Abstract
Mammals have 79 ribosomal proteins (RP). Using a systematic procedure based on sequence-homology, we have comprehensively identified pseudogenes of these proteins in the human genome. Our assignments are available at http://www.pseudogene.org or http://bioinfo.mbb.yale.edu/genome/pseudogene. In total, we found 2090 processed pseudogenes and 16 duplications of RP genes. In relation to the matching parent protein, each of the processed pseudogenes has an average relative sequence length of 97% and an average sequence identity of 76%. A small number (258) of them do not contain obvious disablements (stop codons or frameshifts) and, therefore, could be mistaken as functional genes, and 178 are disrupted by one or more repetitive elements. On average, processed pseudogenes have a longer truncation at the 5′ end than the 3′ end, consistent with the target-primed-reverse-transcription (TPRT) mechanism. Interestingly, on chromosome 16, an RPL26 processed pseudogene was found in the intron region of a functional RPS2 gene. The large-scale distribution of RP pseudogenes throughout the genome appears to result, chiefly, from random insertions with the numbers on each chromosome, consequently, proportional to its size. In contrast to RP genes, the RP pseudogenes have the highest density in GC-intermediate regions (41%–46%) of the genome, with the density pattern being between that of LINEs and Alus. This can be explained by a negative selection theory as we observed that GC-rich RP pseudogenes decay faster in GC-poor regions. Also, we observed a correlation between the number of processed pseudogenes and the GC content of the associated functional gene, i.e., relatively GC-poor RPs have more processed pseudogenes. This ranges from 145 pseudogenes for RPL21 down to 3 pseudogenes for RPL14. We were able to date the RP pseudogenes based on their sequence divergence from present-day RP genes, finding an age distribution similar to that for Alus. The distribution is consistent with a decline in retrotransposition activity in the hominid lineage during the last 40 Myr. We discuss the implications for retrotransposon stability and genome dynamics based on these new findings.
All of the proteins in the cell are synthesized by the ribosomes, large complexes of RNA and protein molecules. A typical mammalian cell has about 4 × 106 ribosomes, and each is composed of four RNA molecules (rRNA) and 79 ribosomal proteins (RPs). In total, ribosomes constitute about 80% of the RNA and 5%–10% of the protein in a cell (Kenmochi et al. 1998). Great progress has been made in recent years in elucidating the structure and mechanism of the ribosome. The peptide sequence of the complete set of mammalian RPs was deduced by Wool and colleagues (1995), and the genes encoding all human RPs have been positioned on the human genetic map (Kenmochi et al. 1998; Uechi et al. 2001; Yoshihama et al. 2002). Moreover, several high-resolution atomic structures are now available for archaeal ribosomes (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000; Yusupov et al. 2001).
Although it is well recognized that rRNA catalyzes the basic biochemistry of protein synthesis, ribosomal proteins are important in facilitating rRNA folding, protecting them from nucleases, and coordinating the multistep process of protein synthesis. Some RPs have substantial extra-ribosomal functions as well (Wool 1996). It is believed that RPs from all three kingdoms of life are related, probably having evolved from the same ancestral set of proteins after the conversion of the ribosome from an RNA complex to a ribonucleoprotein particle (RNP). Among eukaryotes, the number and sequence of cytoplasmic RPs are fairly well conserved. For instance, yeast and rat share all but one RP, and the sequence identity of their RPs ranges from 40% to 88%, with an average of 60%. Among mammals, the amino acid sequences of the RPs are almost identical. For example, for the 72 RPs of which amino acid sequences are available for both human and rat, the average sequence identity is 99%, and 32 of them are perfectly identical (Wool et al. 1995).
In the yeast cell, the 78 RPs are encoded by 137 genes; 59 of the genes are duplicated (Planta and Mager 1998). In all cases, both gene copies are transcribed although their expression levels often differ considerably (Raue and Planta 1991). The proteins encoded by duplicated genes have identical or virtually identical sequences and are functionally indistinguishable. In contrast, it is widely recognized that in mammals a single gene encodes each RP, although most if not all of the RP genes have a number of processed pseudogenes located elsewhere in the genome. The existence of these pseudogenes has greatly hindered the sequencing and mapping efforts of human RP genes, so a special intron-trapping strategy had to be undertaken to differentiate the real transcribed RP gene and pseudogenes (Kenmochi et al. 1998; Uechi et al. 2001). A number of RP genes have also been implicated in various human diseases, such as RPS19 in Diamond-Blackfan anemia (DBA; Draptchinskaia et al. 1999), RPL6 in Noonan syndrome (Kenmochi et al. 2000), and RPS4X gene in Turner's syndrome (Zinn et al. 1993).
In general, pseudogenes are disabled copies of functional genes that do not produce a functional, full-length protein (Vanin 1985; Mighell et al. 2000). The disablements can take the form of premature stop codons or frame shifts in the protein-coding sequence (CDS), or less obviously, deleterious mutations in the regulatory regions that control gene transcription or splicing. There are two main types of pseudogenes: duplicated (nonprocessed) and processed. Duplicated pseudogenes arise from genomic DNA duplication or unequal crossing-over. They have the same general structure as functional genes, with sequences corresponding to exons and introns in the usual locations. Processed pseudogenes result from retrotransposition, that is, reverse-transcription of mRNA transcript followed by integration into genomic DNA, presumably in the germ line. Because of their origin, processed pseudogenes are sometimes considered a special type of retrotransposons just like Alu and long interspersed (LINE) elements, and are sometimes referred to as retro-pseudogenes. They are typically characterized by a complete lack of introns, the presence of small flanking direct repeats, and a polyadenine tract near the 3′ end (provided that they have not decayed). Processed pseudogenes in general are not transcribed, however in very rare cases, transcripts of some pseudogene have been reported, although the functional relevance of these pseudogene transcripts remains unclear (McCarrey et al. 1996; Fujii et al. 1999; Olsen and Schechter 1999).
It is unclear how many pseudogenes exist in the human genome. Estimates for the number of human genes range from ∼22,000 to ∼75,000 (Crollius et al. 2000; Ewing and Green 2000; Lander et al. 2001; Venter et al. 2001; Harrison et al. 2002b). From previous reports, it is thought that up to 22% of these gene predictions may be pseudogenic (Lander et al. 2001; Yeh et al. 2001). It is important to characterize the human pseudogene population, as their existence interferes with gene identification and annotation. They are also an important resource for the study of the evolution of protein families, for example, studies on the human olfactory receptor subgenome (Glusman et al. 2001). Harrison et al. (2002a) performed a detailed analysis of pseudogenes on human chromosomes 21 and 22. It was discovered that the protein family that has the largest number of processed pseudogenes is RPs, a total of 43 of which were found on the two smallest human chromosomes. This extrapolated to over 2000 RP pseudogenes in the whole human genome.
We have developed a pipeline of mostly automatic procedures that enables us to discover and characterize pseudogenes quickly and comprehensively. Here we report the identification of over 2400 processed RP pseudogenes and pseudogenic fragments on the latest human genome draft sequence (Lander et al. 2001). Complete sequence and precise chromosomal location have been obtained for each pseudogene. We provide a comprehensive characterization of the human RP pseudogene population and discuss its implications for retrotransposition and genome dynamics.
RESULTS
Human Genome Has 2090 RP Processed Pseudogenes
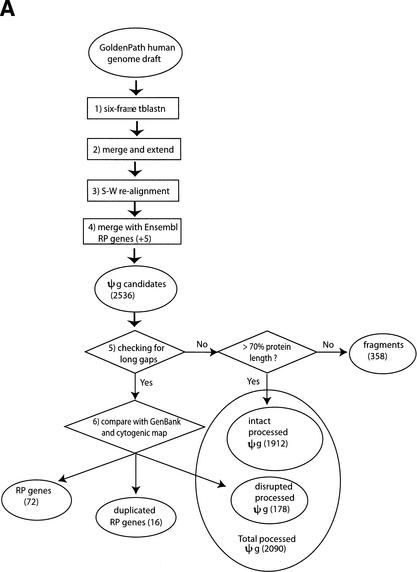
We have conducted a comprehensive search for cytosolic RP pseudogenes on the August 2001 freeze of the human genome draft (Lander et al. 2001). Details of the annotation procedure are described in the Methods section, and a flow chart is shown in Figure 8A below. Table 1 shows the distribution of identified RP pseudogenes among 22 autosomes and two sex chromosomes, together with the length of each chromosome and the number of functional RP genes previously mapped onto it (Kenmochi et al. 1998; Uechi et al. 2001; Yoshihama et al. 2002). Some general statistics of the processed pseudogene population are shown in Table 2. A total of 2090 processed RP pseudogenes were identified in the whole human genome. The substantial majority (1912) of these are termed “intact” pseudogenes because they are continuous in sequence with insertions shorter than 60 bp, whereas the remaining 178 are disrupted by long insertions in the middle of their sequence. The majority (146 of 178) of these disruptions are caused by the insertions of one or more retrotransposons, Alu, or less often, LINE elements.
Figure 8.
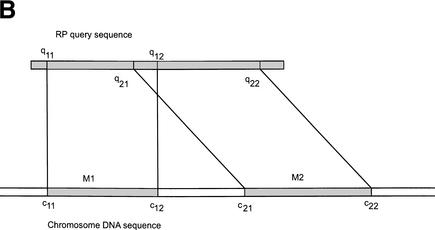
(A) Flow chart of the procedure for searching for RP pseudogenes in the human genome. RP and ΨG denote “ribosomal protein” and “pseudogene”, respectively. S-W., “Smith-Waterman”. The steps are as follows: (1) Six-frame BLAST run searching for RP homologies in the human genome. (2) Merging and extension. BLAST hits were merged and extended on both sides to match the length of RP peptide sequence. (3) Smith-Waterman realignment. Extended homologies were realigned with RP sequence. (4) Comparison with Ensembl annotation. Five RPL41 pseudogenes from Ensembl were added to the set. A total of 2536 PR genes or pseudogenes were identified. (5) Checking for long gaps. Homology sequences that contained gaps shorter than 60 bp were labeled “intact processed pseudogenes” if they were longer than 70% of the full-length RP sequence; otherwise they were labeled “pseudogenic fragments”. (6) Comparison with GenBank and cytogenic mappings. For those RP homologies that contained long gaps (>60 bp), their sequences were compared with the RP exon structure from GenBank and their chromosomal locations were checked with cytogenic mapping. The homology sequences were assigned as functional RP genes, duplicated RP genes, and “disrupted processed pseudogenes.” The latter were processed pseudogenes whose sequences were interrupted by retrotransposons. (B) Schematic graph describing the considerations in merging two adjacent RP matches, M1 and M2. (c11, c12) and (c21, c22) are chromosomal coordinates for M1 and M2. (q11, q12) and (q21, q22) are corresponding regions on the query RP protein that they match.
Table 1.
Number of RP Pseudogenes on Each Chromosome
| Chromosome | Chr. size (Mb) | Chr. GC content | Functional RP genes | Processed pseudogenes | Pseudogenic fragments | Processed + fragments | Pseudogene densityc | ||
|---|---|---|---|---|---|---|---|---|---|
| Intacta | Disruptedb | Total | |||||||
| 1 | 257 | 0.41 | 5 | 202 | 21 | 223 | 35 | 258 | 1.00 |
| 2 | 242 | 0.4 | 4 | 144 | 8 | 152 | 30 | 182 | 0.75 |
| 3 | 205 | 0.39 | 7 | 119 | 6 | 125 | 12 | 137 | 0.67 |
| 4 | 192 | 0.37 | 3 | 86 | 9 | 95 | 19 | 114 | 0.59 |
| 5 | 186 | 0.39 | 3 | 107 | 11 | 118 | 19 | 137 | 0.74 |
| 6 | 179 | 0.39 | 4 | 139 | 6 | 145 | 26 | 171 | 0.96 |
| 7 | 163 | 0.4 | 0 | 94 | 8 | 102 | 22 | 124 | 0.76 |
| 8 | 146 | 0.39 | 4 | 94 | 9 | 103 | 21 | 124 | 0.85 |
| 9 | 132 | 0.41 | 4 | 70 | 6 | 76 | 11 | 87 | 0.66 |
| 10 | 142 | 0.41 | 1 | 92 | 10 | 102 | 10 | 112 | 0.79 |
| 11 | 142 | 0.41 | 6 | 85 | 8 | 93 | 12 | 105 | 0.74 |
| 12 | 141 | 0.4 | 4 | 119 | 7 | 126 | 15 | 141 | 1.00 |
| 13 | 116 | 0.38 | 1 | 43 | 3 | 46 | 10 | 56 | 0.48 |
| 14 | 106 | 0.41 | 1 | 78 | 10 | 88 | 17 | 105 | 0.99 |
| 15 | 100 | 0.42 | 3 | 55 | 7 | 62 | 10 | 72 | 0.72 |
| 16 | 93 | 0.44 | 3 | 44 | 9 | 53 | 16 | 69 | 0.74 |
| 17 | 84 | 0.45 | 6 | 80 | 11 | 91 | 13 | 104 | 1.24 |
| 18 | 82 | 0.39 | 1 | 43 | 2 | 45 | 5 | 50 | 0.61 |
| 19 | 77 | 0.47 | 13 | 66 | 8 | 74 | 16 | 90 | 1.17 |
| 20 | 63 | 0.44 | 1 | 44 | 3 | 47 | 12 | 59 | 0.94 |
| 21 | 45 | 0.41 | 0 | 21 | 0 | 21 | 6 | 27 | 0.60 |
| 22 | 48 | 0.48 | 1 | 25 | 6 | 31 | 6 | 37 | 0.77 |
| X | 152 | 0.39 | 4 | 61 | 8 | 69 | 15 | 84 | 0.55 |
| Y | 59 | 0.39 | 1d | 1 | 2 | 3 | 0 | 3 | 0.05 |
| Total | 3152 | 0.41 | 80 | 1912 | 178 | 2090 | 358 | 2448 | 0.78 |
Processed pseudogenes that are continuous in sequence with insertions ≤60 bp.
Processed pseudogenes that are disrupted by insertions (>60 bp).
Number of processed pseudogenes + pseudogenic fragments per 1 Mb DNA.
RSP4Y on chromosome Y is an isoform of RSP4X on chromosome X.
Correlation (chromosome size, number of processed pseudogenes) = 0.89, P < 1E-8.
Correlation (chromosome size, number of processed pseudogenes + fragments) = 0.89, P < 1E-8.
Table 2.
Overall Statistics of RP Processed Pseudogenes
| Ave. relative sequence lengtha | Ave. a.a.identityb | Ave. nt.identityc | Ave.disablementsd | Ave. 5′ truncatione | Ave. 3′ truncationf | |
|---|---|---|---|---|---|---|
| Processed pseudogenes | 96.5% | 76.2% | 86.8% | 3.6 | 13 | 8 |
| Pseudogenic fragments | 42.1% | 74.2% | 84.1% | 2.0 | 227 | 127 |
| Total | 88.5% | 75.9% | 84.8% | 3.4 | 44 | 26 |
Length of translated pseudogenes divided by the length of RP peptides, averaged over the entire pseudogene population.
Average sequence identity between translated pseudogene and RP peptide sequence.
Average sequence identity between pseudogene sequence and RP cDNA sequence.
Average number of premature stop codons, frame shifts, and repeat insertions in the processed pseudogenes.
Average number of missing nucleotides at 5′ end, CDS only.
Average number of missing nucleotides at 3′ end, CDS only.
358 Pseudogenic Fragments
We also found 358 pseudogenic fragments, which are continuous in sequence but produce transcripts shorter than 70% of a full-length RP peptide. On average these fragments match 40% of the full-length RPs with an average amino acid sequence identity of 74.2% (see Table 2). There are three possible explanations for these short fragments. (1) They could have originally been individual exons of duplicated RP genes. (2) They could have been intact processed pseudogenes and later became truncated by spontaneous DNA deletion or retrotransposon insertion. (3) They could have been caused by premature termination of the reverse transcription process, which would lead to incomplete incorporation of cDNA into the chromosome. Because the reverse-transcription starts at the 3′ end (poly-A tail), such premature truncation would tend to occur at the 5′ end of the cDNA sequence. The first scenario involves duplicated RP genes, and the last two scenarios assume a processed origin for the pseudogenic fragments. We believe the last two are more likely because there is evidence for both hypotheses. For most of these pseudogenic fragments, we could locate a retrotransposon within 300 bp on the chromosome with the average distance between the fragments and the retrotransposon being 108 bp. This close proximity strongly indicates retrotransposon insertion events in past evolution, which caused the RP pseudogene truncation. Also, the average truncation at the 5′ end for these fragments is almost twofold longer than at the 3′ end (227 vs. 127 bp), which is consistent with the mechanism of target-primed reverse transcription (Table 2). Based on these arguments, we counted these pseudogenic fragments as processed when we computed pseudogene density (see Table 1 footnote), but in general these fragments were treated separately from the full-length processed pseudogene population. As the total number of these fragments is much smaller than the number of processed pseudogenes (358 vs. 2090), exclusion of them from the processed pseudogene counts does not affect the conclusions one way or another.
Kenmochi and colleagues sequenced most of the 80 human RP genes and mapped them onto individual cytogenic bands (Kenmochi et al. 1998; Uechi et al. 2001; Yoshihama et al. 2002). In our present search for processed pseudogenes, 72 of these 80 RP genes were located and their cytogenic locations were confirmed. In addition, 16 duplicated copies of these RP genes were identified, mostly in the neighboring region of the original RP genes.
Overall Statistics of the Processed Pseudogenes
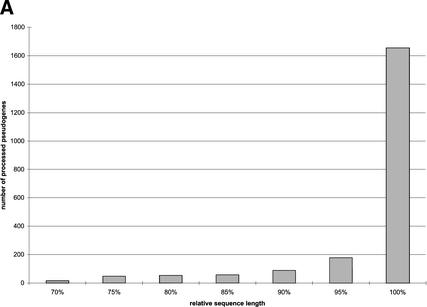
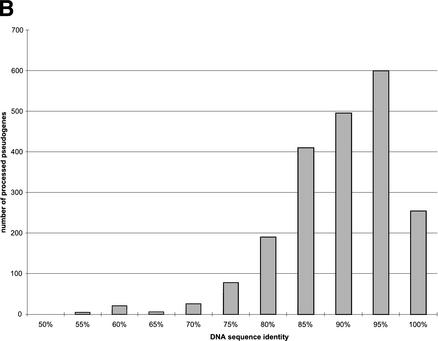
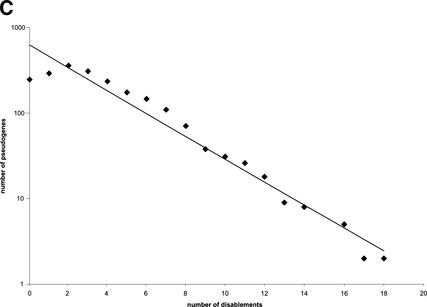
Because the ribosomal proteins are of various lengths, we measure sequence completeness by defining relative length as the ratio between the length of translated pseudogene and the length of the corresponding functional ribosomal proteins. In general, the RP pseudogenes are well preserved, as they tend to be almost full-length in their coding regions (96.5%), with high sequence identity in terms of both translated amino acid sequence (76.2%) and also underlying nucleotides (86.8%). Figure 1A illustrates the distribution of the relative sequence length of processed pseudogenes. Surprisingly, although we used 70% as a threshold to separate the processed pseudogenes from pseudogenic fragments, the CDSs of the majority of the processed pseudogenes (>90% of the set) are practically full-length. It is known that LINE1 reverse-transcriptase (RT) has a low efficiency that often leads to 5′ truncation and thus incomplete insertion of transcripts. It is a little surprising that we have observed such a high percentage of near-complete pseudogenes, but it is probably because RT truncations mostly occurred in the 5′ UTR instead of the protein-coding region. Figure 1B shows the distribution of DNA sequence identity between processed pseudogenes and the RP cDNA sequences. Figure 1C shows the distribution of number of disablements (premature stop codons and frame shifts) per pseudogene, with the y-axis plotted in log scale. Of the 1912 “intact” processed pseudogenes (Table 1), 258 (13%) do not contain any disablements; therefore they could potentially be mistaken as functional genes by some automatic gene prediction algorithms. The graph shows an exponential relationship. A similar exponential relationship was observed in a smaller set of human olfactory pseudogenes (∼600; Glusman et al. 2001), and was interpreted in such a way to support an alternative origin for olfactory receptor pseudogenes other than gene duplication or retrotransposition.
Figure 1.
RP processed pseudogenes statistics. (A) Distribution of relative sequence length among processed pseudogenes. Relative sequence length is the ratio between the length of translated pseudogene and the length of the corresponding functional ribosomal protein. (B) Distribution of the DNA sequence identity between processed pseudogenes and the cDNA sequence of functional RP proteins. (C) Distribution of number of disablements among processed pseudogenes.
We also checked the existence of a polyadenine tail for our processed pseudogene set. Of the 2090 processed pseudogenes, 952 (45.5%) have no obvious polyadenine tail of at least 30 bp detected (see Methods section), 176 (8%) have both a poly-A tail and a polyadenylation signal (mostly AATAAA) within 50 bp of the poly-A tail. Thirty-two pseudogenes (1.5%) have a poly-A tail and a polyadenylation signal 50–100 bp upstream; 903 pseudogenes (44.5%) only have a poly-A tail with no detectable polyadenylation signal. We are confident in our assignment of processed pseudogenes; lack of a poly-A tail for about half of the assigned processed pseudogenes can be explained as decay in genome sequence and nucleotide substitutions. Harrison et al. (2002a) found polyadenylation for only 52% of the processed pseudogenes on chromosomes 21 and 22, which is similar to the ratio we found here for RP pseudogenes.
Distribution of Pseudogenes Among Chromosomes
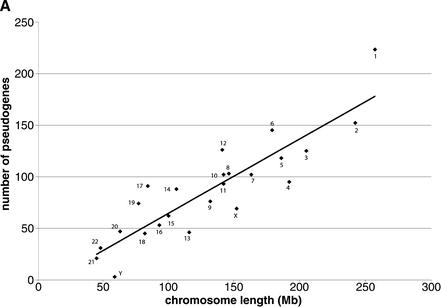
Unlike in prokaryotes, where the RP genes are organized into operons, the distribution of RP genes among human chromosomes is dispersed but not random (Feo et al. 1992; Kenmochi et al. 1998; Uechi et al. 2001; Yoshihama et al. 2002). Every human chromosome except chromosomes 7 and 21 contains at least one or more RP genes. Chromosome 19, one of the smallest chromosomes, contains as many as 13 RP genes (Table 1). Such high density of RP genes on chromosome 19 can be explained by the high chromosome GC content, which results in unusual high gene density (Mouchiroud et al. 1991; Lander et al. 2001; Venter et al. 2001). The distribution of processed RP pseudogenes in the human genome appears more random and uniform than their functional counterparts (Fig. 2). It is obvious that the abundance of processed pseudogenes on each chromosome is proportional to the chromosome length (Fig. 3A), with a correlation coefficient of 0.89 (P<1E-8). Including pseudogenic fragments in the set has no noticeable effect on this result.
Figure 2.
The human RP processed pseudogene population. Twenty-four human chromosomes are shown vertically from left to right. Pseudogenes are represented as short blue horizontal bars; long thick red horizontal bars delimit centromere region. Red dots represent chromosome ends.
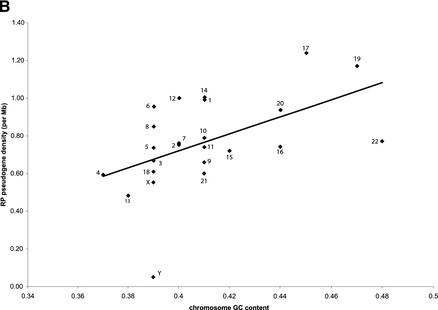
Figure 3.
(A) Correlation between chromosome length and number of processed RP pseudogenes on them. Each ♦ symbol represents a chromosome. The correlation between number of processed pseudogenes on each chromosome and chromosome length is 0.89, P<1E-8. (B) Processed pseudogene density on each chromosome is correlated with the chromosome GC content. The correlation coefficient is 0.51, P<0.01.
We further calculated the RP pseudogene density (number of pseudogenes per Mb) for each chromosome and plotted them against chromosomal GC content (Fig. 3B), which shows a weak positive correlation (correlation coefficient = 0.51, P<0.01). The outlier on the bottom of the graph is the sex chromosome Y, which has the lowest pseudogene density even for its relatively low GC content. Chromosome Y is unusual in many ways, as it also has the lowest density for Alu repeats (Lander et al. 2001); those authors suggested that these phenomena might be related to the high tolerance for DNA insertion and deletion and rapid gene turnover rate on this chromosome. If we weight the chromosome length by its GC content, then the correlation with the pseudogene density increases from 0.89 to 0.91 (P<1E-9). It is likely that the chromosomal GC content reflects the relative stability of the chromosome; that is, pseudogenes are more likely to be preserved on the chromosomes that have a slower gene turnover rate.
Genomic Distribution of Processed Pseudogenes
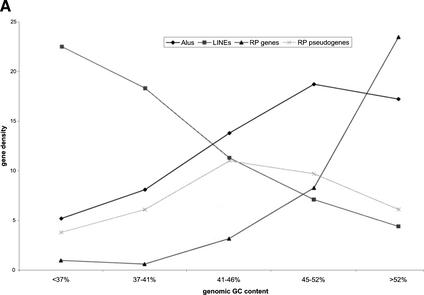
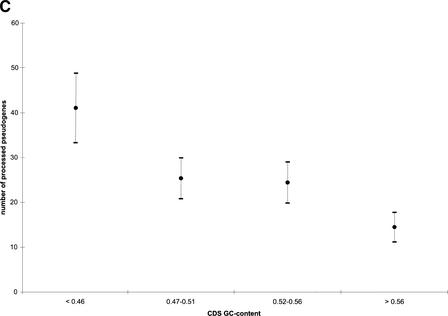
Using a 100-Kb-long nonoverlapping window, we divided the human genome into more than 30,000 segments and assigned them to five classes according to their average GC content. For each class, we also calculated the gene or pseudogene density by dividing the number of genes or pseudogenes by the amount of DNA in that class (Table 3). It is well established that in the human genome, gene density is strongly correlated with local GC content, with the GC-rich regions being mostly gene-dense (Mouchiroud et al. 1991; Lander et al. 2001; Venter et al. 2001). This is clearly the case for functional RP genes, as the GC-rich classes (>46%) contain the majority of the RP genes and have higher RP gene density. In contrast, the RP pseudogenes are enriched in classes with lower GC content; they have the highest density in the genomic region with intermediate GC content (41–46%). In fact, the class that has the highest local GC content (>52%) contains the fewest number of pseudogenes, although it has the highest RP gene density. Similar genomic distributions have been reported for chromosome 22 with a smaller set of 114 pseudogenes (Pavlicek et al. 2001). Our results suggest that this is probably a general rule for all processed pseudogenes in the human genome.
Table 3.
Genomic Distribution of RP Processed Pseudogenes
| Genomic GC-contenta | ||||||
|---|---|---|---|---|---|---|
| <37% | 37–41% | 41–46% | 46–52% | >52% | Total | |
| Number of functional | ||||||
| RP genesb | 8 | 6 | 23 | 27 | 22 | 86 |
| RP gene density | ||||||
| (per 100 Mb) | 0.98 | 0.61 | 3.2 | 8.3 | 23.4 | 2.73 |
| Number of RP | ||||||
| pseudogenes | 310 | 601 | 804 | 318 | 57 | 2090 |
| Pseudogene density | ||||||
| (per 10 Mb) | 3.8 | 6.1 | 11 | 9.7 | 6.1 | 6.63 |
Genomic regions grouped by their average GC content.
Including duplicated copies of the functional RP genes.
It has been proposed that the protein machinery encoded by the LINE1 element is involved in the arising of both the Alu repeats and LINE repeats (Feng et al. 1996; Jurka 1997; Weiner 1999) and the processed pseudogenes (Weiner 1999; Esnault et al. 2000). LINEs and Alus are the most frequent retrotransposons found in the human genome, each occupying about 15% and 10% of the genome respectively. LINEs (long interspersed elements) are about 6-kb long and encode two open reading frames (ORFs). Alus are a major class of SINEs (short interspersed elements), approximately 280 bp in length. Despite their common origin, the Alus in the human genome are predominantly found in GC-rich regions, whereas LINEs and processed pseudogenes are more prevalent in relatively GC-poor regions. In this sense, the distribution of Alus is more similar to that of genes than pseudogenes. In Figure 4A, we plotted the RP pseudogene density along with the densities of functional RP genes, Alus, and LINEs. [The data for Alus and LINEs are from the results of Pavlicek et al. (2001)]. It is obvious that both the functional RP genes and the Alus are enriched in the GC-rich regions and depleted in the GC-poor regions. LINEs are predominantly found in genomic regions with the lowest local GC content. The distribution of RP pseudogenes falls between these extremes, as they have the highest density in the regions with intermediate GC content (41%–46%).
Figure 4.
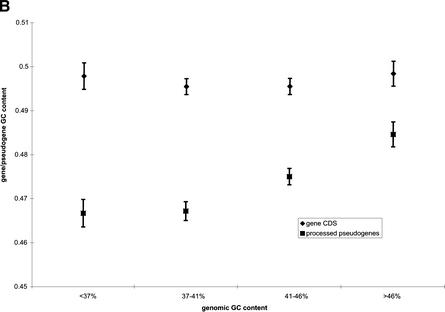
(A) Distribution of Alu elements, LINE elements, processed RP pseudogenes, and functional RP genes among genomic regions of different GC content. Because of their different abundance in genome, these four species are plotted on different scales: number per 10Kb for Alus and LINEs, number per Mb for RP pseudogenes, and number per 100 Mb for functional RP genes. (B) The drift in GC content for RP processed pseudogenes. (♦) The GC content of functional RP gene coding sequence (CDS). (▪) The GC content of processed pseudogenes. The vertical bars are standard errors.
Negative Selection Theory
The puzzling contrast between the genomic distribution of Alus and LINEs was recently explained by comparing the distribution of repeats of different age groups (Lander et al. 2001; Pavlicek et al. 2001). It has been observed that young Alus, similar to LINEs, were more frequently found in the GC-poor region compared to the more ancient Alu elements. Based on such findings, Pavlicek et al. (2001) proposed a negative selection theory, which hypothesized that the enrichment of Alus in the GC-rich region was the result of their higher stability in the compositionally matching environment. It is believed that when the retrotransposons were first integrated into the nuclear genome, both Alus and LINEs preferred a GC-poor (AT-rich) region because the LINE1 reverse-transcriptase/endonuclease specifically targets the TT|AAA insertion site. Because of the conspicuously higher GC content of Alus (∼57%), their existence in GC-poor regions would destabilize the chromosome. Therefore, these Alus would be selected against to be either lost or, perhaps more likely, their nucleotide composition would have drifted towards a lower GC level and decayed into background genomic DNA and become unrecognizable.
We believe that the aforementioned negative selection theory can also explain the pseudogene density distribution illustrated in Figure 4A. The GC content of RP CDS ranges from 42% to 63% with the median at 51%, which is not as high as Alus, but still much higher than the LINE repeats (∼42%) and the genome-wide average (∼41%). The average GC content for the RP pseudogene sequences is 47%, which is intermediate between those of the functional RP genes and genomic DNA. Therefore, at least for RP pseudogenes, we have observed the drift in their GC content, which supports the negative selection hypothesis. We further divided RP processed pseudogenes into four groups according to the average GC content in the 100-Kb genomic region surrounding each pseudogene. For each group, we calculated the average GC content for both the pseudogene sequences and also the CDS of the functional RP genes they originated from. The results are plotted in Figure 4B, which clearly shows a greater drift for pseudogenes in the GC-poor region than in the GC-rich region; therefore, the pseudogenes in GC-poor region appear more decayed than those in the GC-rich region. Such drift in nucleotide composition was previously reported for silent mutation sites in mammalian MHC gene sequences (Eyre-Walker 1999) and interspersed repeats in the human genome (Lander et al. 2001). In both studies, significantly more single nucleotide substitutions from G/C to A/T than from A/T to G/C have been observed. Despite the drift in composition, the majority of the processed RP pseudogenes still have GC content higher than their surrounding genomic sequences.
Age Distribution of Processed Pseudogenes
When mRNA transcripts were reverse-transcribed to become pseudogenes, they were immediately released from selection pressure. Therefore the amount of mutations they accumulated during evolution could be used to infer their ages. Because mammalian RP sequences have stayed almost unchanged since rodents and primates diverged over 100 millions of years (Myr) ago (99% sequence identity between rats and human), we can safely use the present-day human RP sequence as the ancient RP gene sequences to calculate the divergence rate for the processed pseudogenes. The percentage of sequence divergence was converted into approximate age in Myr by using a constant substitution rate of 1.5 × 10−9 per site per year (Li 1997). It is known that substitution rate varies during evolution (Goodman et al. 1998; Lander et al. 2001); however we believe that such simplified treatment is sufficient for our purpose.
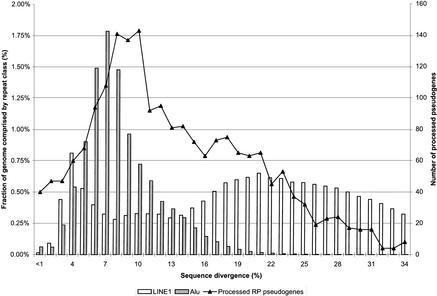
The age distribution of human repetitive sequences has been analyzed (Smit 1999; Lander et al. 2001). Figure 5 shows the distribution of sequence divergences for RP pseudogenes together with LINE1 and Alu repeats; each increment in divergence represents roughly 6.7 Myr. The repeats data are from Arian Smit (pers. comm.). It is obvious that processed pseudogenes have an age distribution much more similar to Alu elements than to LINE1 elements, although they were all processed by the same LINE1 machinery. Note that LINE1s are mammalian-specific and Alus are primate-specific. The distribution for RP pseudogenes peaks at an evolutionary age corresponding to 8%–10% sequence divergence, whereas Alus peak at 7% and LINE1 elements peak at both 4% and 21%. Interestingly, RP pseudogenes also have a shoulder at 17%–18%, which could have been the consequence of the surge of LINE1 retrotransposition activity just a few million years before that. The rate of new processed pseudogenes generated in the human genome has slowed down since ∼40 Myr ago, which was about the time when human species diverged from gibbons. This coincides with the decline of new LINE1 elements and Alus in the genome. It has been proposed that the structure and dynamics of hominid populations are responsible for such decline in retrotransposon activity (Lander et al. 2001).
Figure 5.
Distribution of sequence divergence for RP processed pseudogenes in comparison with Alu and LINE1 repeats. Pseudogenes and repeats were grouped into bins according to their sequence divergence from consensus sequences. Each increment in divergence represents roughly 6.6 million years (Myr). The LINE and Alu data are from A. Smit (pers. comm.).
GC-Poor RP Genes Have More Processed Pseudogenes
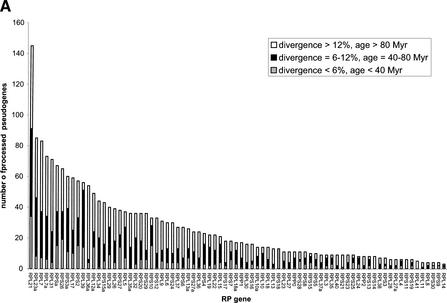
Table 4 lists the number of processed pseudogenes among 79 RPs, sorted in the descending order. The first two columns list the SWISSPROT ID (Bairoch and Apweiler 2000) for the human RPs, and the standard mammalian RP gene nomenclature (Mager et al. 1997). Also listed are the lengths of RP mRNA transcripts, coding sequence (CDS), and the CDS GC content, all retrieved from GenBank. On average, 26 processed pseudogenes are found for each RP gene; however, different RP genes have clearly very different propensities for generating processed pseudogenes. The distribution of numbers of processed pseudogenes among RP genes is strikingly skewed, although presumably for each RP only one functional gene exists (Wool et al. 1995). RPL21 has the most copies of processed pseudogenes at 145, which is about 50% more than that of RPL23A, which has the second-most at 85. Meanwhile, 24 RP genes have less than ten copies of processed pseudogenes each, and MRPL14 has the fewest at three. Regarding the RP genes that have the greatest numbers of processed pseudogenes, we also checked their chromosomal locations to make sure that they were not created from genomic duplication; that is, these processed pseudogenes arose mostly independently.
Table 4.
Distributions of Processed Pseudogenes Among RP Genes
| Gene namea | SWISSPROT IDb | mRNA lengthc | CDS lengthd | CDS GC contente | # Processed | # Fragments |
|---|---|---|---|---|---|---|
| RPL21 | RL21_HUMAN | 568 | 483 | 0.43 | 145 | 13 |
| RPL23A | RL2B_HUMAN | 546 | 471 | 0.49 | 85 | 11 |
| RPL7 | RL7_HUMAN | 838 | 747 | 0.43 | 83 | 19 |
| RPL7A | RL7A_HUMAN | 890 | 801 | 0.54 | 73 | 13 |
| RPL31 | RL31_HUMAN | 442 | 378 | 0.47 | 71 | 5 |
| RPSA | RSP4_HUMAN | 1039 | 888 | 0.53 | 67 | 12 |
| RPS26 | RS26_HUMAN | 459 | 348 | 0.53 | 65 | 5 |
| RPS3A | RS3A_HUMAN | 921 | 795 | 0.43 | 60 | 15 |
| RPL17 | RL17_HUMAN | 898 | 555 | 0.46 | 59 | 11 |
| RPS2 | RS2_HUMAN | 978 | 882 | 0.58 | 57 | 10 |
| RPL39 | RL39_HUMAN | 401 | 156 | 0.42 | 56 | 8 |
| RPL36A | RL44_HUMAN | 425 | 321 | 0.47 | 54 | 6 |
| RPL12A | RL12_HUMAN | 632 | 498 | 0.52 | 49 | 3 |
| RPL34 | RL34_HUMAN | 849 | 354 | 0.45 | 44 | 3 |
| RPS15A | RS1A_HUMAN | 541 | 393 | 0.47 | 43 | 4 |
| RPL29 | RL29_HUMAN | 737 | 480 | 0.56 | 40 | 3 |
| RPL26 | RL26_HUMAN | 525 | 438 | 0.45 | 39 | 3 |
| RPS27 | RS27_HUMAN | 344 | 255 | 0.49 | 38 | 0 |
| RPL5 | RL5_HUMAN | 1033 | 894 | 0.44 | 37 | 8 |
| RPS20 | RS20_HUMAN | 539 | 360 | 0.45 | 36 | 0 |
| RPL35A | R35A_HUMAN | 511 | 333 | 0.47 | 36 | 2 |
| RPL32 | RL32_HUMAN | 521 | 408 | 0.51 | 36 | 7 |
| RPS29 | RS29_HUMAN | 346 | 171 | 0.53 | 36 | 2 |
| RPS12 | RS12_HUMAN | 534 | 399 | 0.46 | 33 | 8 |
| RPS10 | RS10_HUMAN | 598 | 498 | 0.55 | 33 | 4 |
| RPL9 | RL9_HUMAN | 716 | 579 | 0.44 | 31 | 6 |
| RPS24 | RS24_HUMAN | 537 | 402 | 0.44 | 30 | 13 |
| RPL6 | RL6_HUMAN | 950 | 867 | 0.47 | 30 | 5 |
| RPS6 | RS6_HUMAN | 829 | 750 | 0.47 | 27 | 1 |
| RPL37 | RL37_HUMAN | 371 | 294 | 0.51 | 27 | 3 |
| RPL13A | R13A_HUMAN | 1142 | 612 | 0.58 | 26 | 3 |
| RPS27A | R27A_HUMAN | 551 | 471 | 0.44 | 24 | 1 |
| RPL36 | RL36_HUMAN | 428 | 318 | 0.6 | 24 | 2 |
| RPS4 | RS4_HUMAN | 916 | 792 | 0.48 | 23 | 5 |
| RPL22 | RL22_HUMAN | 574 | 387 | 0.44 | 22 | 1 |
| RPL19 | RL19_HUMAN | 698 | 591 | 0.54 | 22 | 4 |
| RPL15 | RL15_HUMAN | 2018 | 615 | 0.54 | 21 | 5 |
| RPS7 | RS7_HUMAN | 729 | 585 | 0.48 | 18 | 5 |
| RPS17 | RS17_HUMAN | 515 | 408 | 0.51 | 18 | 3 |
| RPP1 | RLA1_HUMAN | 512 | 345 | 0.54 | 17 | 3 |
| RPL18A | RL1X_HUMAN | 618 | 531 | 0.6 | 17 | 7 |
| RPL30 | RL30_HUMAN | 524 | 348 | 0.45 | 16 | 1 |
| RPS16 | RS16_HUMAN | 570 | 441 | 0.57 | 16 | 1 |
| RPL10A | R10A_HUMAN | 700 | 654 | 0.51 | 14 | 4 |
| RPL10 | RL10_HUMAN | 2188 | 645 | 0.55 | 14 | 32 |
| RPL18 | RL18_HUMAN | 648 | 567 | 0.59 | 14 | 1 |
| RPS18 | RS18_HUMAN | 549 | 459 | 0.56 | 13 | 3 |
| RPL13 | RL13_HUMAN | 1110 | 636 | 0.62 | 13 | 1 |
| RPL23 | RL23_HUMAN | 493 | 423 | 0.49 | 11 | 1 |
| RPL27 | RL27_HUMAN | 513 | 411 | 0.49 | 11 | 4 |
| RPS8 | RS8_HUMAN | 705 | 627 | 0.52 | 11 | 3 |
| RPP0 | RLA0_HUMAN | 1116 | 954 | 0.54 | 11 | 4 |
| RPS28 | RS28_HUMAN | 398 | 210 | 0.62 | 11 | 2 |
| RPS5 | RS5_HUMAN | 725 | 615 | 0.58 | 10 | 1 |
| RPS15 | RS15_HUMAN | 515 | 438 | 0.63 | 10 | 1 |
| RPS23 | RS23_HUMAN | 506 | 432 | 0.45 | 9 | 1 |
| RPS25 | RS25_HUMAN | 514 | 378 | 0.47 | 9 | 0 |
| RPL37A | R37A_HUMAN | 392 | 279 | 0.52 | 9 | 1 |
| RPL40f | RL40_HUMAN | 501 | 387 | 0.54 | 9 | 1 |
| RPS21 | RS21_HUMAN | 356 | 252 | 0.54 | 9 | 0 |
| RPL3 | RL3_HUMAN | 1311 | 1212 | 0.55 | 9 | 5 |
| RPL35 | RL35_HUMAN | 455 | 372 | 0.57 | 9 | 2 |
| RPS13 | RS13_HUMAN | 529 | 456 | 0.46 | 8 | 4 |
| RPL24 | RL24_HUMAN | 556 | 474 | 0.48 | 8 | 9 |
| RPS14 | RS14_HUMAN | 589 | 456 | 0.54 | 8 | 2 |
| RPP2 | RLA2_HUMAN | 482 | 348 | 0.56 | 8 | 0 |
| RPL38 | RL38_HUMAN | 368 | 213 | 0.46 | 7 | 4 |
| RPS3 | RS3_HUMAN | 843 | 732 | 0.54 | 7 | 3 |
| RPL4 | RL4_HUMAN | 1449 | 1284 | 0.49 | 6 | 5 |
| RPS11 | RS11_HUMAN | 594 | 477 | 0.51 | 6 | 3 |
| RPL27Af | RL2A_HUMAN | 514 | 447 | 0.54 | 6 | 5 |
| RPS19 | RS19_HUMAN | 569 | 438 | 0.58 | 6 | 0 |
| RPL28 | RL28_HUMAN | 500 | 414 | 0.6 | 6 | 1 |
| RPL41 | RL41_HUMAN | 478 | 78 | 0.5 | 5 | 0 |
| RPL11 | RL11_HUMAN | 609 | 537 | 0.51 | 4 | 3 |
| RPS30f | RS30_HUMAN | 574 | 402 | 0.59 | 4 | 1 |
| RPS9 | RS9_HUMAN | 691 | 585 | 0.6 | 4 | 1 |
| RPL8 | RL8_HUMAN | 894 | 774 | 0.61 | 4 | 3 |
| RPL14 | RL14_HUMAN | 843 | 651 | 0.5 | 3 | 3 |
| Total | 685 | 511 | 0.51 | 26 | 4.5 | |
| Correlation | −0.01g | 0.04h | −0.41i | |||
| p-value | 0.93 | 0.73 | 0.0002 |
The SWISSPROT ID (Bairoch and Apweiler 2000) for the human RP protein.
The standard mammalian RP gene nomenclature.
Number of nucleotides for RP mRNA.
Number of nucleotides for RP coding sequence (CDS).
GC content of the RP coding sequence.
RPS27a, RPL40, and RPS30 are carboxyl extensions of ubiquitin or ubiquitin-like proteins.
Correlation between number of processed pseudogenes and mRNA length.
Correlation between number of processed pseudogenes and CDS length.
Correlation between number of processed pseudogenes and CDS GC-content.
We were curious as to whether the differing processed pseudogene abundance among RP genes is correlated with the recent decline in retrotransposition activity. We further divided the processed pseudogenes originated from the same RP gene into three groups according to their ages: <40 Myr, 40–80 Myr, and >80 Myr (Fig. 6A). It is obvious that the age distribution of processed pseudogenes is similar for all 79 RP genes, that is, there were no preferences for a certain group of RP genes in different evolution periods. The correlation between the number of young pseudogenes (<40 Myr) and number of mid-age pseudogenes (40–80 Myr) per RP gene is 0.73 (P<1E-13); the correlation between mid-age pseudogenes and old pseudogenes (>80 Myr) is 0.68 (P<1E-11).
Figure 6.
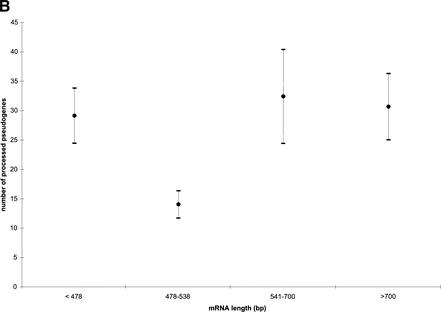
(A) Distribution of processed pseudogenes among RP genes. Bars of different shades represent different age groups. (B) Lack of correlation between mRNA transcript length and number of processed pseudogenes. The pseudogenes are grouped into bins according to the length of their mRNA transcripts. Vertical bars are standard errors. (C) Significant inverse correlation between GC content of RP gene coding sequence (CDS) and number of processed pseudogenes for that RP. The RP genes are grouped into four bins according to their CDS GC content.
It is also plausible that the differences in pseudogene abundance merely reflect the different ages for individual RP genes, as presumably genes that have been around longer will have more chance being reverse-transcribed to generate pseudogenes. To check this, we grouped RP genes into three groups according to their phylogenetic profile, that is, some RP genes are unique to eukaryotes while others have homologs in eubacterial and archaebacterial kingdoms (Wool et al. 1995). There appears to be no correlation between processed pseudogene abundance and the degree of ubiquity. Within eukaryotes, we also looked at the sequence identity between yeast RPs and human RPs; no correlation was found there as well. The pseudogene abundance also has no correlation with the extra-ribosomal function of some of the RP genes (Wool 1996).
Goncalves et al. (2000) analyzed 249 processed pseudogenes, which correspond to 181 functional genes, and concluded that human genes that gave rise to processed pseudogenes in general share four features. They are (1) widely expressed, especially in germ line, (2) highly conserved, (3) short, and (4) GC-poor. The first two criteria are trivial for ribosomal proteins, as RPs are ubiquitous in all cell types, and they are also the most highly conserved among eukaryotes and mammals (Wool et al. 1995). In general, RP genes have short mRNAs and short CDS as seen in Table 4, although there is no significant correlation between the number of processed pseudogenes and the mRNA length (correlation −0.01, P<0.93) (Fig. 6B) or the CDS length (correlation 0.04, P<0.73). We would like to emphasize the lack of obvious correlation between gene length and pseudogene abundance, as it demonstrates that our pseudogene searching procedure did not systematically miss out short pseudogenes; that is, the skewed pseudogene distribution is not an artifact. However, there is a significant inverse correlation between the number of processed pseudogenes and the GC-content of RP gene CDS (correlation −0.41, P<0.0002) as shown in Figure 6C; that is, relatively GC-poorer RP genes tend to have more processed pseudogenes than GC-richer ones. It is not immediately obvious what is the mechanism behind the enrichment for the relatively GC-poor RP genes, since the arising of a processed pseudogene involves multiple steps and the selection for GC-poor RP genes could have occurred at any step along the way. More on this topic will be discussed in the Discussion section.
Nonprocessed Pseudogenes and Duplicated RP Genes
We found only 16 duplicated RP genes in the human genome (Table 5), which share identical exon structure with previously characterized RP genes (Kenmochi et al. 1998; Uechi et al. 2001). This is in sharp contrast to the yeast genome, where most RP genes are duplicated and the duplicated genes are also transcribed and functional. Only one duplicated gene in the human genome (RPL13A) has an obvious disablement in the coding region; it is possible that other duplicated RP genes may have hard-to-detect disablements in the UTR regions or introns. It is not clear whether these duplicated RP genes are transcribed in the cell, although it is generally assumed that only one gene is functional for each ribosomal protein (Wool et al. 1995; Kenmochi et al. 1998). The majority of the duplicated genes are in the vicinity of the original genes, and therefore could not have been resolved from the original genes in the hybridization experiments. There are notable exceptions: RPL26, RPS27, and RPL3 have duplicated copies on separate chromosomes, and RPS4Y has a duplicated copy on the opposite end of chromosome Y. Interestingly, the duplicated copies for RPL26, RPS27, and RPL3 genes have much longer introns than the mapped genes, which were caused by insertion of Alu or LINE repeats (with the exception of RPS27). It is likely that the sequence difference in intron region is the reason that they were missed out in the hybridization experiments, even though they are far apart from the mapped RP genes. Detailed analysis of these duplicated genes will be described in subsequent reports.
Table 5.
Duplicated Human RP Genes
| Gene | Duplicated | Originala | ||||
|---|---|---|---|---|---|---|
| Chr. location | Size (bp) | Ensembl ID | Chr. location | Size (bp) | Ensembl ID | |
| RPS3A | 4q31.23 | 4114 | ENSG00000145425 | 4q31.23 | 4114 | ENSG00000151940 |
| RPL26 | 5q35.3 | 9664 | ENSG00000037241 | 17p13.1 | 4743 | ENSG00000161970 |
| RPL8 (I)b | 8q24.3 | 2287 | ENSG00000130795 | 8q24.3 | 2287 | ENSG00000147785 |
| RPL8 (II) | 8q24.3 | 2287 | ENSG00000161009 | 8q24.3 | 2287 | ENSG00000147785 |
| RPL7A | 9q34.3 | 1814 | ENSG00000160312 | 9q34.3 | 1814 | ENSG00000148303 |
| RLP0 (I)c | 12q24.31 | 3474 | N/Ad | 12q24.23 | 3474 | N/A |
| RLP0 (II)e | 12q24.31 | 2182 | ENSG00000123062 | 12q24.23 | 3474 | N/A |
| RPS27 | 15q21.3 | 912 | N/A | 1q21.3 | 573 | ENSG00000157616 |
| RPS17f | 15q26.3 | 685 | ENSG00000154241 | 15q25.2 | 3329 | ENSG00000103720 |
| RPL3 | 16p13.3 | 8650 | ENSG00000140986 | 22q13.2 | 5402 | ENSG00000100316 |
| RPS15A | 16p13.11 | 6100 | ENSG00000157115 | 16p13.11 | 6100 | ENSG00000134419 |
| RPL17 | 18q21.1 | 2221 | ENSG00000141618 | 18q21.1 | 2220 | ENSG00000154807 |
| RPL13Ag | 19q13.13 | 1016 | N/A | 19q13.33 | 1966 | ENSG00000142541 |
| RPS9 | 19q13.42 | 6385 | ENSG00000131036 | 19q13.42 | 6385 | ENSG00000074164 |
| RPL36 | 19p13.3 | 1108 | ENSG00000130255 | 19p13.2 | 1108 | ENSG00000141995 |
| RPS4Y | Yq11.222 | 24252 | ENSG00000157828 | Yp11.31 | 24727 | ENSG00000129824 |
RP genes identified from hybridation experiments.
b,cTwo copies of duplications are found for the same RP gene.
Not present in Ensembl. (Jan. 2002 release).
This duplicated RP gene starts from amino acid 17.
This duplicated RP gene only contains amino acids 50–138.
This duplicated RP gene has a frame shift.
Our homology matching procedure located at least one intron-containing functional gene for all but eight RP genes: RPP2, RPL4, RPL30, RPL35A, RPL38, RPL41, RPS7, and RPS27A. We did, however, find processed pseudogenes for these RP genes in the genome. These genes either consist of short exons or their protein sequences are predominantly low-complexity, making them difficult to find by homology matching.
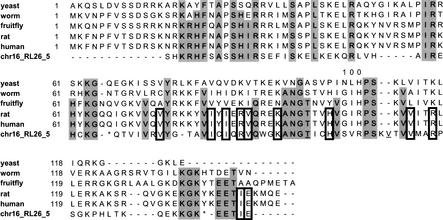
It was surprising to discover a processed RPL26 pseudogene in the intron region of the functional RPS2 gene on chromosome 16 (band p13.3, Contig AC005363.1.1.75108, Ensembl ID ENSG00000140988). RPS2 gene has seven exons; the pseudogene resides in the third intron (1015 bp long), between residues 89 and 90 in the RPS2 protein sequence. Interestingly, there is also an Alu element at the 3′ end of the pseudogene, about 100 bp away. The pseudogene itself is 357 bp long, corresponding to residues 14 to 141 of RPL26, having amino acid sequence identity of 49% and nucleotide sequence identity of 73% (Fig. 7). It appears to be very ancient, has already lost its poly-A tail, and has sequence divergence of 0.28, which corresponds to more than 100 Myr old. Figure 7 shows the alignment of RPL26 sequences from several eukaryotic organisms together with this pseudogene. At 11 positions, the pseudogene has the same residue with the mammalian sequences but not with the invertebrates. Note that rat and human sequences are almost identical except at residue 100, where rat has an arginine and human has a histidine. Interestingly, this RPL26 pseudogene also has a Histidine at that position; this suggests that the pseudogene became part of the intron before the divergence of rodent and hominid species. It has been known that some RP genes contain Alu or LINE elements in the 3′ or 5′ UTR; to our knowledge this is the first case where a processed pseudogene is found in the intron region of another functional gene. This has implications for the origin and evolution of introns.
Figure 7.
Amino acid sequence alignment of RPL26 genes from yeast, worm, fruit fly, rat, and human, and a processed pseudogene (chr16_RL26_5) found in the intron region of the human functional RPS2 gene. The residues highlighted in gray are those present in the pseudogene and also in both the mammalian and invertebrate proteins; the residues outlined in bold are those present in the pseudogene and the mammals but not in invertebrates. In the pseudogene sequence, * represents a stop codon, and an underscored amino acid indicates an adjacent frame shift. Rat and human RPL26 have almost identical sequences except at position 100, where the rat protein and the pseudogene have an Arginine and human protein has a Histidine.
Online Database
The data and results discussed in this report can be accessed online at http://www.pseudogene.org/ or http://bioinfo.mbb.yale.edu/genome/pseudogene/.
DISCUSSION
Significance of RP Pseudogenes
Characterizing ribosomal protein pseudogenes is valuable in many ways. (1) It will be tremendously useful in the study of functional RP genes. RP genes are implicated in many human genetic diseases such as Diamond-Blackfan anemia (Draptchinskaia et al. 1999), Noonan syndrome (Kenmochi et al. 2000), and Turner‘s syndrome (Zinn et al. 1993). The precise nucleotide sequence and chromosomal location of RP pseudogenes will certainly help researchers in designing probes specific to functional genes. (2) Pseudogenes can also serve as genomic milestones, as they provide snapshots of RP sequences existing millions of years back in evolution. Such information will be valuable in studying ribosome biogenesis and the phylogenetic relationships between organisms. The discovery of an RPL26 pseudogene in the intron region of a functional RPS2 gene could certainly shed light on the evolution of both RP genes. (3) From the perspective of studying retrotransposition, processed pseudogenes are just a special type of repetitive elements like Alus. However, processed pseudogenes are much more diverse in terms of sequence length, GC content, and other features than traditional retrotransposons, which makes them useful in studying evolution and dynamics of genomes. To our knowledge, our RP pseudogenes are the largest set ever studied.
Comparing With Ensembl Annotations
The Ensembl database (http://www.ensembl.org/) is an automated system for genome-wide gene prediction and annotation, which has direct links to primary HGP data sources (Birney et al. 2001; Hubbard et al. 2002). The annotation process relies on matching genomic DNA sequence and GenScan peptides (Burge and Karlin 1997) with known proteins, mRNAs, and other sequence information. All of the genes were checked to be transcribed before they were included into the database (Daniel Barker, pers. comm.). As of the end of February 2002, there were approximately 47,000 annotated genes in Ensembl, of which 549 were annotated as ribosomal protein genes. Some of these have more detailed annotations associating them with a particular RP such as “60S RIBOSOMAL PROTEIN L7”, and others were described more loosely such as “60S RIBOSOMAL PROTEIN”. After re-aligning these genes with human RP protein sequences and removing some dubious matches, we derived a set of 481 Ensembl RP entries.
Ensembl does not explicitly differentiate between functional genes and pseudogenes, nor does it aim to (D. Barker, pers. comm.). Consequently, most of these 481 Ensembl RP entries turned out to be pseudogenes instead of functional genes, as only 260 (54%) translate to peptides longer than 95% of full-length ribosomal proteins. For instance, a gene ENSG00000150624 on chromosome 2 was annotated as “60S RIBOSOMAL PROTEIN L17”, but produced a transcript that was only 51.6% of the full-length RPL17, and had sequence identity of 56.2%. Moreover, only 170 of these genes have introns; most of these Ensembl RP genes (64.6%) are single exons. We checked the overlap between our RP pseudogene sets with these Ensembl RP entries: 474 of 481 (98.5%) Ensembl RP entries have significant overlaps with our pseudogenes, and in most cases our pseudogenes were longer than the Ensembl entries. Five RPL41 single-exon processed pseudogenes from Ensembl were the only ones missed by our procedure. The RPL41 is the shortest ribosomal protein, with only 25 amino acids; it also contains 17 near-consecutive Arginine and Lysine residues. It is likely that short length and low complexity caused BLAST to fail to detect these pseudogenes. Note that Ensembl is a database in flux, that is, the sequence and annotation are continuously updated and improved. Therefore some of the examples and statistics given above will probably be out of date when this report is published. Nonetheless, the overlap in annotation of genes and pseudogenes documented above is important as it demonstrates the need to systematically include pseudogene identification in genome annotation efforts.
Automatic gene prediction programs alone do not have the ability to differentiate between functional genes and pseudogenes, especially if the pseudogenes do not contain obvious disablements in the coding sequence (CDS). Furthermore, for those pseudogenes that contain disablements, gene prediction programs either discard them or stop at the disablement and predict the pseudogene as a functional gene but with truncated length. We think this is the reason that so many RP pseudogenes were passed into the Ensembl database as functional genes. The number of genes in the human genome has long been a matter of debate, as different methods such as EST analysis and GenScan (Burge and Karlin 1997) gave different estimates (Harrison et al. 2002b). It is probably not appropriate to extrapolate the overestimation for RP genes onto the whole human proteome, as ribosomal proteins are a very unique protein family in many ways. Nevertheless, special care should be taken in interpreting outputs from automatic gene prediction programs.
Pseudogene Abundance per RP Cannot Be Explained by Positive Selection
As mentioned previously, we found an inverse correlation between RP gene GC content and the pseudogene abundance for that gene (Fig. 6C); that is, the relatively GC-poor RP genes tend to have more processed pseudogenes. Before we further discuss the possible mechanism behind this correlation, it would be well to give a brief overview of the LINE1-mediated retrotransposition process, which is believed to be responsible for generating processed pseudogenes (Kazazian and Moran 1998). LINE1-mediated retrotransposition can be divided into four steps. (1) First, a retrotransposon or gene is transcribed in the nucleus to produce an mRNA transcript. (2) Second, the mRNA transcripts are transported into cytoplasm, and LINE1 mRNA transcripts are translated into two proteins: ORF1 (also known as p40), and ORF2, which is a reverse-transcriptase/endonuclease. (3) Human ORF1 has been demonstrated to be a sequence-specific single-strand RNA binding protein, which binds specifically but not exclusively to LINE1 transcript to form a ribonucleoprotein particle (RNP) which also includes ORF2 protein (Leibold et al. 1990; Martin 1991; Hohjoh and Singer 1996, 1997b; Moran et al. 1996; Kazazian and Moran 1998). (4) Lastly, the RNP particle migrates into the nucleus and undergoes target-primed reverse-transcription, which give rise to a new retrotransposon or processed pseudogene.
If the GC-poor RP genes were selected favorably in retrotransposition (i.e., there is a positive selection for them), it must have occurred in one of the four steps described above. However, we cannot find any evidence for such positive selection in any of the steps. In relation to step 1, we have compared the processed pseudogene abundance per gene with the mRNA expression level in human and yeast cells (see Methods). No significant correlation between the datasets was found, suggesting that the selection could not have occurred at the step of gene transcription. In relation to step 2, the lack of correlation between mRNA length and pseudogene abundance also suggested that the transportation of RP transcript in and out of the nucleus had no effect on retrotransposition. This is based on the idea that longer mRNAs are harder to transport. In relation to step 3, the forming of RNP particle, it has been demonstrated that the binding between ORF1 and mRNA transcript has a cis-preference; that is, ORF1 has higher affinity to wild-type LINE1 transcripts that encode it. However at a much lower level, ORF1 or ORF1 and ORF2 together can also act in trans to retrotranspose mutant LINEs and other mRNA transcripts (Hohjoh and Singer 1997a,b; Esnault et al. 2000; Wei et al. 2001). It is not clear what sequence or structural features on the mRNA transcripts constitute the cis and trans preference, though it is unlikely that the overall GC content is the deciding factor, because Alu elements and LINE elements, the two most populous retrotransposons in human genome, have very different GC content (56.8% for Alus and 42.3% for LINEs). Following the same reasoning, it is also unlikely that the reverse transcription in the fourth step has a preference for GC-poor transcripts.
Negative Selection for GC-Poor RP Genes in Retrotransposition
In the above analysis we found no evidence of a positive selection mechanism in retrotransposition of GC-poor RP genes; however, a negative selection mechanism can readily explain the skewed distribution. In this mechanism, the accumulation of GC-poor RP pseudogenes can be interpreted as the indirect result of a faster decay rate for GC-rich RP pseudogenes in the GC-poor genome region where they were originally inserted.
Analogous to the mechanism of enrichment of Alu elements in the GC-rich region, which we described earlier in this report, the existence of GC-rich RP pseudogenes in the GC-poor genomic region was more unfavorable than GC-poor RP pseudogenes. Thus there would be greater selection pressure against these GC-rich pseudogenes. Pavlicek et al. (2001) divided Alu and LINE elements into different age groups and studied their distribution in genome regions of different GC content. They showed that the young Alus (divergence <2% from consensus sequence) are indeed less depleted in the GC-poor region. This effect is not evident for older Alus (sequence divergence >4%). We did a similar age segmentation analysis on RP pseudogenes, with the results shown in Table 6. (The numbers in the table were not normalized by amount of DNA.) We found different results for young pseudogenes than described above for young Alus. For young pseudogenes, there is no indication of enrichment in the GC-poor region (where “young” here is defined as sequence divergence less than 2% from their parents, the same cutoff as used in the study of the Alus). Note, however, that there is a slight enrichment for the youngest pseudogenes, which have sequence divergence less than 1%, corresponding to roughly 6.7 Myr old. We think that the reason we did not observe the same behavior for young pseudogenes as for young Alus is because of the much smaller sample size for pseudogenes. In addition, the recent decline in retrotransposition activity in the human genome (Fig. 5; Lander et al. 2001) could have further complicated the situation, as fewer fresh pseudogenes were generated in the human genome.
Table 6.
Genomic Distributions of RP Pseudogenes of Different Ages
| Sequence divergence | Number of pseudogenes in the genomic regiona | ||||
|---|---|---|---|---|---|
| <37% | 37–41% | 41–45% | 45–52% | >52% | |
| ≤1% | 4 | 12 | 12 | 8 | 2 |
| ≤2% | 13 | 26 | 31 | 11 | 2 |
| >2% | 303 | 575 | 768 | 306 | 55 |
Genomic regions of 100 Kb long are binned by their average GC-content.
In conclusion, the precise mechanism behind the negative correlation between gene GC content and processed pseudogene abundance remains unsettled until more pseudogene sequences from other protein families are available. As of this writing, based on the analysis of Alu elements and the elimination of positive selection mechanisms for RP pseudogenes, the negative selection mechanism appears attractive.
METHODS
Six-Frame BLAST Search for Raw Fragment Homologies
Figure 8A is a flow chart describing our basic procedure for finding RP pseudogenes. We used the August 6, 2001 freeze of the human genome draft, downloaded from the Ensembl Web site (http://www.ensembl.org). Subsequently, all of the chromosomal coordinates were based on these sequences. The amino acid sequences of the 79 ribosomal proteins were extracted from SWISSPROT (Bairoch and Apweiler 2000). Because the sequence identity between the two RPS4 isoforms (RS4_HUMAN and RS4Y_HUMAN) is very high (91%), only protein RS4_HUMAN was used in the BLAST search. Each human chromosome was split into smaller overlapping chunks of 5.1 million bp, and the tblastn program of the BLAST package 2.0 (Altschul et al. 1997) was run on these sequences. The genome sequence was not repeat-masked (A. Smit and P. Green, unpubl.) because we were concerned that some of the RP pseudogenes may reside in repetitive regions. Default SEG (Wootton and Federhen 1993) low-complexity filter parameters (12 2.2 2.5) were used in the homology search. We then picked the significant homology matches (e-value <1E-4), and reduced them for mutual overlap by selecting the matches in decreasing order of significance and removing any matches that overlap substantially with a picked match (i.e., more than ten amino acids or 30 base pairs).
Merging Adjacent Fragment Homologies Into Single RP Matches
After sorting the BLAST matches according to their starting coordinates on the chromosomes, we found many neighboring matches on the same chromosome that match the same RP. Some of these adjacent matches obviously were separate genes or pseudogenes, whereas others appeared to be part of the same gene or pseudogene. A two-step procedure was developed to determine (1) whether the neighboring matches belong to the same gene structure and (2) whether they should be merged together into a longer homology match.
Step (1): Consider two adjacent homology fragments, M1 and M2, which are on the same chromosomal strand and match the same RP (Fig. 8B). M1 has chromosomal coordinates (c11, c12) and matches amino acid sequence (q11, q12) on the query RP protein. Similarly, M2 has chromosomal coordinates (c21, c22) and matches amino acids (q21, q22) on the query protein. By convention, q21 is always greater than q11 and c21 is always greater than c12. If M1 and M2 satisfy the following two criteria, then we decide they belong to the same gene structure; that is, they are either two exons of the same gene or two fragments of the same pseudogene interrupted by insertions.
(1) | q21−q12 | ≤max (20, 0.2xL) and (2) c21−c12 ≤5000 (L denotes the length of the query RP peptide sequence). The reasoning behind criterion (1) is that if the two homology fragments have too much overlap or have too long a gap between them on the query protein sequence, then they should be considered two separate and independent matches. Criterion (2) sets the maximum length of insertions in the middle of a pseudogene. We checked that the introns in the RP genes are all shorter than 5000 bp, so we would not have accidentally split a gene into two.
Step (2): If two homology fragments are determined to be part of the same gene or pseudogene structure in step (1), then in step (2) the fragments were merged only if the chromosomal distance between the matches was shorter than 60 bp; that is, c22−c21 ≤60. The rationale behind such treatment was that if the gap between the matches were too long, then merging them together would generate errors in the Smith-Waterman realignment procedure described below. In addition, it has been shown that more than 95% of the introns in human are longer than 60 bp (Lander et al. 2001), and thus we would not have accidentally merged two exons together or included introns into the coding sequence.
Optimization From Smith-Waterman Alignment of Merged Matches
After merging, each match was extended on both sides to equal the length of the RP they matched, plus a buffer of 30 bp. For each extended match, the corresponding SWISSPROT protein sequences were then realigned to the genomic DNA sequence following the Smith-Waterman algorithm (Smith and Waterman 1981) by using the program FASTA (Pearson 1997). The reason for such an extension procedure is that BLAST may have skipped low-complexity segments in the query RP sequence; also, BLAST does not recognize frame shifts. After the realignment, the matches are “cleaned up”: any redundant matches were removed, and matches that contain gaps longer than 60 bp were split up into two individual matches. Because sequence alignment programs sometimes tend to pick up some extra residues at the ends of the alignment, each alignment was filtered to remove dubious matches at the ends. At this step, we had a total of 2531 pseudogene candidates in the whole genome that matched the human RPs. Most of these were potential pseudogenes, but there could also be real functional RP genes in this set, because we did not exclude any matches based on disablement.
Deriving a Set of RP Genes From the Ensembl Database
We wanted to compare our pseudogene sets with the RP genes from the Ensembl database (http://www.ensembl.org; Birney et al. 2001; Hubbard et al. 2002). As of the end of February 2002, there were approximately 47,000 confirmed genes, each with an annotated function. (Details regarding the Ensembl annotation procedure can be found in the aforementioned references.) We searched the Ensembl database and picked out 549 genes that have been annotated as ribosomal proteins. We then reannotated these genes by aligning them pairwise with human RP protein sequences, and picked out those Ensembl genes that had FASTA e-values lower than 0.0001. After removing a few remaining mitochondrial ribosomal protein genes, we had a set of 481 Ensembl nuclear RP genes.
In our examination of these Ensembl RP entries, it became obvious that most of these were pseudogenes other than real functional RP genes, because they do not contain introns. We found that 474 (98.5%) of the 481 Ensembl RP genes have significant overlaps with our pseudogene sets. Five single-exon RPL41 pseudogenes from Ensembl were added to our pseudogene sets.
Assessing for Processing by Checking for Exon Structures
We divided our pseudogene population into two subsets based on whether they contained long gaps in the middle of the sequence (Fig. 8A). We labeled those pseudogenes as “processed” if they met two criteria: (1) they contained gaps of shorter than 60 bp, that is, c21−c12 ≤60 in Figure 8B, and (2) they produced transcripts longer than 70% of the ribosomal protein they matched. Venter et al. (2001) also used the last criterion. We also checked in GenBank that all 79 ribosomal protein genes contain introns longer than 60 bp. The remaining single-exon pseudogenes, which are shorter than 70% of the full-length protein, were labeled “fragments”. A total of 1912 “intact” processed pseudogenes and 358 pseudogenic fragments were identified at this step.
For those pseudogene candidates that contained multiple segments separated by gaps longer than 60 bp (total of 266), it was not straightforward to determine whether they were of processed or nonprocessed origin because the gaps could be either introns or repeat insertions. It is also likely that there were real functional ribosomal protein genes in this group. The cytogenetic locations of the 80 human RP genes (including the isoform gene RPS4Y on chromosome Y) were previously mapped (Kenmochi et al. 1998; Uechi et al. 2001; Yoshihama et al. 2002). Using the cytogenetic map as reference and comparing the position of the gaps in the sequence with the exon structure of the functional RP genes, we identified 72 functional RP genes and 16 duplicated genes, and assigned the remaining 178 as “disrupted” processed pseudogenes. In summary, at the end of this process we had 2090 processed pseudogenes, 358 pseudogenic fragments, 72 functional RP genes, and 16 duplicated RP genes.
Further Verification of Processing by Poly-A Signal
When processed pseudogenes were integrated into genome from mRNA, a polyadenine tail at the 3′ end would also be included (Vanin 1985; Mighell et al. 2000). This polyadenine tail is at least 15–20 nucleotides long and is preceded by a polyadenylation signal (mostly AATAAA; Wool et al. 1995). We were interested to survey how many of the ribosomal pseudogenes still had the polyadenine tail. Following the procedure described by Harrison et al. (2001), we searched a 1000-bp region that was 3′ to the pseudogene homology segment, with a sliding window of 50 nucleotides for a region of elevated polyadenine content (>30 bp), and picked the most adenine-rich 50-bp segment as the most likely candidate. An interval of 1000 nucleotides was used because of the possible existence of 3′-untranslated regions (3′-UTRs); 90% of 3′-UTRs are of length less than 942 bp (Makalowski et al. 1996). In addition, we searched in the same 1000-bp region for candidate AATAAA or other polyadenylation signals and checked whether they were upstream of the candidate polyadenine tail site.
Dating Processed Pseudogenes
Processed pseudogene sequences are aligned together with the corresponding functional RP gene sequences using program ClustalW (Thompson et al. 1994). For each pseudogene, we calculated sequence divergence from the present-day RP gene with the program MEGA2 (Kumar et al. 2001), using the Kimura two-parameter model and pairwise deletion. Kimura’s two-parameter model (Kimura 1980) corrects for transitional and transversional substitution rates while assuming that the four nucleotide frequencies are the same and rates of substitution do not vary among sites. Evolutionary ages were calculated by the formula T = D/k, where D is the corrected divergence rate and k is the mutation rate per year per site for nonfunctional sequences. A mutation rate of 1.5 × 10–9 per site per year (Li 1997) was used.
Calculating Pseudogene Density In Different GC Regions
Each human chromosome was divided into consecutive 100K bp-long, nonoverlapping segments. The GC content for each segment was calculated and the segment was assigned to one of the five groups according to their GC content: <37%, 37%–41%, 41%–46%, 46%–52% and >52%. The number of processed pseudogenes in each group was counted, and the pseudogene density for each group was calculated. Note that we used the same GC content that was used for isochore classification (Macaya et al. 1976; Bernardi 2000), although the validity of the isochore definition has been under debate (Bernardi 2001; Lander et al. 2001).
Expression Analysis
To investigate the possible correlation between the pseudogene abundance and the mRNA expression level, we compared the number of processed pseudogenes for each functional RP gene with its cellular mRNA expression level in the human cell (Yuval Kluger, pers. comm.) and the yeast cell (Cho et al. 1998). No significant correlation was found. Ribosomal protein genes are the most highly expressed genes in the cell; it is likely, in this case, that the overabundance of mRNA transcripts has made the expression level a nondeciding factor for RP pseudogene retrotransposition.
WEB SITE REFERENCES
http://www.pseudogene.org/; Pseudogene database.
http://bioinfo.mbb.yale.edu/genome/pseudogene; Pseudogene database.
http://www.ensembl.org/; Ensembl database.
Acknowledgments
We thank Adam Pavlicek for carefully reading the manuscript and Arian Smit for providing the data on Alu/LINE sequence divergence. Mark Gerstein acknowledges NIH CEGS grant (P50 HG02357-01) for financial support. Zhaolei Zhang acknowledges Ted Johnson for doing the BLAST runs and thanks Paul Bertone, Ronald Jansen, Nick Luscombe, Yuval Kluger, and Jiang Qian for helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Mark.Gerstein@yale.edu; FAX (360) 838-7861.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.331902.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz T. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Nature. 2000;400:841–847. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. doi: 10.1016/s0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- ————— Misunderstandings about isochores. Part 1. Gene. 2001;276:3–13. doi: 10.1016/s0378-1119(01)00644-8. [DOI] [PubMed] [Google Scholar]
- Birney E, Bateman A, Clamp ME, Hubbard TJ. Mining the draft human genome. Nature. 2001;409:827–828. doi: 10.1038/35057004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Crollius HR, Jaillon O, Bernot A, Dasilva C, Bouneau L, Fisher C, Fizames C, Wincker P, Brottier P, Quetier F. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat Genet. 2000;25:235–238. doi: 10.1038/76118. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Analysis of expressed sequence tags indicates 35,000 human genes. Nat Genet. 2000;232:232–233. doi: 10.1038/76115. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. Evidence of selection on silent site base composition in mammals: Potential implications for the evolution of isochores and junk DNA. Genetics. 1999;152:675–683. doi: 10.1093/genetics/152.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, LKazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Feo S, Davies B, Fried M. The mapping of seven intron-containing ribosomal protein genes shows they are unlinked in the human genome. Genomics. 1992;13:201–207. doi: 10.1016/0888-7543(92)90221-d. [DOI] [PubMed] [Google Scholar]
- Fujii GH, Morimoto AM, Berson AE, Bolen JB. Transcriptional analysis of the PTEN/MMAC1 pseudogene, psiPTEN. Oncogene. 1999;18:1765–1769. doi: 10.1038/sj.onc.1202492. [DOI] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- Goncalves I, Duret L, Mouchiroud D. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000;10:672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Harrison PM, Hegyi H, Balasubramanian S, Luscombe NM, Bertone P, Echols N, Johnson T, Gerstein M. Molecular fossils in the human genome: Identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 2002a;12:272–280. doi: 10.1101/gr.207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Kumar A, Lang N, Snyder M, Gerstein M. A question of size: The eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 2002b;30:1083–1090. doi: 10.1093/nar/30.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Hegyi H, Bertone P, Echols N, Johnson T, Balasubramanian S, Luscombe N, Gerstein M. Molecular fossils in the human genome: Identification and analysis of processed and non-processed pseudogenes in chromosomes 21 and 22. Genome Res. 2001;12:272–280. doi: 10.1101/gr.207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- ————— Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J Mol Biol. 1997a;271:7–12. doi: 10.1006/jmbi.1997.1159. [DOI] [PubMed] [Google Scholar]
- ————— Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997b;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Camero G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- Kenmochi N, Kawaguchi T, Rozen S, Davis E, Goodman N, Hudson TJ, Tanaka T, Page DC. A map of 75 human ribosomal protein genes. Genome Res. 1998;8:509–523. doi: 10.1101/gr.8.5.509. [DOI] [PubMed] [Google Scholar]
- Kenmochi N, Yoshihama M, Higa S, Tanaka T. The human ribosomal protein L6 gene in a critical region for Noonan syndrome. J Human Genet. 2000;45:290–293. doi: 10.1007/s100380070018. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Leibold DM, Swergold GD, Singer MF, Thayer RE, Dombroski BA, Fanning TG. Translation of LINE-1 DNA elements in vitro and in human cells. Proc Natl Acad Sci. 1990;87:6990–6994. doi: 10.1073/pnas.87.18.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. Molecular Evolution. Sunderland, MA: Sinauer Associates, Inc.; 1997. [Google Scholar]
- Macaya G, Thiery JP, Bernardi G. An approach to the organization of eukaryotic genomes at a macromolecular level. J Mol Biol. 1976;108:237–254. doi: 10.1016/s0022-2836(76)80105-2. [DOI] [PubMed] [Google Scholar]
- Mager WH, Planta RJ, Ballesta JG, Lee JC, Mizuta K, Suzuki K, Warner JR, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski W, Zhang J, Boguski MS. Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res. 1996;6:846–857. doi: 10.1101/gr.6.9.846. [DOI] [PubMed] [Google Scholar]
- Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR, Kumari M, Aivaliotis MJ, Wang Z, Zhang P, Marshall F, Vandeberg JL. Analysis of the cDNA and encoded protein of the human testis-specific PGK-2 gene. Dev Genet. 1996;19:321–332. doi: 10.1002/(SICI)1520-6408(1996)19:4<321::AID-DVG5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Mouchiroud D, D'Onofrio G, Aissani B, Macaya G, Gautier C, Bernardi G. The distribution of genes in the human genome. Gene. 1991;100:181–187. doi: 10.1016/0378-1119(91)90364-h. [DOI] [PubMed] [Google Scholar]
- Olsen MA, Schechter LE. Cloning, mRNA localization and evolutionary conservation of a human 5-HT7 receptor pseudogene. Gene. 1999;227:63–69. doi: 10.1016/s0378-1119(98)00588-5. [DOI] [PubMed] [Google Scholar]
- Pavlicek A, Jabbari K, Paces J, Paces V, Hejnar J, Bernardi G. Similar integration but different stability of Alus and LINEs in the human genome. Gene. 2001;276:39–45. doi: 10.1016/s0378-1119(01)00645-x. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Raue HA, Planta RJ. Ribosome biogenesis in yeast. Prog Nucleic Acid Res Mol Biol. 1991;41:89–129. doi: 10.1016/s0079-6603(08)60007-0. [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uechi T, Tanaka T, Kenmochi N. A complete map of the human ribosomal protein genes: Assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics. 2001;72:223–230. doi: 10.1006/geno.2000.6470. [DOI] [PubMed] [Google Scholar]
- Vanin EF. Processed pseudogenes: Characteristics and evolution. Annu Rev Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Jr, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM. Do all SINEs lead to LINEs? Curr Biol. 1999;9:842–844. [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:323–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wool IG. Extraribosomal functions of ribosomal proteins. TIBS. 1996;21:164–165. [PubMed] [Google Scholar]
- Wool IG, Chan YL, Gluck A. Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol. 1995;73:933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Comput Chem. 1993;17:149–163. [Google Scholar]
- Yeh RF, Lim LP, Burge C. Computational inference of homologous gene structures in the human genome. Genome Res. 2001;11:803–816. doi: 10.1101/gr.175701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama M, Uechi T, Asakawa S, Kawasaki K, Kato S, Higa S, Maeda N, Minoshima S, Tanaka T, Shimizu N, et al. The human ribosomal protein genes: Sequencing and comparative analysis of 73 genes. Genome Res. 2002;12:379–390. doi: 10.1101/gr.214202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:868–869. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zinn AR, Page DC, Fisher EM. Turner syndrome: The case of the missing sex chromosome. Trends Genet. 1993;9:90–93. doi: 10.1016/0168-9525(93)90230-f. [DOI] [PubMed] [Google Scholar]