Abstract
To identify candidate genes encoding QTLs in baboons, we have developed a novel strategy that integrates comparative mapping, bioinformatics, and expression arrays. A genome-wide scan, performed previously on pedigreed baboons to localize QTLs for phenotypes that are known risk factors for atherosclerosis, revealed a QTL on chromosome 18q that influences high-density lipoprotein cholesterol (HDL-C) phenotypes. After ruling out the only two biologically relevant positional candidate genes in this chromosomal region, we combined information from baboon pedigrees and HDL-C phenotypes with a baboon microsatellite marker map, human microsatellite marker maps, and human genome maps to develop a chromosomal region expression array (CREA). The CREA was screened with heterologous liver cDNA from sib-pairs of contrasting HDL-C phenotypes on two different diets, and genes were prioritized for further study by expression profiles. Analysis of gene expression in this restricted chromosomal region, combined with HDL-C phenotypic information, yielded a list of candidate genes for the QTL regulating HDL-C in baboons. Our data demonstrate the power of this strategy for identifying candidate genes encoding QTLs for multigenic traits. This strategy is applicable to many species that serve as models for human diseases and can even be used with human subjects.
Our laboratory is using baboons as an animal model to search for genes controlling serum profiles of lipoproteins to better understand cholesterol metabolism and its relationship to atherogenesis. Genetic effects on serum cholesterol concentrations in humans have been demonstrated in many studies (for review, see Fruchart et al. 1998). Previous studies in baboons fed diets enriched in fat and cholesterol have also demonstrated genetic effects on serum lipoprotein concentrations, suggesting common genetic pathways in regulation of lipid metabolism (Mott et al. 1978; Flow et al. 1984; Kammerer et al. 1984; McGill et al. 1987).
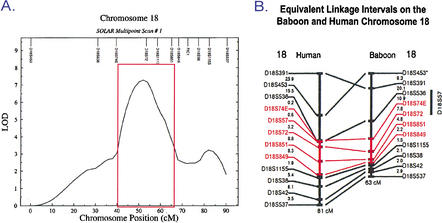
A baboon genetic linkage map including 331 random microsatellite markers that were typed for 694 pedigreed baboons (Rogers et al. 2000) was used to perform a genome scan for HDL-C. Strong evidence of a QTL regulating HDL1-C (a size fraction of HDL-C) (Cheng et al. 1988) was detected on baboon chromosome 18. Two-point linkage analysis showed a peak LOD score at D18S72 for the QTL of 7.32 (Mahaney et al. 1998). A multipoint genome scan upon which the published data (Mahaney et al. 1998) were based is shown in Figure 1A (M.C. Mahaney, L.A. Cox, D.L. Rainwater, J. Blangero, C.M. Kammerer, J. Rogers, and J.L. VandeBerg, unpubl.). An additional 270 baboons in the pedigree were genotyped for chromosome 18 markers, and the chromosomal interval was fine mapped by adding 12 markers to the region. Two of the 12 markers were in genic DNA sequences of known candidate genes in the homologous human chromosomal region. These efforts confirmed the previous findings showing strong evidence for linkage between serum levels of HDL1-C and a region of chromosome 18q (L.A. Cox, M.C. Mahaney, C.M. Kammerer, D.L. Rainwater, J. Rogers, and J.L. VandeBerg, unpubl.).
Figure 1.
(A) Chromosome 18 map based on SOLAR multipoint scan of HDL phenotypes and microsatellite marker genotypes from 694 pedigreed baboons. Region of interest is indicated by the red box. Work published in Mahaney et al. (1998) was based on this scan (M.C. Mahaney, L.A. Cox, D.L. Rainwater, J. Blangero, C.M. Kammerer, J. Rogers, and J.L. VandeBerg, unpubl.) (B) Alignment of baboon chromosome 18 and human chromosome 18 by microsatellite markers (Rogers et al. 2000). The region of interest is shown in red.
Only two biological candidate genes, Familial Intrahepatic Cholestasis-1 (FIC1) and endothelial lipase (LIPG), are located near the homologous human chromosomal region. Both are located outside of the 95% confidence interval for the peak LOD score, and neither locus showed evidence for variation that contributed to variation in serum HDL1-C (L.A. Cox, M.C. Mahaney, C.M. Kammerer, D.L. Rainwater, J. Rogers, and J.L. VandeBerg, unpubl.). Therefore, both were ruled out as the gene encoding the QTL.
Consequently, we have begun to positionally clone the gene of interest. The commonly used methods for positionally cloning novel genes are very labor and time intensive. In addition, these methods are complicated when localizing and identifying genes regulating multigenic traits. As we continue to integrate the baboon phenotypic data with the baboon genome map, we will identify numerous QTLs associated with multigenic phenotypes in our baboons. To identify all of these novel genes in a reasonable amount of time, we have developed an efficient strategy to identify candidate genes for all identified baboon genome QTLs.
The Chromosomal Region Expression Array (CREA) strategy is based on the fact that the gene regulating the QTL must be encoded within the region of the QTL signal. In developing our strategy, we needed to ensure that we would evaluate any DNA sequences that could encode the gene. Therefore, we did not limit our approach to analysis of known genes, ESTs (expressed sequence tags), and predicted genes. We created an array that was all inclusive for all genes that might lie within the QTL region of interest by assembling a contig of BAC clones for the region of interest. Because human and baboon are highly conserved in this chromosomal region, we used human BACs and basic bioinformatics tools to assemble our contig. To interrogate the arrays, we used heterologous liver cDNA from sibling baboons of contrasting HDL1-C phenotypes. From the CREAs, we have identified 138 BAC clones containing expressed genes. In addition, we categorized expressed genes as similarly expressed and differentially expressed with the 53 differentially expressed genes consistent with the phenotype given top priority. We have cloned and sequenced four of the differentially expressed genes and found that they are all novel genes. We are currently characterizing the remaining differentially expressed genes. The data presented here demonstrate the power of combining CREA as target with tissue-specific, heterologous cDNA from animals with significantly different phenotypes as probe to identify candidate genes encoding a QTL.
RESULTS
Defining the Chromosomal Region of Interest
To isolate the gene encoding the QTL, we chose to use a conservative approach and include all DNA in the chromosomal region with multipoint LOD scores of 3.0 or greater (Fig. 1A). In addition, to reduce the likelihood that we would miss the gene in the chromosomal region of interest, we analyzed all available DNA using a contig of BAC clones.
Comparison of the baboon linkage map (Rogers et al. 2000) for this region of baboon chromosome 18 with the homologous region of the human chromosome 18 linkage map (Fig. 1B) shows that the chromosomal region is conserved between human and baboon. In addition, we used human BAC clones containing microsatellite markers mapped to the region of interest to isolate baboon BAC clones from a library on high-density filters. We confirmed that each baboon BAC clone contained the same microsatellite marker as the corresponding human BAC clone by PCR. In addition, we end-sequenced the baboon BAC clones and compared these sequences with the human genome database. In all cases, the baboon BAC end-sequences aligned within the corresponding human BAC clone or aligned within a human BAC clone overlapping the microsatellite-containing clone. Sequence similarity for these regions of DNA ranged from 92% to 96% (data not shown).
Optimizing Baboon to Human DNA Array Conditions
To demonstrate that human and baboon DNA sequences were similar enough for hybridizations in which specific signals were greater than background noise, we conducted a number of dot blot experiments. In the first experiment, we spotted a series of dilutions (100 ng to 100 pg) of human and baboon BAC clones onto nylon membranes. The human and baboon BAC clones contained LIPG sequences. We probed the dot blots with (1) human endothelial lipase (LIPG) gene fragment, (2) baboon LIPG gene fragment, and (3) human genomic fragment from the human BAC clone containing LIPG. As negative controls, we included human and baboon BAC clones that did not contain LIPG sequences. In all cases, we detected specific signals with only 100 pg of target DNA and very low signals (background) with the negative controls (data not shown).
Another component of these experiments was to define the amount of target DNA necessary to spot onto the array. We needed to ensure that the amount of target DNA was not the limiting factor in detecting differential gene expression, that is, the target must be in excess, so that all of the cDNA probe for that gene will hybridize. Because each BAC clone contains a high percentage of noncoding sequences, we could not use cDNA or oligonucleotide array target DNA amounts. Using the same dot blots as before, we hybridized increasing amounts of cDNA probes. We found that 50 ng of BAC DNA in a 4-mm diameter spot was excess target for highly expressed genes. To be conservative, we chose to use 100 ng of BAC DNA target for the 4-mm diameter spots. For our high density CREAs, we spotted 1-mm diameter spots with 12.5 ng of BAC DNA. From these experiments, we defined optimum conditions for interrogating human BAC arrays with baboon cDNA probes (data not shown).
The CREA Strategy
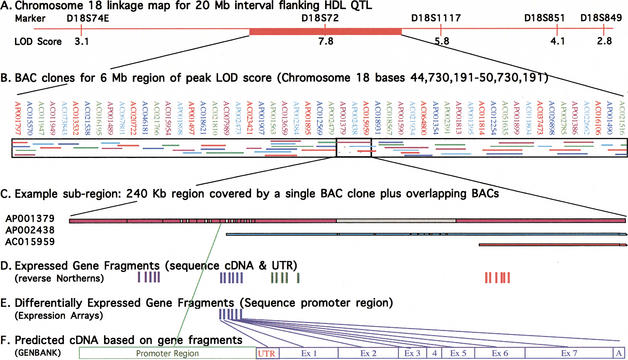
After demonstrating the feasibility of baboon to human cross-species hybridization, we implemented a strategy for discovery of genes encoding QTLs using the chromosomal region expression array. Figure 2 illustrates how genes that are expressed in baboon liver samples and are encoded in human BAC clones are identified, and how the gene fragments are isolated and used to identify the full-length cDNA. The example presented here represents one region within the indicated linkage interval. To produce a complete chromosomal region expression array, the method was applied to the entire 20-Mb chromosomal interval. As mentioned previously, human and baboon linkage map comparisons (Fig. 1B; Rogers et al. 2000) and human and baboon BAC sequence comparisons (L.A. Cox, S. Birnbaum, and J.L. VandeBerg, unpubl.) show that this region of chromosome 18 is highly conserved between human and baboon. Thus, for the QTL on chromosome 18 regulating HDL1-C levels, we can use human sequence data as a first step to design the expression arrays. The 20-Mb interval of human genome sequence flanking the position of the equivalent microsatellite marker that gave the highest LOD score in baboons for the QTL regulating HDL1-C levels is represented at the top of this figure (Fig. 2A). For this figure, we selected one 6-Mb portion of the indicated interval to exemplify representative coverage with human BAC clones (Fig. 2B). We used the Draft Human Genome Browser (Kent et al. 2001; Lander et al. 2001) to align BAC clones within this selected region. This alignment was confirmed using NCBI electronic PCR. As shown in Figure 2B, BAC coverage in this region of human chromosome 18 is very nearly complete and many regions contain overlapping BAC clones. After construction of the array with BAC clones that redundantly cover the region of interest, we interrogated the BAC array with heterologous cDNA from liver, the tissue likely responsible for the phenotype. BAC clones containing expressed genes were identified from the array (Fig. 2C). The clones were restriction digested, subcloned, size fractionated, and subjected to reverse Northern Blot analyses to identify gene fragments that were expressed (Fig. 2D,) and of those, genes that were differentially expressed (Fig. 2E). Sequences of gene fragments were analyzed in silico to identify known or predicted cDNAs (Fig. 2F). In future work, promoters of differentially expressed candidate genes will be sequenced in baboon for polymorphisms, and cDNAs and UTRs of similarly expressed candidate genes will be sequenced for polymorphisms.
Figure 2.
Experimental design for prioritization of genes within a linkage interval. (A) Chromosome 18 linkage map of the 20-Mb interval in the human genome flanking the equivalent position of the indicated HDL1-C QTL in the baboon genome. The top labels show the microsatellite markers, and the bottom labels indicate the multipoint LOD score for each equivalent marker in the baboon genome rounded to the nearest integer. (B) Human BAC clones covering a 6-Mb portion of the 20-Mb region of interest. Designations of the clones are shown on the top line, and coverage by each clone is shown within the boxed area. Association between clone position and designation is indicated by colors of clone names corresponding to colors of clone fragments. (C) A sample 240-Kb subregion (47,885,905–48,125,905) covered by a single BAC clone plus additional overlapping clones. Colored regions indicate complete DNA sequence data; gray regions indicate draft sequence data. Only the 5′ portions of BAC clones AP002438 and AC015959 are shown (i.e., the portions that overlap AP001379). (D) Expressed gene fragments for this region are denoted by solid vertical lines as determined by reverse Northern blot analysis. Fragments are grouped into predicted cDNAs based on GenBank homologies. Expressed genes showing no differential expression will be sequenced in cDNA and untranslated regions (UTRs) for the panel of sib-pairs. (E) For expressed gene fragments showing differential expression between animals of contrasting phenotypes, baboon BAC clones will be isolated using the gene fragments. The BAC clone promoter will be sequenced and the sequence data used to sequence promoters for the panel of sib-pairs. (F) cDNA and promoter region structures will be predicted based on sequence data comparison with GenBank database homologies.
Selection of Animals on the Basis of Quantitative Trait and Genotypic Data
Baboons were chosen for this study on the basis of phenotypic and genotypic analysis of the pedigreed baboon population. We used pairs of sibling baboons with contrasting phenotypes for the trait of interest; the selected sib-pairs differed by at least one standard deviation for the quantitative trait value (Table 1). The sib-pairs chosen for this study of the chromosome 18 QTL regulating HDL1-C levels have serum HDL1-C measurements on the HCHF diet ranging from 42 to 122 mg/dL for the high responders and from 8 to 31 mg/dL for the low responders. In addition to being selected for contrasting phenotypes, members of each sib-pair were selected to share no IBD alleles, or for some markers, to share only one IBD allele, in the chromosomal region of interest. The use of these selected sib-pairs with contrasting phenotypes and contrasting genotypes provides a powerful tool because the contrasting sibs have common genetic backgrounds but differ at the locus responsible for the trait.
Table 1.
Serum HDL1-C Measures of Contrasting Sib-pairs Used in Chromosome 18 CREAs
Values are given in mg/dl.
One low HDL1 responder baboon is a sibling of three different high-responder HDL1 baboons.
Prioritization of Expressed Genes
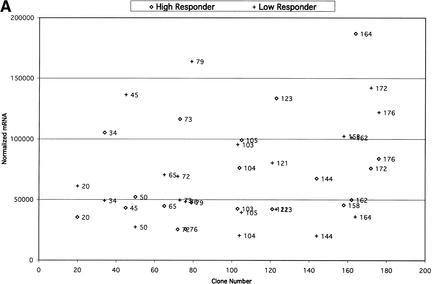
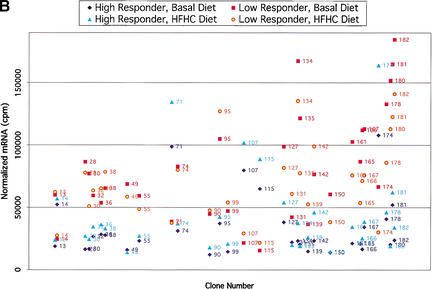
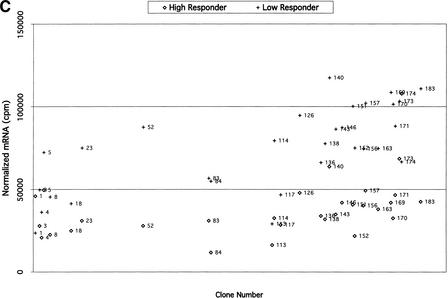
For comparison of expression in BAC clones, samples from low responders on basal diet were grouped together, samples from low responders on challenge diet were grouped together, samples from high responders on basal diet were grouped together, and samples from high responders on challenge diet were grouped together. For each clone in each group, average mRNA values (on the basis of the normalized duplicate values for each individual) and standard deviations were calculated. Clones were considered differentially expressed between groups on the basis of the average mRNA values and standard deviations (Fig. 3).
Figure 3.
(A) Comparisons of expression patterns in differentially expressed BAC clones for high and low HDL1-C responders on challenge diet. (B) Comparisons of BAC clones that show similar expression patterns on either diet for high and low HDL1-C responders. (C) Comparisons of expression patterns in BAC clones for high and low HDL1-C responders on basal diet. Eight baboons were screened for gene expression from liver biopsies sampled while the animals were on the basal diet and while the animals were on the challenge diet. Each data point represents an average of duplicate measures for all animals in that phenotypic group for that BAC clone. BAC clone numbers represent the series number assigned to that BAC clone in the contig. Clone number 1 is the most p-ter clone in the contig and clone 183 is the most q-ter clone in the contig. Clones showing expression differences greater than ± one standard deviation were considered differentially expressed. Clones that were not expressed above levels of the negative controls and clones that did not contain differentially expressed genes are not shown.
To prioritize differentially expressed and similarly expressed genes, clones were divided into four categories. (1) Differentially expressed genes, differing between sib-pairs in response to the challenge diet, consistent with the sib-pair phenotypes. High HDL1-C levels are observed only when animals are on the challenge diet; therefore, differences in gene expression between contrasting groups on the challenge diet are of highest priority; (2) Differentially expressed genes, differing between sib-pairs in response to both diets, consistent within the sib-pair phenotypes. It is possible that the gene responsible for the phenotype is constitively expressed, but the phenotype is manifested only with the challenge diet. Genes in this group are our second priority. (3) Similarly expressed genes; genes in this group show no significant differences in expression between groups and no significant differences comparing diets within each group. It is possible that the gene responsible for the phenotype varies in translational regulation or protein structure. Genes in this group are our third priority. (4) Differentially expressed genes, differing between sib-pairs by response to the basal diet, inconsistent within the sib-pair phenotypes. As mentioned previously, the phenotype is manifest on the challenge diet with little phenotypic difference between groups on the basal diet. Therefore, genes with differential expression in basal diet samples are inconsistent with the phenotype and are of low priority. The first and second categories of genes were given primary consideration for analysis and characterization. In the absence of viable candidates in the first two categories, we will analyze genes in the third category, which could include genes that differ by translational activity or structure. Because the gene responsible for the phenotype should have similar expression profiles across sib-pairs, top-priority candidates are genes that exhibit expression and sequence differences between sib-pair members and do so in several sib-pairs.
Using these criteria, we prioritized BAC clones for further analysis. We found 46 BAC clones that did not contain any expressed genes and 138 BAC clones that contained expressed genes. Of these, 83 BAC clones contained differentially expressed genes. Although the percentage of differentially expressed clones may seem high, this result is based on four different comparisons as follows: (1) low responder on basal versus challenge diet, (2) high responder on basal versus challenge diet, (3) low responder versus high responder on basal diet, and (4) low responder versus high responder on challenge diet. With 4 categories of comparisons, we are evaluating 138 clones times 4, or 552 events. Thus, we observed 83 differences in 552 events (15%).
In the group of 83 clones containing differentially expressed genes, 53 clones showed expression patterns consistent with the HDL1-C phenotype (groups 1 and 2; Fig. 3A,B). Of the 53 BAC clones with differentially expressed genes, only 3 of these were due to expression differences by diet within a phenotypic group. Two of these clones overlap with the clones that showed differential expression between high HDL responders versus low HDL responders on either the challenge diet or the basal diet, but not both. The 53 clones in groups 1 and 2 were placed in the high-priority group. Thirty clones showed differential expression patterns inconsistent with the HDL1-C phenotype (group 4; Fig. 3C) and were given low priority. The 53 group 1 and group 2 clones were analyzed further using reverse Northern blotting methods.
Expression Profile Analysis of Expressed Gene Fragments
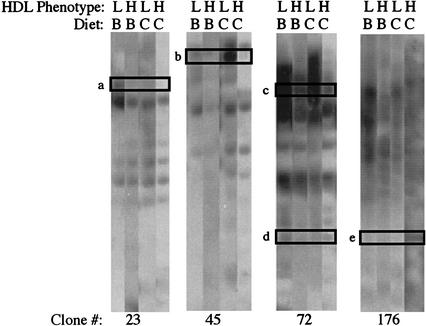
To identify gene fragments within the 53 BAC clones showing differential expression, we performed reverse Northern blots screening replicate blots with heterologous liver cDNA from one pair of sibling baboons for each diet. The results for 4 of the 15 clones subjected to this procedure are shown in Figure 4. For each clone, data from both animals of a sib-pair on each diet are shown. Data values for each band, normalized against vector sequence, are shown in Table 2. From these data, we found at least one gene fragment with differential expression for each BAC clone. Each differentially expressed gene profile in a sib-pair on two diets was consistent with the corresponding BAC array profile.
Figure 4.
BAC Reverse Northern blot of BAC clones containing differentially expressed genes. BAC DNA was digested with PstI, the fragments were subcloned into pBluescript SK+, linearized with NotI, and agarose gel size-fractionated in four replicate gels. After transfer to charged nylon membranes, each membrane was hybridized with heterologous liver cDNA from one baboon in the sib-pair on a specified diet. The HDL phenotype L indicates the low HDL responder and the H indicates the high HDL responder of the sib-pair. The diet is denoted by B for basal and C for challenge. The BAC clone number in the contig is indicated on the bottom line of the figure. DNA fragments pursued in subsequent expression array and sequencing experiments are boxed and assigned a letter (a, b, c, d, and e).
Table 2.
Quantification of Reverse Northern Blot Gene Fragments
| Band | Clone | Low responder baseline | High responder baseline | Low responder HCHF | High responder HCHF |
|---|---|---|---|---|---|
| a | 23 | 465,000 | 168,000 | 333,000 | 60,600 |
| b | 45 | 270,000 | 379,000 | 507,000 | 90,100 |
| c | 72 | 644,000 | 568,000 | 508,000 | 247,000 |
| d | 72 | 120,000 | 104,000 | 10,600 | 32,500 |
| e | 176 | 45,500 | 63,400 | 49,200 | 95,200 |
Values were normalized against vector sequence and are given in cpm.
The results reveal several patterns that support the feasibility of this strategy to screen for candidate genes. First, it is apparent that a number of genes are differentially expressed. The results of box a in Figure 4 illustrate that for this sib-pair, the low expressor (L) exhibited high expression of the gene, from which the boxed fragment was derived when fed the basal diet (B). However, when the same baboon (L) was fed the challenge diet (C), the gene for which the fragment was derived was down-regulated (Fig. 4, box a, LC). In contrast, the gene was not expressed in the high-responding sibling (H) on either diet (B or C) (Fig. 4, box a). This expression pattern suggests that the gene from which this fragment was derived is probably not responsible for the differential expression of HDL1-C levels, because differential expression is elicited on the challenge diet (C). The results in box b are much more likely to apply to the gene we are seeking. In this case, the two sibs differ in expression of the gene from which the boxed fragment was derived only when fed the challenge diet (C). Because the gene is expressed only in the low expressing sib, this gene, if it were the gene being sought, would be a gene whose expression is elicited by the dietary challenge and whose product prevents the accumulation of HDL1-C. The results in box c reflect a gene that is expressed at a high level in the low expressor (L) on both diets, with higher expression on the basal diet (B, Fig. 4, box c) than the challenge (C, Fig. 4, box c) diet. In addition, the gene is expressed at a lower level in the high expressor (H) on both diets than the low expressor. This pattern might also be characteristic of the gene for which we are searching, that is, it could be a gene whose expression is constitutively different between low and high expressors (i.e., regardless of diet), but whose phenotypic effects are manifested only on the challenge diet. The results in box d reflect a gene that exhibits a similarly low level of expression in both baboons on both diets. The results in box e are similar to those in box b, except that in this case, it is the high responder (H) that shows up-regulation of gene expression on the challenge diet (C). This pattern would be characteristic of a gene whose expression is elicited by the dietary challenge and whose product causes the accumulation of HDL1-C or prevents its catabolism.
Expression Profile Confirmation for Differentially Expressed Candidate Genes
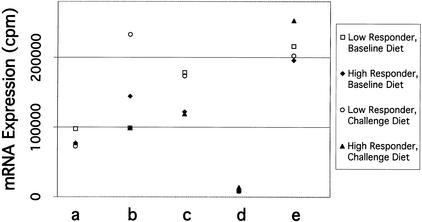
To confirm expression profiles observed in the reverse Northern blots, we selected four types of genes for analysis as follows: (1) moderate level of expression and differential expression (band a); (2) high level of expression and differential expression (bands b and c), (3) low level of expression and similar expression (band d), and (4) low level of expression and differential expression (band e). DNA bands, each containing vector sequence, were isolated from agarose gels and arrayed onto charged nylon membranes. Each array was probed with the same heterologous cDNAs that were used in the reverse Northern blot experiment. The quantitative results from the gene expression arrays (Fig. 5; Table 3) showed profiles that were consistent with the results from the reverse Northern blots. As has been observed typically in other array experiments, the magnitude of expression differences in the gene expression arrays was less than the quantified magnitude of differences observed in the reverse Northern blots.
Figure 5.
Analysis of gene expression arrays. DNA fragments, containing gene fragments plus pBluescript SK+ vector, denoted in Fig. 4 (a, b, c, d, and e), were gel isolated, recircularized, and the plasmid DNA for each clone was spotted onto charged nylon membranes in duplicate. For each clone, 3–4 different plasmids were prepared and spotted onto the membrane. Replicate membranes were hybridized, quantified, and normalized as described in Fig. 4. The gene fragment in the array is displayed on the bottom line, corresponding to the gel band (a, b, c, d, or e) in Fig. 4. For each data series, the standard error of the sample mean (i.e., 6–8 values) is indicated in Table 3. Some symbols cannot be seen in the figure because they are partially or completely superimposed on other symbols
Table 3.
Quantification of Gene Expression Array
| Band | Low responder baseline | Low responder HCHF | High responder baseline | High responder HCHF |
|---|---|---|---|---|
| a | 97545 ± 9703 | 72518 ± 4520 | 76972 ± 2988 | 77415 ± 4946 |
| b | 98510 ± 6082 | 232617 ± 10393 | 144343 ± 6658 | 99000 ± 4720 |
| c | 178110 ± 11495 | 173205 ± 10871 | 122081 ± 9920 | 118922 ± 18735 |
| d | 8406 ± 535 | 9625 ± 1076 | 7872 ± 2442 | 13843 ± 1865 |
| e | 215599 ± 13683 | 202271 ± 8597 | 195542 ± 7495 | 252521 ± 11310 |
Values are given in cpm ± S.D.
Sequence Analysis of Differentially Expressed Gene Fragments
After subcloning differentially expressed gene fragments and confirming expression profiles by array, we end-sequenced these DNA fragments and used BLAT to align the sequences with the human genome. Comparisons of bands denoted in the reverse Northern analysis (Fig. 4) showed that (1) we have not identified any expressed genes that align with known genes, (2) we have identified three expressed genes that align with expressed sequence tags (ESTs) and/or cDNAs predicted by the algorithms Genscan or Fgenesh++, and (3) we have identified two expressed genes that align with DNA sequences that are not known or predicted to harbor sequences encoding cDNAs (Table 4). These data underline the importance of using BAC clones for our expression arrays. Use of arrays based on known genes, ESTs, and predicted genes may not include DNA sequences encoding the QTL in this chromosomal interval.
Table 4.
Alignment of Selected Genes From Reverse Northern Blotsa With Human Genome Data
| Band | Clone | Human genome alignment |
|---|---|---|
| a | 23 | Genscan and Fgenesh++ predicted genes, and human ESTs |
| b | 45 | Genscan predicted gene |
| c | 72 | Genscan and Fgenesh++ predicted genes, and human ESTs |
| d | 72 | No known genes, predicted genes or human ESTs |
| e | 176 | No known genes, predicted genes or human ESTs |
See Figure 4.
DISCUSSION
To identify the gene(s) encoding a QTL regulating HDL cholesterol on chromosome 18, we have (1) designed a chromosome 18 regional BAC array using in silico methods, (2) used baboon liver mRNA generated probes from contrasting HDL1-C phenotypes to show hybridization and differential hybridization to some BAC clones in the array, (3) size-fractionated the BAC clones to identify DNA fragments that showed hybridization and differential hybridization with contrasting HDL1-C phenotypes consistent with the BAC array expression profiles, (4) probed gene fragment arrays of contrasting HDL1-C phenotypes with baboon liver mRNA-generated probes and confirmed differential expression, and (5) end-sequenced DNA fragments showing differential expression and compared the data with the human genome database to align the sequences with known genes, known ESTs, or predicted genes.
These data demonstrate the feasibility of using chromosomal region-specific expression arrays to identify expressed genes in a defined chromosomal interval. In addition, using sib-pairs of contrasting phenotypes, we can identify fragments of genes that are differentially expressed in the interval. Finally, we have determined the presence of a number of genes in which expression varies in response to diet. Now that we have demonstrated the feasibility of the chromosomal region expression array strategy, we are beginning to analyze all five sib-pairs for differentially expressed genes identified in the 53 BAC clones (categories 1 and 2). The criterion that we established for selection of differentially expressed candidate genes (i.e., regulatory variants) is that at least four of the five high expressors must display the same expression pattern and at least two of the three low expressors must display the contrasting expression pattern. We expect that many of the genes that are differentially expressed in any one sib-pair will not be consistently differentially expressed in four of five sib-pairs. Therefore, this strategy is expected to yield a limited number of genes that will receive highest priority as positional candidates. If none of these clones proves to harbor the gene encoding the QTL regulating HDL1-C, we will analyze the similarly expressed BAC clones (category 3) in order to determine the translational status of each mRNA, and we will prioritize the genes for detailed investigation according to translational profiles among sib-pairs and between diets.
On the basis of the current assembly of the human genome, there are 42 known genes in the region of interest. In addition, using current gene prediction algorithms, there are 322 predicted genes ranging from genes with only one exon to genes with 56 exons. By designing an array specific for a chromosomal interval, we have greatly reduced the number of candidate genes to be investigated. From the preliminary data presented here, we predict that there are ∼40 genes in the interval that are differentially expressed (the BAC clones overlap, so some differentially expressed genes in BAC clones are duplicates of overlapping BAC clones). We expect only a minority of the differentially expressed genes to be expressed in a manner consistent with all or most sib-pair phenotypes. Therefore, the number of differentially expressed genes to investigate will be relatively small.
Our data suggest that ∼90 similarly expressed genes are expressed above baseline levels. This estimate is based on the results of the CREAs of the 5 sib-pairs; 55 of the BAC clones exhibited gene expression above the negative controls (i.e., genes expressed only in bacteria). In addition, it is possible that the 83 BAC clones containing differentially expressed genes will contain 1 similarly expressed gene. With an average of 2 genes encoded in each similarly expressed BAC clone and 1 similarly expressed gene encoded in each differentially expressed BAC clone, we have a maximum of 193 genes to investigate. However, in some cases, a gene expressed in one BAC clone will also be expressed from an overlapping BAC clone. Furthermore, as 25% the 184 BAC clones did not express any gene above baseline levels, we can predict that 25% of the encoded genes in similarly expressed BAC clones and differentially expressed BAC clones will not be expressed. Taken together, these data suggest that there may well be <90 genes expressed above baseline levels that will require investigation if the candidate gene encoding the QTL regulating HDL1-C is not among the differentially expressed genes.
This strategy demonstrates the power of combining genomic data with quantitative phenotypic data to identify candidate genes encoding a QTL. Additional power is added to the candidate gene prioritization process when the observed phenotype is the result of a response to an environmental stimulus, such as diet, and appropriate tissues can be collected before and after the stimulus. For any species with a comparative map that relates to the human genome map, this strategy can be applied for identifying genes expressed in the chromosomal region of interest and in the tissue relevant to the phenotype. This same strategy can be used for species with mapped QTLs in which a physical map of the chromosomal region exists. Furthermore, this strategy can be applied to identification of candidate genes encoding QTLs in humans for phenotypes that can be studied by use of easily accessible tissues such as lymphocytes and fibroblasts.
METHODS
Defining the Chromosomal Region of Interest and Assembly of the BAC Contig
The region of interest was defined as the region including microsatellite markers with multipoint LOD scores of 3.0 or greater for the HDL1-C phenotype while baboons were fed the challenge diet. Using the same microsatellite markers in human as baboon, we assembled the contig in silico using the human genome draft assembly for August 2001. This region includes bases 40,941,413–60,217,852 (UCSC Human Genome Project Working Draft, August 6, 2001 assembly) (Kent et al. 2001; Lander et al. 2001). Overlapping BAC clones were included for redundancy in the contig. However, when multiple BAC clones aligned to the same region, BAC clones with the greatest amount of finished sequence were included in the contig. BAC clones were purchased from BACPAC Resources.
BAC DNA Preparation and Spotting Nylon Membranes
For each BAC clone, single colony inoculants were grown in a sterile 96-well block in 300 μL of 2xYT medium and 20 μg/mL of chloramphenicol at 37°C for 16 h, 250 rpm. A total of 2.5 μL of preculture was used to inoculate a 48-well block containing 2.5 mL of 2xYT medium with 20 μg/mL chloramphenicol. Cultures were grown for 18 h at 37°C, 225 rpm, covered with AirPore Tape Sheets (QIAGEN) for aeration. Bacterial cells in the block were harvested by centrifugation. BAC DNA was prepared using QIAGEN R.E.A.L prep 96 reagents. DNA was collected in 96-well plates. BAC DNA was desalted and concentrated by isopropanol precipitation and washed in 75% EtOH. After air drying the samples, the BAC DNA was resuspended in 15 μL of dH2O, incubated at 4°C overnight, and quantified with a Dynaquant (Hoefer). Twelve and a half nanograms of denatured BAC DNA was spotted with a 384-pin replicator (Nalge Nunc) onto Hybond XL nylon membrane (Amersham) and UV-cross-linked to the membrane. Escherichia coli gene DNA (Sigma Genosys) was included as a negative control.
Selection of Animals and Diet Screening
On the basis of phenotypic and genotypic analysis of the pedigreed baboon population, five sib-pairs with contrasting phenotypes for HDL1-C were chosen. The sib-pairs differed by at least one standard deviation for HDL1-C serum concentrations. In addition, members of each selected sib-pair did not share IBD (identical-by-descent) alleles, or for some markers shared only one IBD allele, in the chromosomal region of interest. Baboons were fed commercial monkey chow (basal diet; Teklad) for at least 7 wk. Blood and liver biopsies were collected. Baboons were then fed a high-cholesterol, high-fat diet (challenge diet; 1.7 mg/kcal cholesterol and 40% of calories as fat from lard) (McGill et al. 1981) for 7 wk, and blood and liver biopsies were again collected. HDL-C and sub-fractions were measured from serum. Liver biopsies were quick frozen at the time of collection and later used for RNA extractions.
mRNA Extractions, Heterologous cDNA Synthesis, and Probe Labeling
Baboon liver mRNA was isolated from liver biopsies using Microfast track mRNA kit (Ambion) according to the manufacturer's instructions. Messenger RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen Life Technologies), random hexamers, and radioactively labeled [α-33P]dCTP (Perkin Elmer), according to Research Genetics' protocol (www.resgen.com/products/GF200_protocol_pf.php3).
Interrogating Arrays With Heterologous cDNA Probes
Each array was probed with heterologous liver cDNA for each animal on each of two diets. E. coli genes were included as negative controls in each blot. In addition, baboon endothelial lipase (LIPG) cDNA fragment, and human transferrin (TF) cDNA fragment, baboon BAC clones containing LIPG, human BAC clones containing LIPG, and human BAC clones containing FIC1 were included as positive controls. Membranes were prehybridized for 2 h at 42°C in 9 mL Ultrahyb hybridization buffer (Amersham) with 5 μL of Cot-1 DNA (1 μg/μL), 5 μL of poly d(A). For hybridization, 107 cpm denatured probe was added to the prehybridization buffer and incubated 12–18 h at 42°C. Filters were washed four times as follows: (1) 15 min at room temperature in 2× SSC, 0.1% SDS; (2) 15 min at room temperature in 2× SSC, 0.1% SDS; (3) 15 min at 45°C in 1× SSC, 0.1% SDS; (4) 15 min at 45°C in 0.1× SSC, 0.1% SDS. Filters were exposed to PhosphorImager cassettes and signals quantified using IP Lab Gel software (Molecular Dynamics). After decay of the signal for CREAs screened with heterologous liver cDNA, blots were reprobed with BAC vector DNA fragment to normalize BAC DNA quantities in each spot. All mRNA expression values were normalized with BAC vector signal data for each spot on the filter. Clones were considered to contain expressed genes if the normalized signal was greater than the E. coli gene negative control. Clones with signals below the negative control were considered unexpressed and not pursued.
BAC Clone Expression Analysis
Duplicate values of BAC clone expression data for all high HDL responders on basal diet were averaged and the standard deviation for each clone was calculated. The same was done for high HDL responders on challenge diet, low HDL responders on basal diet, and low HDL responders on challenge diet. For each BAC clone, expression was compared for diet response, that is, high HDL responders on basal diet versus high HDL responders on challenge diet and low HDL responders on basal diet versus low HDL responders on challenge diet. Expression was also compared for differences by phenotype, that is, high HDL responders on basal diet versus low HDL responders on basal diet and high HDL responders on challenge diet versus low HDL responders on challenge diet. Expression values differing by at least one standard deviation were considered significant.
Shotgun Cloning and Size Fractionation of BAC DNA Fragments
For shotgun cloning of BAC clone inserts, 100 ng of BAC DNA and the 37.5 ng of pBluescript SK+ plasmid (Stratagene) were restriction digested with PstI restriction enzyme (New England Biolabs) for 3 h at 37°C. BAC DNA and plasmid were ligated overnight at 4°C using T4 DNA ligase, 10 mM ATP, 10× ligation buffer [50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg/mL BSA] (New England Biolabs). Ligation reactions were diluted 1:2, and 5 μL was transformed into 40 μL XLI-Blue-competent cells for 30 min on ice (Stratagene). After the transformation reaction was heat shocked for 50 sec at 42°C, cells were incubated for 1 h at 37°C with 225 rpm in 960 μL SOC medium. To check transformation efficiencies, 200 μL of each 1000-μL transformation was spread on LB agar plates with 50 μg/mL ampicillan, 40 μg/mL X-gal, and 100 μM IPTG. A total of 200 μL of transformed cells were added to 2 mL of LB with 50 μg/mL ampicillan, and cultures were grown for 24 h at 37°C with 225 rpm. Subcloned DNA in pBluescript SK+ plasmids (Stratagene) was extracted from overnight cultures using Qia REAL 96 prep (QIAGEN) according to manufacturer's instructions. DNA was linearized with NotI (New England Biolabs) and size fractionated in a 1% SeaKem LE agarose gel (BMA) with 1× TAE buffer. Gels were subjected to electrophoresis for 20 h at 6 V/cm with the buffer recirculated at 14°C.
Reverse Northern Blot Analyses
Gels containing size-fractionated subcloned BAC DNA fragments were depurinated in 0.125 M HCl for 10 min, denatured in 1.5 M NaCl, 0.5 M NaOH for 30 min, and neutralized in 1.5 M NaCl, 0.5 M Tris (pH 7.5) for 30 min. DNA was transferred by capillary action to Hybond XL membranes (Amersham) using 20× SSC (Southern 1975). DNA was UV-cross-linked to the membrane. One sib-pair was chosen using cDNA probes for each animal on each diet. Heterologous, α-33P-labeled liver cDNA (Research Genetics; http://www.resgen.com/ products/GF200_protocol_pf.php3) was hybridized with the BAC DNA filters using Ultrahyb buffer (Ambion) overnight at 42°C. Filters were washed twice in 2× SSC, 0.1% SDS at room temperature and twice in 0.1× SSC, 0.1% SDS at 45°C. Filters were exposed to PhosphorImager cassettes, and signals quantified using IP Lab Gel software (Molecular Dynamics). After decay of the signal for filters screened with heterologous liver cDNA, blots were reprobed with vector DNA to normalize DNA quantities in each band.
Gel Isolation of Gene Fragments
After reverse Northern blotting, DNA fragments of interest were isolated by size fractionation in 1% SeaPlaque Agarose (BMA) with 1× TAE buffer and excision from the gel. Isolated fragments, each containing vector sequence, were recircularized in gel by mixing 10 μL of gel/vector isolated fragment with a 40 μL ligation reaction containing T4 DNA ligase, 10 mM ATP, 10× ligation buffer [50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg/mL BSA] (New England Biolabs). Two microliters of each ligation reaction was transformed into XLI-Blue competent cells (Stratagene) and plated onto LB agar plates containing 50 μg/mL ampicillan.
Gene Fragment Arrays
For each DNA fragment, DNA was isolated from 12 separate colonies (Qia REAL 96 prep, QIAGEN). Plasmid DNA containing differentially expressed gene fragments were spotted onto Hybond XL membranes (Amersham) and bound by UV-cross-linking. Arrays were screened with heterologous liver cDNA used with the reverse Northern blots as described in reverse Northern blotting. Those clones with expression profiles consistent with reverse Northern blots were considered positive for the gene fragment of interest.
End-Sequencing Gene Fragments
Gene fragments were end-sequenced using 100 μM sequencing primer (T7, 5′-TAATACGACTCACTATAGGGAGA-3′ and T3, 5′-ATTAACCCTCACTAAAGGGA-3′), Big Dye buffer [80 mM Tris-HCl (pH 9.0), 2 mM MgCl2] and 2 μL of Big Dye enzyme mix in a 10-μL reaction using an ABI 377 automated sequencer (Applied Biosystems). End-sequence data was compared with human genome data base sequence using BLAT (Kent 2002; Kent et al. 2002) to identify the region of human genome aligning with the cloned fragment. In addition, the alignment was used to determine whether known genes, ESTs, or predicted genes by Genscan (Burge et al. 1997) or by Fgenesh++ (Salamov et al. 2000) aligned with the gene fragment.
WEB SITE REFERENCES
http://www.chori.org/BACPAC/home.htm; BACPAC Resources, purchase of human and baboon BAC clones.
http://www.ncbi.nlm.nih.gov/genome/sts/epcr.cgi; NCBI electronic PCR.
http://www.resgen.com/products/GF200_protocol_pf.php3; Research Genetics, synthesis and radioactive labeling of heterologous cDNA from mRNA.
http://genome.cse.ucsc.edu/; UCSC Human Genome Project Working Draft (August 6, 2001) assembly (hg8), contig assembly for CREA.
Acknowledgments
We thank Jane F. VandeBerg for technical assisitance, Drs. Karen S. Rice and K.D. Carey for managing the diet experiment, and Dr. Michelle Leland for performing the baboon liver biopsies. We also thank Drs. Michael C. Mahaney and Jeffrey Rogers for baboon linkage map and genome screen data that were used to define the chromosome 18 region of interest and Dr. Candace M. Kammerer who provided advice on statistical analysis of array data. This work was supported by NIH grants P01 HL28972 and P51 RR13986.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lcox@darwin.sfbr.org; FAX (210) 670-3344.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.333502. Article published online before print in October 2002.
REFERENCES
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cheng ML, Kammerer CM, Lowe WF, Dyke B, VandeBerg JL. Method for quantitating cholesterol in subfractions of serum lipoproteins separated by gradient gel electrophoresis. Biochem Genet. 1988;26:657–681. doi: 10.1007/BF02395514. [DOI] [PubMed] [Google Scholar]
- Flow BL, Mott GE. Relationship of high density lipoprotein cholesterol to cholesterol metabolism in the baboon (Papio sp.) J Lipid Res. 1984;25:469–473. [PubMed] [Google Scholar]
- Fruchart JC, Duriez P. High density lipoproteins and coronary heart disease. Future prospects in gene therapy. Biochimie. 1998;80:167–172. doi: 10.1016/s0300-9084(98)80023-0. [DOI] [PubMed] [Google Scholar]
- Kammerer CM, Mott GE, Carey KD, McGill HC., Jr Effects of selection for serum cholesterol concentrations on serum lipid concentrations and body weight in baboons. Am J Med Genet. 1984;19:333–345. doi: 10.1002/ajmg.1320190216. [DOI] [PubMed] [Google Scholar]
- Kent WJ. The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Haussler D. Assembly of the working draft of the human genome with GigAssembler. Genome Res. 2001;11:1541–8. doi: 10.1101/gr.183201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Mahaney MC, Rainwater DL, Rogers J, Cox LA, Blangero J, Almasy L, VandeBerg JL, Hixson JE. A genome search in pedigreed baboons detects a locus mapping to human chromosome 18q that influences variation in serum levels of HDL and its subfractions. Circulation. 1998;98:I-5. (abstract). [Google Scholar]
- McGill HC, Jr, McMahan CA, Kruski AW, Mott GE. Relationship of lipoprotein cholesterol concentrations to experimental atherosclerosis in baboons. Arteriosclerosis. 1981;1:3–12. doi: 10.1161/01.atv.1.1.3. [DOI] [PubMed] [Google Scholar]
- McGill HC, Jr, McMahan CA, Mott GE, Marinez YN, Kuehl TJ. Effects of selective breeding on the cholesterolemic responses to dietary saturated fat and cholesterol in baboons. Arteriosclerosis. 1987;8:33–39. doi: 10.1161/01.atv.8.1.33. [DOI] [PubMed] [Google Scholar]
- Mott GE, McMahan CA, McGill HC., Jr Diet and sire effects on serum cholesterol and cholesterol absorption in infant baboons. Circ Res. 1978;43:364–371. doi: 10.1161/01.res.43.3.364. [DOI] [PubMed] [Google Scholar]
- Rogers J, Mahaney MC, Witte SM, Nair S, Newman D, Wedel S, Rodriguez LA, Rice KS, Slifer SH, Perelygin A, et al. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics. 2000;67:237–247. doi: 10.1006/geno.2000.6245. [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]