. We wondered whether a short region of sequence homology could mediate a heterologous recombination between the promoter sequences of ECM34 and SSU1 genes. When comparing the sequences of both gene promoters at the translocation breakpoint, we realized that the best alignment contained a short sequence of 9–13 bp of microhomology (Fig. 3), which includes the recombination site as deduced from the sequences of the recombinant and nonrecombinant alleles (Fig. 2). Taken together, these results suggest that the SSU1-R allele present in T73 and Y-9 wine yeast strains was generated by a reciprocal translocation between chromosomes VIII and XVI due to unequal crossing-over between a short region of microhomology located in the promoter region of SSU1 and ECM34 genes.
Figure 3.
Microhomology regions, located in the ECM34 and SSU1 promoters, that were involved in the crossing-over generating the reciprocal translocation between chromosomes VIII and XVI. Strains where sequences were obtained from are indicated in parentheses. Black and gray lines highlight ECM34 and SSU1 promoter sequences, respectively. Perfect sequence matches are shown in capitals, and dots correspond to gaps required to align the sequences.
Frequency and Origin of the Translocation (VIII;XVI) in S. cerevisiae
The fact that two geographically distant but naturally occurring wine yeast strains, Y-9 and T73, exhibited the same translocation prompted us to investigate whether this rearrangement was present in other S. cerevisiae strains isolated from different sources (wine and nonwine), in diverse geographic origins, and during several periods of time. A total of 30 strains (Table 1, 18 isolated from wine and 12 from other sources) were analyzed by PCR amplification with the different combinations of primers. The characteristic 450-bp PCR fragment of the recombinant SSU1-R allele (ECM34-derived promoter region+SSU1 coding region) was found in five additional strains, all of them also isolated from wines in different geographical areas. Four of them (CECT 10120, 1485, 10557, and 11827) were homozygous for the translocation as Y-9, and the other one (CECT 10233) corresponded to a translocation heterozygote as T73 (Table 1). The recombinant SSU1-R promoters from four of these additional strains were sequenced (EMBL accession numbers AJ458364–AJ458367), and the results showed they were all identical, except for the number of 76-bp repeats, indicating that the translocation (VIII; XVI) was a rare and unique event.
Table 1.
Saccharomyces “sensu stricto” Strains Analyzed, Origins, Sources Whence They Were Isolated, PCR Patterns Obtained by Amplification With Primers Specific of the SSU1 and ECM34 Regions, and ECM34 Promoter Sequence Type Exhibited
| Strain designation | Other designations | Source, origin, and year of isolation | PCR pattern | pECM34 type |
|---|---|---|---|---|
| S. cerevisiae strains | ||||
| A364a | Laboratory | 207, (568) | C | |
| CEN-PK 113 17A | Laboratory | 207, (568) | C | |
| S288c | Laboratory | 207, 568 | D | |
| SK1 | Laboratory | 207, (568) | C | |
| W303 | Laboratory | 207, (568) | D | |
| T73 | CECT 1894 | Wine, Spain (1987) | 220, 450, 568, 573 | E and S |
| Y-9 | Wine, Japan | 450, 573 | S | |
| CECT 1383 | CBS 2978 | Distiller's yeast | 207, (568) | A |
| CECT 1462 | NCYC 963 | Ale beer, UK | 207, (568) | C |
| CECT 1475 | UCD 519 | Sherry wine, Jerez, Spain | 207, 568 | B |
| CECT 1476 | UCD 522 | Montrachet, California, USA | 207, 568 | A |
| CECT 1477 | UG5 | Sparkling wine, Bordeaux, France | 380, 568 | G |
| CECT 1485 | González Byass wineries, Spain | 450, 573 | S | |
| CECT 1882 | Wine pellicle, Huelva, Spain | 207, 568 | B | |
| CECT 10009 | CBS 4054 | Red wine, Spain (1958) | 207, (568) | B |
| CECT 10095 | CBS 5835 | Wine, Spain (1959) | 207, (568) | n.d. |
| CECT 10120 | Fruit of Arbutus unedo, Spain | 450, 573 | S | |
| CECT 10131 | Flower of Centaurea alba, Spain | 207, (568) | A | |
| CECT 10233 | CBS 2247 | Grape must, South Africa (1955) | 207, 450, (568), 750 | D and T |
| CECT 10392 | CBS 3081 | Alpechin, Spain (1958) | 220, (568) | E |
| CECT 10557 | CBS 5112 | Grape must, Spain (1962) | 450, 497 | R |
| CECT 10691 | CBS 400 | Palm wine, from Ivory Coast (1927) | 207, 568 | A |
| CECT 10692 | CBS 429 | Fermenting Champagne grapes (1899) | 207, 280, (568) | B |
| CECT 11032 | CBS 459 | Grape must, Italy (1938) | 220, (568) | F |
| CECT 11827 | Dry wine yeast, Switzerland (1984) | 450, (573) | n.d. | |
| CECT 11833 | CBS 423 | Wine, Switzerland (1924) | 207, (568) | C |
| CECT 11834 | CBS 4070 | Red wine, Spain (1958) | 207, (568) | D |
| CECT 11835 | CBS 1250 | Sherry, Spain (1936) | 207, (568) | D |
| CECT 11837 | CBS 405 | Bili wine, West Africa (1925) | 207, (568) | B |
| CECT 11838 | CBS 5287 | Grape, Russia (1961) | 207, (568) | A |
| Other Saccharomyces “sensu stricto” species | ||||
| CECT 1969 | CBS 395 | S. bayanus (type of S. uvarum) | No amplification | – |
| CECT 12635 | S. bayanus isolated from wine | No amplification | – | |
| CECT 12636 | S. bayanus isolated from wine | No amplification | – | |
| CECT 1939NT | CBS 432 | S. paradoxus neotype strain | 568 | – |
| CECT 11152 | IFO 1804 | S. paradoxus from tree exhudate, in Japan | 568 | – |
| CECT 11158 | CBS 2980 | S. paradoxus from Drosophila, in California | 568 | – |
| CECT 1970 | CBS 1503 | S. pastorianus (type of S. monacensis) | 207 | A |
| CECT 11037 | CBS 1513 | S. pastorianus (type of S. carlsbergensis) | 207 | A |
Between parentheses are indicated those expected fragments that did not amplify. 450 is the diagnostic band for the translocation, n.d., not determined. CBS (Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands), CECT (Spanish Type Culture Collection, University of Valencia, Spain), IFO (Institute for Fermentation, Osaka, Japan), UCD (Hermann Phaff Collection, University of California, Davis, USA), NCYC (National Collection of Yeast Cultures, Norwich, UK).
The 76-bp repeats present in the recombinant SSU1-R promoters contain some nucleotide substitutions and insertions/deletions (indels) compared to the original promoter of the ECM34 allele (see Fig. 2). Therefore, to deduce the origin of the recombinant promoter, we decided to amplify and sequence the nonrecombinant ECM34 promoter region from the two heterozygous strains T73 and 10233, from 23 additional S. cerevisiae strains, and from eight strains from other Saccharomyces “sensu stricto” species (Table 1, EMBL accession numbers AJ458340–AJ458363). The ECM34 promoter region could only be amplified in the S. cerevisiae strains and in the two S. pastorianus strains. This species is a partial allotetraploid originated from an S. cerevisiae × S. bayanus hybridization (Vaughan-Martini and Kurtzman, 1985; Casaregola et al. 2001; de Barros Lopes et al. 2002), and the amplified ECM34 promoter region probably corresponds to the S. cerevisiae fraction of the S. pastorianus genome. The results showed seven different sequence variants of the nonrecombinant ECM34 promoter (Fig. 4, variants A to G). The most frequent variants, A and B, and the related variants E to G, are mainly present in wine strains, with the exception of strains 10131 and 10392, which were isolated from the plant Centaurea alba and from alpechin (olive residues after oil extraction), respectively, in Spain, a wine-producing country, and 10691, isolated from palm wine in West Africa. In contrast, variants C and D are present in laboratory strains, but also in strains 1462, 11837, and 11838 isolated from ale beer, bili wine and grapes, respectively.
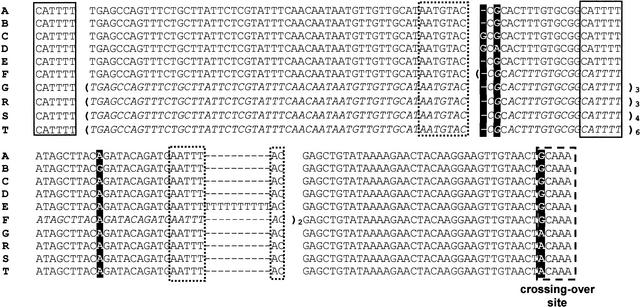
Figure 4.
Sequence alignment of the seven nonrecombinant and three recombinant variants of the ECM34 promoter region found in Saccharomyces strains (see Table 1). Variable positions are shown in negative. Italic sequences in parentheses correspond to repeated regions, and subscript numbers after the parentheses indicate the number of repeats. Continuous rectangles highlight a small direct repeated sequence flanking a large 76-bp sequence repeat that could be involved in its duplication. Dotted rectangles indicate a small imperfect repeat that could be involved in the generation of a 47-bp duplication found in the CECT11032 strain. The discontinuous rectangle indicates the crossing-over site involved in the reciprocal translocation t(VIII,XVI).
We obtained a maximum parsimonious tree that minimizes the number of changes (nucleotide substitutions, indels, and repeat insertions) required to connect these sequences (Fig. 5). In this tree, variant A occupies a central position from which all the other variants can be derived. Variant A and its closest related variant B, which differs in a single nucleotide substitution, are exhibited by a heterogeneous group of wine-related strains and by the hybrid S. pastorianus strains.
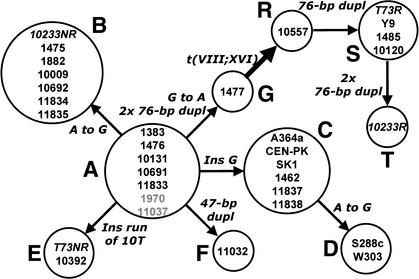
Figure 5.
Maximum parsimony tree that minimizes the number of mutational events required to connect all the sequence variants of the ECM34 gene promoter from different Saccharomyces strains N to N, nucleotide substitutions; ins, insertions; dupl; sequence duplications or repeats; 2x, a double event. A to G are nonrecombinant sequence variants, and R to T corresponds to the recombinant variants (pECM34-SSU1 or SSU1-R) generated by the unequal crossing-over involved in the translocation t(VIII;XVI). The translocation event due to unequal crossing-over between microhomology regions located in the ECM34 and SSU1 promoters is indicated by a thick arrow. Strains in gray correspond to S. pastorianus. Strains in italics are heterozygotes for the translocation and contain both a recombinant (R) and a nonrecombinant (NR) variant.
Variant C, exhibited by three laboratory and three natural strains, differs from variant A by a single G nucleotide insertion. Variant D differs in a single nucleotide substitution from variant A and is present only in laboratory strains S288c and its derivative W303 (Rogowska-Wrzesinska et al. 2001). S288c, the most popular genetic background in yeast research laboratories, is a derivative of a natural heterothallic diploid strain isolated from rotting figs in California in 1938 (Mortimer and Johnston 1986). It is quite likely that this yeast strain was carried from cellars by insects. Taken together, these observations suggest that wine strains could probably be the ancestors of the domesticated laboratory strains (Mortimer and Johnston 1986).
The other variant sequences (E, F, G, and the recombinant R, S, and T), which include strains containing the promoter of the recombinant SSU1-R allele, differ from variant A by the presence of a series of tandem repeats of 1, 47, or 76 bp (see Fig. 4). The nonrecombinant allele of the translocation heterozygote T73 (variant E, T73NR) exhibits an insertion of 10 T that was probably generated by replication slippage within a region of 3 T. The same insertion is also present in strain 10392. Strain 11032 (variant F) contains two tandem repeats of a 47-bp region that could have been generated either by unequal crossing-over between two almost identical 7-bp regions flanking the repeated region (Fig. 4), or by replication slippage also favored by the pairing of the 7-bp flanking regions and the possible secondary structure of the repeat. A different repeated region, although overlapping with that from strain 11032, is present in the promoter region of the recombinant SSU1-R alleles (variants R, S, and T). These promoter variants contain three, four, or six tandem repeats of a 76-bp region, respectively. The first duplication event, which occurred either by unequal crossing-over or by replication slippage, could be favored by the presence of a 6-bp identical sequence flanking the repeated region (Fig. 4), and also by a potential secondary structure of three hairpin-loops.
The nonrecombinant ECM34 promoter from strain 1477 (variant G) also contains three tandem repeats of the same 76-bp region and a G to A substitution located at the putative crossing-over site shared with the recombinant promoter variants of the SSU1-R allele, R, S, and T (Figs. 2–4). This sequence organization strongly suggests that the first duplication of the 76-bp region occurred at the ECM34 promoter before the illegitimate crossing-over between ECM34 and SSU1 promoters produced the translocation (VIII;XVI). Once the rare first duplication event took place, subsequent duplications could, with a higher probability, be extended by unequal crossing-over (either meiotic or mitotic) between any repeats, or by slipped-strand mispairing between contiguous repeats. It is likely that one subsequent duplication also occurred before the translocation.
The translocation between chromosomes VIII and XVI by the illegitimate crossing-over between ECM34 and SSU1 promoters took probably place in a strain with three 76-bp repeats in its ECM34 promoter, similar to 1477, giving rise to a recombinant promoter with three repeats as that found in strain 10577 (Fig. 2). However, the presence of heterozygotes for the translocation (strains 10233 and T73), which exhibit nonrecombinant alleles (variants B and E, respectively) quite different from that of strain 1477 (variant G), can only be explained by subsequent sexual reproduction. Convergent evolution as an alternative explanation is discarded because it implies convergent changes in three characters: the A-G substitution, the gain of a run of Ts, and the loss of 76-bp repeats. Finally, a heterozygote for the translocation should also give rise to homozygotes such as strains Y-9, CECT1485, 10120, 10557, and 11827. The simplest explanation is that the fixation in homozygosis of this advantageous translocation conferring a higher resistance to sulfite occurred after sporulation and homothallic conjugation.
The observed selection for strains that contain several repeats of the 76-bp sequence suggests that the presence of these repeats increases sulfite tolerance. This fact was demonstrated by Goto-Yamamoto et al. (1998) with natural strains, and by Park and Bakalinski (2000) with genetically modified strains. In the present study, we corroborated this hypothesis by measuring sulfite tolerance of several wine strains containing different numbers of 76-bp repeats in their recombinant SSU1 promoters (Table 2). From these results, it can be concluded that there is a direct relationship between the number of 76-bp repeats and sulfite tolerance, irrespective of the homozygotic or heterozygotic nature of the locus.
Table 2.
Sulfite Tolerance of Yeast Strains Exhibiting Different Numbers of Repeats of a 76-bp Region in the Recombinant SSU1 Promoter
| Strain | Number of 76-bp repeats in recombinant SSU1 promoter | Sulfite concentration (mM) | ||||
|---|---|---|---|---|---|---|
| 0 | 1 | 3 | 6 | 8 | ||
| S288c | 0 (nonrecombinant control) | + | + | − | − | − |
| T73 | 4 | + | + | + | − | − |
| CECT10233 | 6 | + | + | + | +/− | − |
| CECT10557 | 3 × 2a | + | + | + | +/− | − |
| Y-9 | 4 × 2a | + | + | + | + | − |
Sulfite sensitivity was determined on YPD+TA plates containing 0–8 mM Na2SO3 as described in Methods. +, growth; −, no growth; +/−, poor growth scored after 24 h. Similar results were obtained in liquid medium in microtiter plates after 40 h.
These strains are homozygous for the recombinant SSU1 promoter.
DISCUSSION
Translocations have been shown to be very common in S. cerevisiae; however, most of them are mediated through Ty or subtelomeric Y‘ element recombination (Kupiec and Petes 1988; Casaregola et al. 1998), especially in wine strains (Bidenne et al. 1992; Rachidi et al. 1999). Ectopic translocations through subtelomeric repetitive elements have also been proposed as the mechanism involved in the origin of some subtelomeric gene families such as SUC, MAL, RTM, or MEL, and have been correlated with the improvement of the features of some industrial yeast strains (discussed in Ness and Aigle 1995). However, in the present study we showed that the most probable cause of the reciprocal translocation between chromosomes VIII and XVI in wine yeasts is an illegitimate recombination mediated by microhomology. This kind of nonhomologous recombination is extremely rare in wild-type strains, occurring at a frequency of 3.5× 10–10. This frequency is only increased in double-strand break (DSB) repair-deficient mutants (Chen and Kolodner 1999). Thus, it seems that the new VIIIXVI and XVIVIII chromosomes in wine yeast strains were probably generated by a spontaneous reciprocal translocation mediated by the fortuitous appearance of a broken chromosome end produced by a DSB in either of the two gene promoters, ECM34 or SSU1. This end likely facilitated recombination with the other promoter through a very short homologous region. Whether this presumed recombination has been produced during mitosis or meiosis is currently unknown. This is the first time that such a rare event has been described in the evolution of yeast strains in the wild.
The enhanced expression of SSU1 gene enabled wine yeast strains carrying the translocation to resist higher sulfite concentrations (Goto-Yamamoto et al. 1998, Park and Bakalinsky 2000; our results, Table 2). The new chromosomes, which did not lack any essential element, contained a new SSU1 promoter with putative binding sites for the Fzf1p transcription activator within the 76-bp repeats (Avram et al. 1999). According to this hypothesis and the evolutionary analysis of DNA sequences performed, it seems likely that the 76-bp sequence was already repeated before the translocation event, giving rise to a higher expression of SSU1 since the very beginning. We observed a clear relationship between the number of 76-bp repeats in the SSU1 promoter and the level of sulfite resistance. This 76-bp sequence is partially palindromic and has a direct 6-bp repeat at both ends that may easily promote tandem repeat formation (Fig. 4). Similarly, a 147-bp repeated element found in MAL promoters from baker’s yeast strains also alters gene expression (Bell et al. 1997).
The equilibrated chromosome pair VIIIXVI and XVIVIII is quite frequent in wine yeasts. Thus, Goto-Yamamoto et al. (1998) and Bidenne et al. (1992) also observed that among different wine yeast strains other than those studied here, eight were heterozygous and two homozygous for the SSU1-R allele (and hence, as demonstrated in the present study, heterozygous for the translocation), and that the SSU1-R allele (and hence, the translocation) was absent in the different nonwine strains analyzed. This higher frequency of the SSU1-R allele generated by the t(VIII;XVI) translocation in wine yeasts can be explained by its adaptive value in wine-making environments where sulfite is widely used as a preservative. Wine strains of S. cerevisiae tolerate relatively high concentrations of sulfur dioxide as a result of adaptation and natural selection. This is due to the fact that sulfur dioxide is an antioxidant and antimicrobial agent that has been used in winemaking for millennia (Romano and Suzzi 1993). The Egyptians, and later the Greeks and Romans, made use of burning sulfur fumes to clean their wine containers. During the Middle Age, SO2 became a widely used preservative, obtained originally by burning sulfur but later by adding sulfite or bisulfite to musts. Nowadays, the use of SO2 in winemaking is a common practice that is permitted by all wine-producing countries, in concentrations varying from 160 to 400 mg/L of total SO2 or 20 to 100 mg/L of free SO2. Moreover, sulfur dioxide resistance is an enological character used for the selection of commercial wine yeast, which is why many of the commercial strains analyzed exhibit the t(VIII, XVI) translocation (T73, this study and Goto-Yamamoto et al. 1998).
Finally, another conclusion that can be drawn from the present study is the role of sexual reproduction during wine fermentation. Sexual reproduction in wine S. cerevisiae yeasts has been a matter of controversy. Wine yeasts are prototrophic, homothallic, highly heterozygous and aneuploid, and exhibit low sporulation rates and spore viability (Bakalinsky and Snow 1990; Barre et al. 1993; Guijo et al. 1997). These characteristics, along with the observation of sexual isolation in yeast population during wine production (Guijo et al. 1997), are evidence favoring sexual reproduction as very rare, or even absent, in wine yeasts. However, the observation in the present study of translocation heterozygotes with very different nonrecombinant alleles supports the conclusion that sexual reproduction may be present in natural S. cerevisiae strains.
METHODS
Yeasts Strains and Culture Conditions
Forty-four strains of genus Saccharomyces were examined, all of them obtained from the Spanish Type Culture Collection (CECT). Three strains belong to the species S. bayanus, 30 to S. cerevisiae, three to S. paradoxus and two to S. pastorianus. The sources from which they were isolated are shown in Table 1. For laboratory culture, yeast cells were grown at 30°C in YPD (1% yeast extract, 2% bacteriological peptone, 2% glucose).
Sulfite tolerance was scored in YPD+TA (tartaric acid) agar plates as described by Park et al. (1999) by replicating cells grown in YPD plates. Alternatively, liquid YPD+TA containing 0–12 mM Na2SO3 was distributed in 100 μL aliquots in multiwell plates, and 2 μL of YPD-exponentially growing cells (0.2 OD600) was added to every well. Growth was scored after 40 h.
PCR Reaction and Sequencing
A single yeast colony, taken using a micropipet yellow tip, was suspended in 100μL of a PCR reaction mix containing 100 μM deoxynucleotides, 1× reaction buffer, and 1 μM of these primers: ECM34D (5′-TCGAACATCGAGCATGCA-3′), ECM34R (5′-CCATATTTGTGATGATATCG-3′), SSU1MD (5′-ACCTATCGAGTCTCCCAC-3′), SSU1R (5′-GACACCCAT GACCATCAC-3′). The mixture was heated at 95°C for 15 min in a thermocycler, and 1 U of Biotools II DNA Polymerase (Biotools) was added to each tube. The PCR conditions were as follows: denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 52.5°C for 1 min, and polymerization at 72°C for 1 min. The polymerization was completed by one additional cycle of 5 min at 72°C.
The PCR product was separated on 3 % (w/v) agarose gels with 1×TAE (40 mM Tris-Acetate, 1mM EDTA) buffer. After electrophoresis, gels were stained with ethidium bromide, visualized under UV light, and photographed. Molecular weights were estimated by comparison against a 100-bp DNA ladder. PCR bands were purified with a GeneClean II Kit (Bio101) and directly sequenced using the Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin Elmer), following the manufacturer's instructions, in an Applied Biosystems automatic DNA sequencer model 3700.
Sequences were deposited in the EMBL database under accession numbers AF239757, AF239758, and AJ458340 to AJ458367.
Acknowledgments
We thank Drs. F.J. Ayala and M. Lau for their valuable comments that improved the manuscript. Dr. Goto-Yamamoto kindly provided the Y-9 wine yeast strain. We also thank M.J. Peris for her technical assistance. This work was funded by grant BIO4-CT97-2294 (EUROFAN 2) from the European Union to J.E. P.-O. and by grant GV01-268 from “Generalitat Valenciana”, Spain, to E.B. and A.Q.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jose.e.perez@uv.es; FAX 34 96 386 4635.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.436602.
REFERENCES
- Aguilera, A., Chávez, S., and Malagón, F. 2000. Mitotic recombination in yeast: Elements controlling its incidence. Yeast 16: 731–754. [DOI] [PubMed]
- Avram D, Leid M, Bakalinsky AT. Fzf1p of Saccharomyces cerevisiae is a positive regulator of SSU1 transcription and its first zinc finger region is required for DNA binding. Yeast. 1999;15:473–480. doi: 10.1002/(SICI)1097-0061(199904)15:6<473::AID-YEA388>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bakalinsky AT, Snow R. The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast. 1990;6:367–382. doi: 10.1002/yea.320060503. [DOI] [PubMed] [Google Scholar]
- Barre P, Vézinhet F, Dequin S, Blondin B. Genetic improvement of wine yeasts. In: Fleet GH, editor. Wine microbiology and biotechnology. Chur, Switzerland: Harwood Academic Publishers; 1993. pp. 421–447. [Google Scholar]
- Bell PJL, Higgins VJ, Dawes IW, Bissinger PH. Tandemly repeated 147-bp elements cause structural and functional variation in divergent MAL promoters of Saccharomyces cerevisiae. Yeast. 1997;13:1135–1144. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1135::AID-YEA162>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Bidenne C, Blondin B, Dequin S, Vézinhet F. Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Curr Genet. 1992;22:1–7. doi: 10.1007/BF00351734. [DOI] [PubMed] [Google Scholar]
- Casaregola S, Nguyen HV, Lapathitis G, Kotyk A, Gaillardin C. Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. Int J Syst Evol Microbiol. 2001;51:1607–1618. doi: 10.1099/00207713-51-4-1607. [DOI] [PubMed] [Google Scholar]
- Casaregola S, Nguyen HV, Lepingle A, Brignon P, Gendre F, Gaillardin C. A family of laboratory strains of Saccharomyces cerevisiae carry rearrangements involving chromosomes I and III. Yeast. 1998;14:551–564. doi: 10.1002/(SICI)1097-0061(19980430)14:6<551::AID-YEA260>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Codón AC, Gasent-Ramírez JM, Benítez T. Factors which affect the frequency of sporulation and tetrad formation in Saccharomyces cerevisiae baker's yeasts. Appl Environ Microbiol. 1995;61:630–638. doi: 10.1128/aem.61.2.630-638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barros Lopes M, Bellon JR, Shirley NJ, Ganter PF. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 2002;1:323–331. doi: 10.1111/j.1567-1364.2002.tb00051.x. [DOI] [PubMed] [Google Scholar]
- Goto-Yamamoto N, Kitano K, Shiki K. SSU1-R, a sulphite resistance gene of wine yeast, is an allele of SSU1 with a different upstream sequence. J Ferm Bioengineer. 1998;86:427–433. [Google Scholar]
- Groth C, Hansen J, Piškur J. A natural chimeric yeast containing genetic material from three species. Int J Syst Bacteriol. 1999;49:1933–1938. doi: 10.1099/00207713-49-4-1933. [DOI] [PubMed] [Google Scholar]
- Guijo S, Mauricio JC, Salmon JM, Ortega JM. Determination of the relative ploidy in different Saccharomyces cerevisiae strains used for fermentation and ‘flor’ film ageing of dry sherry-type wines. Yeast. 1997;13:101–117. doi: 10.1002/(SICI)1097-0061(199702)13:2<101::AID-YEA66>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hauser NC, Fellenberg K, Gil R, Bastuck S, Hoheisel JD, Pérez-Ortín JE. Whole genome analysis of a wine yeast strain. Comp Funct Genom. 2001;2:69–79. doi: 10.1002/cfg.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- Kupiec M, Petes TD. Allelic and ectopic recombination between Ty elements in yeast. Genetics. 1988;119:549–559. doi: 10.1093/genetics/119.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo E, Vézinhet F. Chromosomal rearrangements during vegetative growth of a wild strain of Saccharomyces cerevisiae. Appl Environ Microbiol. 1993;59:322–326. doi: 10.1128/aem.59.1.322-326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Polsinelli M. On the origins of wine yeast. Res Microbiol. 1999;150:199–204. doi: 10.1016/s0923-2508(99)80036-9. [DOI] [PubMed] [Google Scholar]
- Ness F, Aigle M. RTM1: A member of a new family of telomeric genes in yeast. Genetics. 1995;140:945–956. doi: 10.1093/genetics/140.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Bakalinsky AT. SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast. 2000;16:881–888. doi: 10.1002/1097-0061(200007)16:10<881::AID-YEA576>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Park H, Lopez NH, Bakalinsky AT. Use of sulfite resistance in Saccharomyces cerevisiae as a dominant selectable marker. Curr Genet. 1999;36:339–344. doi: 10.1007/s002940050508. [DOI] [PubMed] [Google Scholar]
- Pretorius IS. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Puig S, Querol A, Barrio E, Pérez-Ortín JE. Mitotic recombination and genetic changes in Saccharomyces cerevisiae during wine fermentation. Appl Environ Microbiol. 2000;66:2057–2061. doi: 10.1128/aem.66.5.2057-2061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi N, Barre P, Blondin B. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:841–850. doi: 10.1007/s004380050028. [DOI] [PubMed] [Google Scholar]
- Rogowska-Wrzesinska A, Larsen PM, Blomberg A, Görg A, Roepstorff P, Norbeck J, Fey SJ. Comparison of the proteomes of three yeast wild type strains: CEN.PK2, FY1679 and W303. Comp Funct Genom. 2001;2:207–225. doi: 10.1002/cfg.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P, Suzzi G. Sulfur dioxide and wine microorganisms. In: Fleet GH, editor. Wine microbiology and biotechnology. Chur, Switzerland: Harwood Academic Publishers; 1993. pp. 373–394. [Google Scholar]
- Salmon JM. Enological fermentation kinetics of an isogenic ploidy series derived form an industrial Saccharomyces cerevisiae strain. J Ferment Bioeng. 1997;83:253–260. [Google Scholar]
- Vaughan-Martini A, Martini A. Facts, myths and legends on the prime industrial microorganism. J Ind Microbiol. 1995;14:514–522. doi: 10.1007/BF01573967. [DOI] [PubMed] [Google Scholar]
- Vaughan-Martini A, Kurtzman CP. Deoxyribonucleic acid relatedness among species of the genus Saccharomyces sensu stricto. Int J Syst Bacteriol. 1985;35:508–511. [Google Scholar]