Abstract
Viruses are intracellular parasites that use many cellular pathways during their replication. Large DNA viruses, such as herpesviruses, have captured a repertoire of cellular genes to block or mimic host immune responses, apoptosis regulation, and cell-cycle control mechanisms. We have conducted a systematic search for all homologs of herpesvirus proteins in the human genome using position-specific scoring matrices representing herpesvirus protein sequence domains, and pair-wise sequence comparisons. The analysis shows that ∼13% of the herpesvirus proteins have clear sequence similarity to products of the human genome. Different human herpesviruses vary in their numbers of human homologs, indicating distinct rates of gene acquisition in different lineages. Our analysis has identified new families of herpesvirus/human homologs from viruses including human herpesvirus 5 (human cytomegalovirus; HCMV) and human herpesvirus 8 (Kaposi's sarcoma–associated herpesvirus; KSHV), which may play important roles in host-virus interactions.
Viruses are obligate intracellular parasites and, as such, use many normal cellular pathways and components during their replication cycle. Large DNA viruses may contain up to a few hundred open reading frames (ORFs). Among the proteins they encode, we can distinguish between those that have essential viral functions, such as genome replication and capsid assembly, and those that are involved in direct interaction with the host, effecting immune evasion, cell proliferation, and apoptosis control (Ploegh 1998; Tschopp et al. 1998). Many of the latter genes are likely to have been acquired from the host to mimic or block normal cellular functions ( Moore et al. 1996; Alcami and Koszinowski 2000; McFadden and Murphy 2000). Identifying and understanding the functions of such “acquired” viral proteins may lead to the development of therapeutic strategies to combat persistent viral infection.
An approach to the identification of virus proteins that interfere with the host system is to search for homologs in the host genome. Until recently, the fraction of host genome sequence data available for analysis, and the quality of annotation of such data, has limited the identification of such homologs. The publication of the draft of the human genome and conceptual translated products (Lander et al. 2001) enables us to conduct, for the first time, a comprehensive assessment of homologous proteins between a vertebrate genome and viral ORFs. There are two methods particularly applicable to mass analysis of sequence databases. The first involves searching of individual protein sequences against a database using pair-wise sequence comparison algorithms, and has previously been used to identify individual virus/host homologs. Viral proteins, however, are subject to high mutation rates, and that may cloud or mask true homology. A second, more sensitive approach is to search databases with amino acid sequence motifs that are conserved between related proteins. Motifs can be defined as regions of amino acid sequence that are more highly conserved than the rest of the protein owing to functional constraints. An accurate representation of such motifs can be obtained by constructing position-specific scoring matrices (PSSMs) that store the frequency of occurrence of different amino acids along the motif.
In the present study, we focus on the analysis of herpesviruses, one of the best-characterized large DNA virus families. Typically, each herpesvirus genome contains between 70 and 120 ORFs, with the exception of human cytomegalovirus (HCMV), which codes for up to 220 ORFs. The herpesviruses infect a wide range of animal hosts and—on the basis of differences in genome content, organization, and cellular tropism—have been divided into three subfamilies: the alphaherpesviruses, betaherpesviruses, and gammaherpesviruses. There are a number of herpesviruses that have yet to be categorized in a herpesvirus subfamily, including channel catfish herpesvirus, and these are classified as “other” in this study (see Table 1; ICTV 2000). Eight different herpesviruses, encompassing all three subfamilies, are known to infect humans. Herpesviruses persist and replicate their genomes in the nucleus and acquire host genes by an ill-defined process (Brunovskis and Kung 1995; Chaston and Lidbury 2001). Most of these acquired genes are located in regions outside the five gene blocks common to all herpesvirus genomes. Previous work by others and ourselves has identified a set of 26 ORFs that are conserved across all herpesviruses (McGeoch and Davison 1999; Albà et al. 2001a). The remaining herpesvirus genes are present in all members of a virus subfamily, present in a subset of viruses in a subfamily, or unique to a particular virus. Many of these potentially important proteins, however, remain uncharacterized.
Table 1.
Herpesvirus-Human Homologs
| Function class | Viral function (VIDA) | HPF1 | Virus2 | GenBank3 | Human function |
|---|---|---|---|---|---|
| DNA replication | DNA polymerase | 1 | a,b,g | 8393995 | polymerase (DNA-directed), α |
| 293 | o | 15303524 | polymerase (DNA directed), δ 1 | ||
| helicase/primase | 16 | a,b,g | 5523990 | DNA helicase | |
| Nucleotide repair/ | uracil–DNA glycosylase | 8 | a,b,g | 6224979 | uracil–DNA glycosylase |
| metabolism | ribonucleotide reduct. large sub. | 24 | a,b,g | 4506749 | ribonucleotide reductase M1 polypeptide |
| ribonucleotide reduct. small sub. | 33 | a,g | 4557845 | ribonucleotide reductase M2 polypeptide | |
| thymidylate synthase | 92 | a-,g- | 15297069 | thymidylate synthetase | |
| dihydrofolate reductase | 141 | g-,b- | 15297069 | dihydrofolate reductase | |
| dUTP pyrophosphatase | S | CCHV ORF49 | 4503423 | dUTP pyrophosphatase | |
| S | SaHV-1 ORF49 | 14756895 | dUTP pyrophosphatase | ||
| thymidine kinase | S | CCHV ORF5 | 11430716 | thymidine kinsae 2, mitochondrial | |
| DNA methyltransferase | S | RaHV-1 54_21 | 4503351 | DNA (cytosine-5-)–methyltransferase 1 | |
| Enzyme | protein kinase | 29 | a,b,g- | 14746991 | serine/threonine-protein kinase PRP4 |
| 40 | a,o | 4505649 | protein kinase cdc2-related PCTAIRE-2 | ||
| 214 | o | 9994197 | G protein-coupled receptor kinase 7 | ||
| S | RaHV-1 54_2 | 14741902 | CamKI–like protein kinase | ||
| phospholipase-like protein | 328 | a- | 5174497 | endothelial cell–derived lipase precursor | |
| b-1,6-N-acetylglucosaminyltransf. | S | BoHV-4 ORF3-4 | 11431963 | glucosaminyl (N–acetyl) transferase 3 | |
| serine protease | S | CCHV ORF47 | 4505577 | paired basic amino acid cleaving system 4 | |
| Gene expression | transcriptional activator | 74 | a | 5174653 | ring finger protein (C3H2C3 type) 6 |
| regulation | bZIP domain | 174 | a- | 4504809 | jun B proto–oncogene |
| Glycoprotein | glycoprotein OX-2-like | 194 | b- | 730246 | OX-2 membrane glycoprotein precursor |
| glycoprotein OX-2-like | 242 | g- | 730246 | OX-2 membrane glycoprotein precursor | |
| Host-virus interaction | TNFR receptor | 13 | HHV-5 UL144 | 4507571 | tumor necrosis factor receptor, member 14 |
| virion–assoc. host shutoff factor | 48 | a | 14738228 | flap structure–specific endonuclease 1 | |
| viral interferon regulatory factor | 89 | g- | 4504723 | interferon regulatory factor 2 | |
| 243 | g- | 13629153 | interferon consensus seq. binding prot. 1 | ||
| S | HHV-8 vIRF-3 | 4505287 | interferon regulatory factor 4 | ||
| G protein-coupled receptor | 27 | b,g- | 13643500 | chemokine (C–C motif) receptor 2 | |
| 248 | b- | 4758468 | G protein–coupled receptor 50 | ||
| S | EHV-2, ORF 74 | 4502639 | chemokine (C–C motif) receptor 5 | ||
| complement binding protein | 10 | g- | 10835143 | decay accelerating factor for complement | |
| viral cyclin | 102 | g- | 14767736 | cyclin D1 | |
| viral interleukin 10 | 140 | g- | 10835141 | interleukin 10 | |
| viral interleukin 6 | 273 | g- | 10834984 | interleukin 6 (interferon, β 2) | |
| viral interleukin 17 | S | HVS-2 ORF13 | 4504651 | interleukin 17 | |
| vBcl-2 | 161 | g- | 4502363 | BCL2–antagonist-killer 1 | |
| 259 | g- | 4557355 | B–cell lymphoma protein 2 α | ||
| 850 | MeHV-1 ORF1 | 11433559 | BCL2-like 10 (apoptosis facilitator) | ||
| MHC I downregulation | 150 | g- | 8923613 | hypothetical protein FLJ20668 | |
| viral FLICE–inhibitory protein | 256 | g- | 14731507 | CASP8 and FADD–like apoptosis regulator | |
| S | EHV-2 E8 | 4505229 | Fas (TNFRSF6)–associated via death domain | ||
| CxC chemokine vIL8 | 531 | a- | 10834978 | interleukin 8 | |
| vMIP-I | 225 | g- | 5174671 | small inducible cytokine subf. A, member 26 | |
| α chemokine | 321 | b- | 4885589 | small inducible cytokine subf. B, member 9B | |
| β chemokine | 387 | b- | 5174671 | small inducible cytokine subf. A, member 26 | |
| vMIP-III | S | HHV-8 K4.1 | 4506829 | small inducible cytokine subf. A, member 17 | |
| signal transduction protein | 316 | RRV, R1 | 12056967 | Fc fragment of IgG, receptor for (CD16) | |
| CARD–like apoptotic protein | 355 | EHV-2, E10 | 4502379 | CARD–like apoptotic protein | |
| U-PAR antigen CD59 | 352 | HVS-2, ORF15 | 13639271 | CD59 antigen p18-20 | |
| natural killer (NK) cell decoy pr. | S | HHV-5 UL18 | 5031745 | major histocompatibility complex, class I, E | |
| colony-stimulating factor I | S | HHV-4 BARF1 | 4885123 | CD80 antigen | |
| C-type lectin-like protein | S | RCMV lectin | 4504883 | killer cell lectin–like receptor subf. C, member 2 | |
| semaphorin homolog | S | AIHV-1 A3 | 4504237 | sema domain, Ig domain, GPI memb. anchor | |
| MHC1 heavy chain | S | RCMV R144 | 9665232 | major histocompatibility complex, class I | |
| Unknown | unknown | 258 | a- | 4504883 | killer cell lectin–like receptor subf. C, member 2 |
| Unknown | S | GaHV-1 UL45 | 4504883 | killer cell lectin–like receptor subf. C, member 2 | |
| Unknown | S | HHV-5 UL1 | 14764567 | pregnancy specific beta-1glycoprotein 5 | |
| Unknown | S | HHV-5 US21 | 6912468 | lifeguard |
HPF: homologous protein family no. S indicates singleton. HPF details can be visualised by searching VIDA by HPF number in http://www.biochem.ucl.ac.uk/bsm/virus_database/VIDA.html (Herpesviridae link).
a indicates alphaherpesvirus; b, betaherpesvirus; g, gammaherpesvirus; o, other; — only a subset of subfamily members are represented. For singletons, virus abbreviation and gene name are given: CCHV, channel catfish herpesvirus; SaHV-1, salmonid herpesvirus 1; RaHV-1, ranid herpesvirus 1; BoHV-4, bovine herpesvirus 4; HHV-8, human herpesvirus 8; EHV-2, equine herpesvirus 2; HVS-2, saimiriine herpesvirus 2; MeHV-1, meleagrid herpesvirus 1; HHV-5, human herpesvirus 5; HHV-4, human herpesvirus 4; RCMV, rat cytomegalovirus; AHIV-1, alcelaphine herpesvirus 1; and GaHV-1, gallid herpesvirus 1.
GenBank protein accession no. (GI number). Only the human that hit with the lowest E-value is shown.
We have recently developed a virus database, VIDA (Albà et al. 2001b), in which all herpesvirus ORFs are grouped together into homologous protein families (HPFs), each defined by one or more conserved amino acid regions (motifs). To identify human proteins that are related to the herpesvirus protein families, we have constructed PSSMs for all HPF-defining motifs and used them to perform sensitive searches of the translated human genome products. Mapping of homologs in the human genome has been complemented by BLAST-based pair-wise sequence comparison searches (Altschul et al. 1990, 1997). Our analysis has resulted in the identification of protein families or singleton proteins that show clear homology with gene products in the human genome, including new host-virus homologs in human herpesvirus (HHV) 5 (HCMV) and HHV-8 (Kaposi's sarcoma–associated herpesvirus; KSHV).
RESULTS
Herpesvirus Proteins With Human Homologs
The identification of herpesvirus/human homologs was undertaken by searching the set of conceptual and known protein sequences derived from the public Human Genome Project (Lander et al. 2001) against herpesvirus protein sequences in the virus database VIDA (Albà et al. 2001b) using two different sequence-similarity search methods. The first method was based on PSSMs derived from predefined viral protein motifs in VIDA. The second used BLAST-based pair-wise sequence comparisons with the collection of singleton viral proteins and a representative set of viral proteins that share <95% sequence identity (N95-rep, see Methods).
Careful examination of putative homologs showed that 39 herpesvirus HPFs and 20 singleton proteins had significant sequence similarity to human gene products (Table 1). This represented 13% of all herpesvirus ORFs in GenBank. Sequence similarity between herpesvirus and human proteins was clearly related to functional similarity, based on previous experimental data. However, functional similarity is defined here in a broad sense, meaning the viral proteins participate in the given functional network. This is because viral proteins can change from the precise mechanistic function of the host homolog in subtle ways after acquisition by the virus while still maintaining the broader function. For example, the HHV-8 viral cyclin participates in the cell cycle as a cyclin D homolog but, unlike the host cyclin D, is not negatively regulated (Swanton et al. 1997). The use of PSSMs to perform database searches was more sensitive than using N95-reps with BLASTP, as six of the 39 HPF homologs could only be detected by the first method. One homolog, however, complement binding protein, could only be identified using BLASTP.
Approximately 54% of the combined HPF and singleton hits corresponded to proteins classified in VIDA as being involved in host-virus interaction, primarily effecting immune and/or apoptosis controls. Of the remaining homologs, 32% have functions that can be generally termed metabolic (being “enzymes,” involved in “DNA replication,” or involved in “nucleotide repair/metabolism”). Homologs to capsid constituents or capsid assembly proteins were not detected. Approximately 42% of the HPFs and singletons that showed homology with human proteins did not contain any HHV ORF members. This method can therefore be used to annotate gene products from non-HHVs for which complete host genome sequence information is still unavailable.
Identification of New Virus-Human Homologs
Of special interest was the identification of human homologs for herpesvirus protein families and singletons of unknown function. The new homologs may provide putative functional annotations for several herpesvirus and/or human proteins. New herpesvirus/human protein families were found for the US12 (unique short) HCMV protein family, the UL1 (unique long) HCMV protein, the gallid/meleagrid herpesvirus UL45 protein family, and the K3/K5 HHV-8 family (Fig. 1).
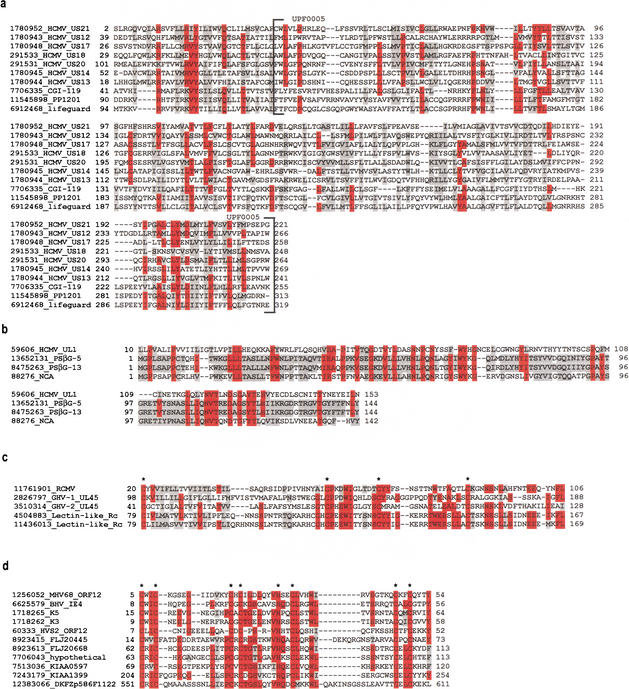
Figure 1.
Alignment of new herpesvirus/human homologs. Proteins are labeled with GenBank identification number (GI) and a short description. Amino acids that are shaded red share identity across ≥50% of the alignment; amino acids shaded grey share similarity across ≥50% of the alignment. (a) Herpesvirus US12 protein family members, human lifeguard protein, and two additional human proteins. The Pfam UPF0005 domain is indicated. (b) HCMV UL1, two PSG proteins (PSβG 5 and 13), and one member of the carcinoembryonic antigen subfamily (NCA, nonreacting antigen). (c) A representative from each of the herpesvirus protein families found to contain C-type lectin domains and two natural killer receptors (NKG2-A). The four conserved cysteines, important for disulphide bond formation in the carbohydrate recognition domain, are indicated. (d) K3/K5 herpesvirus protein family with six human homologs. Cysteine/histidine conserved residues in the BKS (BHV-4 [bovine herpesvirus 4], KSHV, and swinepox) motif are indicated.
HCMV US21 is a distant member of a larger HCMV protein family, the US12 protein family, encompassing gene products US12 to US21 (Chee et al. 1990). The US21 showed significant overall sequence similarity to three human proteins: lifeguard, CGI-119, and PP1201. Other members of the US12 protein family, including an HPF that groups six of them in VIDA, did not initially hit any human proteins, but multiple sequence alignments revealed the true extent of amino acid similarity between all these proteins (Fig. 1a). The herpesvirus and human proteins also matched the protein family domain UPF0005 in the Pfam database (Bateman et al. 2000), a putative seven-transmembrane region domain. Lifeguard is the human homolog of the rat protein neuromembrane protein 35, proposed to protect against Fas-mediated apoptosis (Somia et al. 1999).
HCMV UL1 showed sequence similarity to the pregnancy-specific glycoprotein 5 (PSG-5) and other members of the human carcinoembryonic antigen (CEA) protein family. The PSGs, a subgroup of the CEA family, are mainly expressed in the placenta and are secreted into the maternal circulation, possibly regulating immune system responses. The region of sequence similarity covered about two thirds of the UL1 protein and the N-terminal region of PSG and CEA subgroup proteins (Fig. 1b).
The protein family represented by UL45 in gallid (includes Marek's disease herpesvirus) and meleagrid herpesviruses shows homology with human C-type (calcium-dependent) lectin domain containing natural killer (NK)–cell receptor proteins. Two other herpesvirus proteins, from rat cytomegalovirus (RCMV) and from a different gallid herpesvirus strain (GenBank accession no. Y14300), also show significant sequence similarity to C-type lectin domain containing NK-cell receptors. The presence of C-type lectin domain in the RCMV protein was recently reported (Voigt et al. 2001) which now clearly extends to homologs in some avian herpesviruses. NK-cell receptors interact with HLA (human leukocyte antigen) class I antigens and facilitate triggering or inhibition of NK cell–mediated cytotoxicity (Biassoni et al. 2001). C-type lectins contain a carbohydrate recognition domain, which includes four conserved cysteine residues forming two disulphide bonds. These conserved cysteines are also present in the herpesvirus C-type lectin-like homologs (Fig. 1c).
The K3/K5 protein family in VIDA contains a highly conserved zinc finger motif identified in the proteins K3 and K5 from HHV-8, IE1 in bovine herpesvirus 4 (BHV-4), and ORF12 in murine herpesvirus 68 (MHV-68). An additional gene, ORF 12 in saimiriine herpesvirus 2 (HVS-2), a singleton in VIDA, did not initially hit any human gene product. However, it also contains the same conserved motif and should therefore be considered a member of the family (Nicholas et al. 1997). The motif is known as the BKS (BHV-4, KSHV, and swinepox) motif, a member of the PHD/LAP zinc finger class (C4HC3), but clearly differing from PHD/LAP zinc fingers owing to its distinct spacing of the cysteine/histidine residues. K3 and K5 from HHV-8 have been recently discovered to down-regulate MHC class 1 molecules in infected cells (Coscoy and Ganem 2000). We identified six unannotated human proteins, including three identified by pair-wise searches (Jenner and Boshoff 2002), that contain this highly conserved BKS finger motif (Fig. 1d). In the herpesvirus proteins, the motif is always found in the N terminus, but in one human protein, it appeared in the central part of the peptide, whereas in another, the counterpart of murine axotrophin, at the C terminus.
Human Homologs in HHVs
Our analysis provides an estimate of the number of homologs between the eight different HHVs and the translated products from their host genome. A total of 34 different HHV proteins, including HPFs and singletons, showed significant homology with human proteins (Fig. 2). This represents a minimum estimate, as some proteins may still be functionally homologous but not show significant sequence similarity, and the total number of genes in the human genome is still uncertain (Lander et al. 2001).
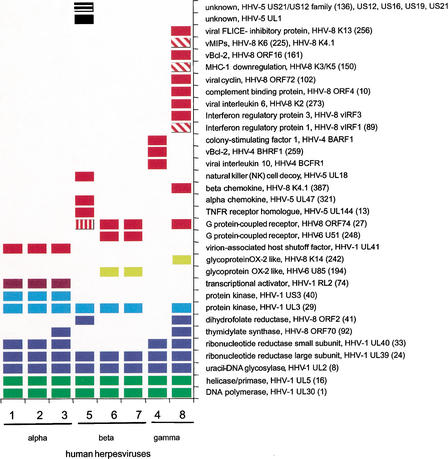
Figure 2.
Human herpesvirus (HHV) proteins with human homologs. Alternative names for the HHVs are HHV-1, human simplex virus 1; HHV-2, human simplex virus 2; HHV-3, varicella zoster virus; HHV-4, Epstein-Barr virus; HHV-5, human cytomegalovirus; and HHV-8, Kaposi's sarcoma–associated herpesvirus. Labels show the virus protein function, the name of a member of the HPF (homologous protein family) or singleton, and, for HPFs, the corresponding number in brackets. All the annotations and HPF numbers are taken from VIDA. Note that in some cases more than one HPF/singleton, shown as separate rows in Table 1, are shown together here. This corresponds to highly divergent families. The graph is color coded according to functional class: light green, DNA replication; dark blue, nucleotide repair/metabolism; light blue, enzyme; purple, gene expression regulation; yellow, glycoprotein; red, host-virus interaction; and black, unknown. Diagonal lines within a box indicate two gene copies (per viral genome); vertical lines, three copies; and horizontal lines, 10 copies.
Four human homologs are known to be present in all HHVs (i.e., DNA-dependent DNA polymerase, helicase/primase, uracil-DNA glycosylase, and ribonucleotide reductase large subunit), and these were all correctly identified by our methods. An additional protein family, protein kinase HHV-1 UL13, is present in all HHVs except in HHV-4. It is known that the gammaherpesviruses share a common evolutionary branch with the betaherpesvirus, and that the alphaherpesvirus forms a separate lineage (McGeoch and Davison 1999; Albà et al. 2001a). One of the human homologs, ribonucleotide reductase small subunit, is found in the alpha- and gammaherpesviruses, but not in the betaherpesviruses, indicating that it has been lost in the latter lineage. There are three human homologs that appear to be alphaherpesvirus-specific: protein kinase HHV-1 US3, transcriptional activator HHV-1 ICP0 (infected cell protein), and host shutoff factor HHV-1 UL41. This compares to seven homologs that are betaherpesvirus specific and 14 that are gammaherpesvirus specific. Of particular interest are two human homologs that appear in disparate positions in the herpesvirus evolutionary tree: thymidylate synthase in HHV-3 (varicella zoster virus) and in HHV-8 (Kaposi's sarcoma–associated herpesvirus); dihydrofolate reductase in HHV-5 (HCMV) and HHV-8. Independent acquisition of these genes from the host genome, multiple gene loss events in different herpesvirus lineages, or gene transfer between virus genomes could explain their distribution.
The total proportion of human homologs in the different HHVs varies. Using the number of gene products in the corresponding herpesvirus genome GenBank entries (Table 1 in Albà et al. 2001a), this percentage is 11% to 16% of the genes in human alphaherpesviruses, 9% to 11% in the human betaherpesviruses, 10% of the genes in HHV-4, and 30% in the HHV-8 genome. HHV-8 contains a markedly higher proportion of human homologous genes, indicating a higher degree of recent gene transfer from the host genome.
Dynamics of Host Gene Acquisition in the Gammaherpesviruses
Human homologs that are present in all or a large proportion of the herpesvirus genomes, such as DNA polymerase or uracil-DNA glycosylase, are likely to have been acquired from a distant host by an ancestral herpesvirus. Other genes appear to have been acquired more recently, appearing only in a subset of viruses. From the 59 HPFs and singletons that showed homology with human proteins, only 16 were present in alphaherpesviruses, 17 in betaherpesviruses, and 32 in gammaherpesviruses. More than half (54%) of these homologs have host-virus interaction functions. Gammaherpesvirus genomes are particularly rich in genes that have a human counterpart. Therefore, a more detailed analysis of the distribution of gammaherpesvirus-specific human homologs in complete gammaherpesvirus genomes was undertaken (Fig. 3).
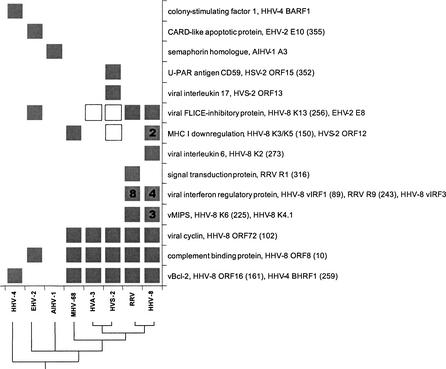
Figure 3.
Gammaherpesvirus-specific proteins involved in host-virus interactions that have human homologs. Boxes indicate the presence of a particular gene(s) in a virus genome. Numbers in boxes represent copies within a genome. Labels show the virus protein function, the name of a member of the HPF (homologous protein family) or singleton, and, for HPFs, the corresponding number in brackets. All the annotations and HPF numbers are taken from VIDA. Note that in some cases more than one HPF/singleton, shown as separate rows in Table 1, is shown together. This corresponds to highly divergent families. The HPF/singletons that are not present in Table 1 are represented as unfilled boxes. These are herpesvirus proteins for which we did not identify human homologs in the database searches but that, nevertheless, can be grouped together, by function and residue conservation, with other herpesvirus HPF/singletons for which we could detect human homologs. A consensus phylogenetic tree of the gammaherpesvirus is shown at the bottom. This was generated as described for all HPFs from complete herpesvirus genomes (Albà et al. 2001a).
Phylogenetic reconstruction of the fully sequenced gammaherpesvirus subfamily members (McGeoch et al. 2000; Montague and Hutchison 2000; Albà et al. 2001a) has established that HHV-4 forms a separate lineage, the lymphocrytpo or gamma-1-herpesviruses 1. The remaining fully sequenced gammaherpesviruses, which include HHV-8, form the rhadino or gamma-2-herpesviruses lineage. The relative positions of alcelaphine herpesvirus 1 (AIHV-1), equine herpesvirus 2 (EHV-2), and MHV-68 within the gammaherpesvirus 2 are still ill-defined, although recent work shows that MHV-68 is probably more closely related to the primate herpesvirus (Fig. 3; McGeoch et al. 2000; Albà et al. 2001a). The presence of human homologs in the different genomes is consistent within the different gammaherpesvirus groups defined by gene-content phylogenetics (Fig. 3); however, some of the homologs show a complex distribution. For example, ORF12, a homolog of the K3/K5 HHV-8 genes, is also present in MHV-68 and HVS-2 but not in the HHV-8 closely related primate herpesviruses ateline herpesvirus 3 (AtHV-3) and Macaca mulatta rhadinovirus (RRV). Therefore, the gene may have been lost on several occasions. Another explanation would be independent acquisition from the host genome in HHV-8, MHV-68, and HVS-2, although the fact that the gene is in equivalent positions in these genomes would favor the former. In other homolog cases, a single event of gene acquisition is easier to delineate; for example, the interferon regulatory factor and the macrophage inflammatory protein families are only found in RRV and HHV-8; they are at the same loci in both genomes and hence were presumably captured before host speciation by an ancestor of these two viruses.
DISCUSSION
The publication of the human genome has provided the opportunity to analyze host-parasite interactions in a new light. Herpesviruses capture genes from their host and use them to their own advantage. In the present study, we have analyzed virus-host protein homology using consistent cross-comparative methods for herpesviruses proteins and gene products of the human genome. The study has allowed us to derive a global picture of cellular functions for which herpesviruses have captured and evolved their own counterparts.
Sequence similarity alone revealed a minimum estimate of human homologs in different HHV genomes to be ∼9% to 16% of virus genes, with the exception of HHV-8, which is ∼30% of viral genes. The reason for a higher percentage of homologs in this virus, and in gammaherpesviruses in general, is unclear but may relate to properties of the cell types infected by this subfamily of herpesviruses. Most of the herpesvirus/human homologs identified correspond to proteins involved in immune modulation and apoptotic control. These proteins are normally specific to one or a few viruses, and they often show a complex distribution across the herpesvirus phylogeny tree (Fig. 3). They are, therefore, likely to contribute to the adaptation of the virus to different hosts or different cellular tropisms. This is in contrast to a more stable group of homologs, composed of proteins involved in DNA replication and nucleotide metabolism, components of the well-conserved virus (and host) DNA genome replication machinery.
In our analysis, we have used PSSMs representing herpesvirus protein motifs to increase sensitivity over pair-wise sequence comparison-based searches. The method has allowed us to identify a number of new herpesvirus/human homologs. The new putative functions require experimental testing but are of interest. The HCMV US12 protein family, composed of 10 members, has homology with lifeguard and related human proteins (CGI-119). Lifeguard is known to inhibit the apoptosis signal mediated by the Fas receptor, and therefore, the related HCMV proteins may also have an antiapoptotic role. Viral proteins that interfere with Fas-mediated apoptosis have already been described in gammaherpesviruses (Belanger et al. 2001) but not in betaherpesviruses. This is surprising as HCMV also replicates in cells of the haematopoietic system, namely, monocytes/macrophages. From our analysis, HCMV potentially encodes a repertoire of anti-Fas apoptosis homologs distinct form the gammaherpesvirus FLIP homologs. Interestingly, in the cowpox virus, a member of the Poxviridae family, a gene termed SR1, of unknown function but similar to the CGI-119 protein, was also identified (Shchelkunov et al. 1998).
Homology was found between the HCMV UL1 gene product and the CEA/PSG human protein family. Known functions for the CEA family include involvement in cell adhesion, signal transduction, and possibly innate immunity (Hammarstrom 1999). The PSGs, a subgroup of the CEA family, are mainly expressed in the placenta and are secreted into the maternal circulation, possibly regulating immune system responses. HCMV infection, which is usually benign in immunocompetent individuals, can have catastrophic consequences during pregnancy (Fisher et al. 2000). Infection of the placenta has a 30% to 40% risk of intrauterine virus transmission to the foetus. Similarity of UL1 to PSGs could subsequently be related to the pathology of HCMV during pregnancy or to general immune modulation in the host.
In the present study, we have also detected human gene products that contain the virus BKS ring finger domain, characteristic of K3 and K5 HHV-8 proteins, indicating a possible common origin and shared function for proteins containing this domain. The BKS domain has not previously been reported in mammals. K3 and K5 from HHV-8 have been recently discovered to down-regulate MHC class 1 molecules in infected cells (Coscoy and Ganem 2000; Coscoy et al. 2001); therefore, the BKS domain may be common to virus and host proteins involved in regulating cellular membrane proteins.
We have detected sequence homology with human proteins for ∼13% of all known herpesvirus proteins. The question remains whether the remaining 87% can be considered exclusively viral. It is likely that a fraction may still be functional homologs with global sequence similarity too limited to be detectable by the methods used here. In addition, our methods will not detect very small sequence motifs such as phosphorylation and protein binding sites. Therefore, viral proteins such as HHV8 K15, which contains a tumour necrosis factor receptor–associated factor binding domain (Glenn et al. 1999), or EBV LMP-2A, which contains immunoreceptor tyrosine-based activation motif sequences (Fruehling and Longnecker 1997), are not detected here.
A further confounding factor for detection of viral homologs is the rapid evolution of some viral sequences. It has been estimated that herpesvirus proteins typically evolve one or two orders of magnitude more rapidly than host proteins (McGeoch and Cook 1994), and this may quickly mask any common sequence identifiable ancestry of two proteins. For example, one known human/herpesvirus homolog, thymidine kinase, is present in all known herpesviruses. Because of very limited sequence similarity, however, it could not be identified using our methods; although a human thymidine kinase mitochondrial homolog of the channel catfish herpesvirus thymidine kinase protein was detected. Human homologs of the MHV-68 serpin (serine protease inhibitor) M1 were similarly not identified using sequence similarity searches.
For proteins with viral structural functions, such as capsid constituents and capsid assembly proteins, which make a large proportion of herpesvirus genome coding capacity (20% of the genes of HHV-1), no resemblance to any human protein could be found. This is perhaps not surprising, as these have “viral-only” functions. Recently, however, another method of formulating functional hypotheses of viral proteins, in silico protein structure prediction using threading techniques, has been applied to herpesvirus proteins. This was performed for all proteins of HCMV, yielding complete structural identifications for 36 viral proteins, only eight of which were previously known. These included some HCMV structural proteins (Novotny et al. 2001).
The relative number of homologs between herpesviruses and the human genome may also increase as the prediction methods and number of human gene products from the human genome becomes more accurate. This is highlighted by failure to detect the sequence-based homology between human and herpesvirus α-N-formylglycineamide ribonucleotide aminotransferase (FGARAT), or between human dUTPase and the dUTPase protein family found in all alpha- and gammaherpesviruses (HPF 43). Neither of the human predicted protein data sets contains FGARAT, even though a human FGARAT gene was recently reported (Patterson et al. 1999), and until recently neither contained the human homolog dUTP pyrophosphatase (GenBank accession no. 18583771), which shares homology with its human herpesvirus counterparts. Additional homologs for non-HHV may be identified when their host genome sequence becomes available. The reverse of this argument applies equally to herpesvirus proteins. Many of the ORFs in the herpesvirus genomes are only conceptual translations from the virus genome sequence and are, therefore, predicted hypothetical proteins. Most of the hypothetical proteins are singletons, of which only 4% showed homology with human proteins, in contrast to 10% of the herpesvirus protein families. The analysis of the expression of all ORFs using methods such as DNA array-based profiling (Chambers et al. 1999; Stingley et al. 2000; Jenner et al. 2001) will establish if these potential products are expressed during the virus cycle. Overall, the continued, virus-focused searching of constantly growing protein databases using cross-comparable methods is likely to increase our understanding of the relationship between virus and host.
METHODS
Initial Data Sets
All complete herpesvirus ORFs are available in the viral database VIDA (Albà et al. 2001b). In VIDA, the ORFs are organized into HPFs according to amino acid sequence motifs shared between the proteins, as determined by the XDOM algorithm (Gouzy et al. 1997). In some instances, HPFs contain several proteins from the same virus species. This is owing to the existence of proteins from different strains or to the presence of more than one copy of the gene in the virus genome. Each HPF is annotated with a functional description and functional class, and can contain proteins from any or all of the three herpesvirus subfamilies. The functional descriptions in VIDA include a representative gene name (e.g., “protein kinase, HHV-1 UL13” is a protein kinase family that includes gene UL13 product from HHV-1), and they are used throughout this paper to designate HPFs. When no homology with other herpesvirus proteins can be found, ORFs are represented as singleton proteins in VIDA. A total of 393 homologous multiprotein families (HPFs) and 494 singleton proteins were used in the analysis. This comprises all herpesvirus ORFs from VIDA (4054 nonredundant proteins), including all eight HHVs. VIDA can be accessed at http://www.biochem.ucl.ac.uk/bsm/virus_database/VIDA.html.
The conceptual protein translations of two human genome databases were searched in this study: The collection of human genome gene products at the National Centre for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/genome/guide/human/) and the Ensembl Project at the European Bioinformatics Institute (http://www.ensembl.org/).Both databases were downloaded by anonymous FTP and stored locally. The two databases were concatenated into a single library, and low-complexity protein segments were masked using the SEG program with default parameters (Wootton and Federehen 1993).
Construction of Motif PSSMs
Herpesvirus HPFs containing two or more proteins are defined by one or more amino acid motifs conserved across all members of the family. The large majority of HPFs are identified by a single motif (371 out of 393). However, there are 11 HPFs that contain two conserved motifs, eight HPFs that contain three conserved motifs, and three HPFs that share four motifs. The motifs, in the form of multiple alignments, were used to construct PSSMs using the program PSI-BLAST (Altschul et al. 1997). Taking into account that some families contain more than one motif, the total number of PSSMs we constructed was 429.
Construction of a Herpesvirus Protein Data Set at the 95% Identity Level
A data set of all individual herpesvirus proteins with <95% sequence identity was constructed. The representative proteins were selected by computing the global amino acid identity of each protein in each of the HPFs and grouping the proteins into subsets that shared ≥95% sequence identity using the programs HOMOL and SEQCLUSTER, respectively (Orengo et al. 1997). An ORF was then selected at random from each 95% subset (an N95-rep) and used to perform pair-wise sequence similarity searches of the human protein databases. For example, nine proteins from HPF 13 (protein kinase, HHV-1 UL13) were selected to represent the 33 proteins it comprised.
Database Searches and Sequence Analysis
The IMPALA program (Schaffer et al. 1999) was used to run searches against the 429 PSSMs derived from the motifs in VIDA. An E-value cutoff of 0.01 and default parameters were used. The collection of singleton protein sequences was searched with both BLASTP (Altschul et al. 1990) and PSI-BLAST (Altschul et al. 1997), with default parameters and an E-value cutoff of 0.01. PSI-BLAST uses iterative profile construction and is more computationally expensive but generally more sensitive. As PSI-BLAST did not reveal any additional singleton homologs, N95-reps were then searched against the human protein library using BLASTP with the same parameters as above.
All database hits were examined and curated manually based on sequence alignments, conserved domain regions, functional annotation, and reference to the literature. The manual inspection of putative homologs led to the removal of some of the initial hits, which appeared to be caused by compositional bias rather than true homology. When appropriate, additional proteins from different organisms were retrieved from GenBank for sequence alignment construction. The alignments were produced by the program MULTALIN (Corpet 1988) and, when necessary, manually edited using JALVIEW (http://www2.ebi.ac.uk/∼michele/jalview/contents.html/) and further visualized using BOXSHADE (http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html/). Analysis of homologous families also included searching the domain database at the NCBI, which is linked to the Pfam (Bateman et al. 2000) and SMART (Schultz et al. 2000) domain databases, using reverse position-specific BLAST (RPS-BLAST; Altschul et al. 1997).
Phylogenetic Tree Construction
Herpesvirus phylogenetic trees based on the gene content of 19 complete herpesvirus genomes were previously constructed (Albà et al. 2001a). For this type of reconstruction, phylogenetic profiles were obtained by considering the protein families as molecular function characters for which different viruses were positive (1) or negative (0). Maximum parsimony and distance methods (neighbor-joining) were applied to the phylogenetic profiles to construct phylogenetic trees. The tree shown in Figure 3 represents a consensus tree from such methods (Albà et al. 2001a).
WEB SITE REFERENCES
http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html; BOXSHADE.
http://www.ensembl.org; Ensembl Project at the European Bioinformatics Institute.
http://www2.ebi.ac.uk/∼michele/jalview/contents.html; JALVIEW
http://www.ncbi.nlm.nih.gov/genome/guide/human; National Centre for Biotechnology Information.
http://www.biochem.ucl.ac.uk/bsm/virus_database/VIDA.html; VIDA.
Acknowledgments
We thank Robin Weiss for support and critical reading of the manuscript. This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC; R.H. and M.M.A) and the Medical Research Council (MRC; C.O. and P.K.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL p.kellam@ucl.ac.uk; FAX 44-020-7679-9555.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.334302. Article published online before print in October 2002.
REFERENCES
- Albà MM, Das R, Orengo CA, Kellam P. Genomewide function conservation and phylogeny in the Herpesviridae. Genome Res. 2001a;11:43–54. doi: 10.1101/gr.149801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albà MM, Lee D, Pearl FM, Shepherd AJ, Martin N, Orengo CA, Kellam P. VIDA: A virus database system for the organization of animal virus genome open reading frames. Nucleic Acids Res. 2001b;29:133–136. doi: 10.1093/nar/29.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami A, Koszinowski UH. Viral mechanisms of immune evasion. Trends Microbiol. 2000;8:410–418. doi: 10.1016/S0966-842X(00)01830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger C, Gravel A, Tomoiu A, Janelle ME, Gosselin J, Tremblay MJ, Flamand L. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J Hum Virol. 2001;4:62–73. [PubMed] [Google Scholar]
- Biassoni R, Cantoni C, Pende D, Sivori S, Parolini S, Vitale M, Bottino C, Moretta A. Human natural killer cell receptors and co-receptors. Immunol Rev. 2001;181:203–214. doi: 10.1034/j.1600-065x.2001.1810117.x. [DOI] [PubMed] [Google Scholar]
- Brunovskis P, Kung H J. Retrotransposition and herpesvirus evolution. Virus Genes. 1995;11:259–270. doi: 10.1007/BF01728664. [DOI] [PubMed] [Google Scholar]
- Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan JS, Bittner A, Frueh K, Jackson MR, Peterson PA, et al. DNA microarrays of the complex human cytomegalovirus genome: Profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73:5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston TB, Lidbury BA. Genetic “budget” of viruses and the cost to the infected host: A theory on the relationship between the genetic capacity of viruses, immune evasion, persistence and disease. Immunol Cell Biol. 2001;79:62–66. doi: 10.1046/j.1440-1711.2001.00973.x. [DOI] [PubMed] [Google Scholar]
- Chee MS, Satchwell SC, Preddie E, Weston KM, Barrell BG. Human cytomegalovirus encodes three G protein–coupled receptor homologs. Nature. 1990;344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L, Ganem D. Kaposi's sarcoma–associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci. 2000;97:8051–8056. doi: 10.1073/pnas.140129797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L, Sanchez DJ, Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol. 2001;155:1265–1273. doi: 10.1083/jcb.200111010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Genbacev O, Maidji E, Pereira L. Human cytomegalovirus infection of placental cytotrophoblasts in vitro and in utero: Implications for transmission and pathogenesis. J Virol. 2000;74:6808–6820. doi: 10.1128/jvi.74.15.6808-6820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruehling S, Longnecker R. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology. 1997;235:241–251. doi: 10.1006/viro.1997.8690. [DOI] [PubMed] [Google Scholar]
- Glenn M, Rainbow L, Aurad F, Davison A, Schulz TF. Identification of a spliced gene from Kaposi's sarcoma–associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J Virol. 1999;73:6953–6963. doi: 10.1128/jvi.73.8.6953-6963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy J, Eugene P, Greene EA, Kahn D, Corpet F. XDOM: A graphical tool to analyze domain arrangements in any set of protein sequences. Comput Appl Biosci. 1997;13:601–608. doi: 10.1093/bioinformatics/13.6.601. [DOI] [PubMed] [Google Scholar]
- Hammarstrom S. The carcinoembryonic antigen (CEA) family: Structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses (ICTV) Virus taxonomy: The classification and nomenclature of viruses. The seventh report of the International Committee on Taxonomy of Viruses. San Diego, CA: Academic Press; 2000. [Google Scholar]
- Jenner RG, Boshoff C. The molecular pathology of Kaposi's sarcoma–associated herpesvirus. Biochim Biophys Acta. 2002;1602:1–22. doi: 10.1016/s0304-419x(01)00040-3. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Albà MM, Boshoff C, Kellam P. Kaposi's sarcoma–associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol. 2001;75:891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- McFadden G, Murphy PM. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr Opin Microbiol. 2000;3:371–378. doi: 10.1016/s1369-5274(00)00107-7. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Cook S. Molecular phylogeny of the Alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Mol Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Davison AJ. The descent of human herpesvirus 8. Semin Cancer Biol. 1999;9:201–209. doi: 10.1006/scbi.1999.0093. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Dolan A, Ralph AC. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J Virol. 2000;74:10401–10406. doi: 10.1128/jvi.74.22.10401-10406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague MG, Hutchison CA., III Gene content phylogeny of herpesviruses. Proc Natl Acad Sci. 2000;97:5334–5339. doi: 10.1073/pnas.97.10.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo HG, Reitz MS, Hayward GS. A single 13-kilobase divergent locus in the Kaposi sarcoma–associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny J, Rigoutsos I, Coleman D, Shenk T. In silico structural and functional analysis of the human cytomegalovirus (HHV5) genome. J Mol Biol. 2001;310:1151–1166. doi: 10.1006/jmbi.2001.4798. [DOI] [PubMed] [Google Scholar]
- Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH: A hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Patterson D, Bleskan J, Gardiner K, Bowersox J. Human phosphoribosylformylglycineamide amidotransferase (FGARAT): Regional mapping, complete coding sequence, isolation of a functional genomic clone, and DNA sequence analysis. Gene. 1999;239:381–391. doi: 10.1016/s0378-1119(99)00378-9. [DOI] [PubMed] [Google Scholar]
- Ploegh HL. Viral strategies of immune evasion. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Wolf YI, Ponting CP, Koonin EV, Aravind L, Altschul SF. IMPALA: Matching a protein sequence against a collection of PSI-BLAST–constructed position-specific score matrices. Bioinformatics. 1999;15:1000–1111. doi: 10.1093/bioinformatics/15.12.1000. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelkunov SN, Safronov PF, Totmenin AV, Petrov NA, Ryazankina OI, Gutorov VV, Kotwal GJ. The genomic sequence analysis of the left and right species-specific terminal region of a cowpox virus strain reveals unique sequences and a cluster of intact ORFs for immunomodulatory and host range proteins. Virology. 1998;243:432–460. doi: 10.1006/viro.1998.9039. [DOI] [PubMed] [Google Scholar]
- Somia NV, Schmitt MJ, Vetter DE, Van Antwerp D, Heinemann SF, Verma IM. LFG: An anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc Natl Acad Sci. 1999;96:12667–12672. doi: 10.1073/pnas.96.22.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingley SW, Ramirez JJ, Aguilar SA, Simmen K, Sandri-Goldin RM, Ghazal P, Wagner EK. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J Virol. 2000;74:9916–9927. doi: 10.1128/jvi.74.21.9916-9927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C, Mann DJ, Fleckenstein B, Neipel F, Peters G, Jones N. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature. 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- Tschopp J, Thome M, Hofmann K, Meinl E. The fight of viruses against apoptosis. Curr Opin Genet Dev. 1998;8:82–87. doi: 10.1016/s0959-437x(98)80066-x. [DOI] [PubMed] [Google Scholar]
- Voigt S, Sandford GR, Ding L, Burns WH. Identification and characterization of a spliced C-type lectin-like gene encoded by rat cytomegalovirus. J Virol. 2001;75:603–611. doi: 10.1128/JVI.75.2.603-611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Federehen S. Statistics of local complexity in amino acid sequences and sequence databases. Computational Chem. 1993;17:179. [Google Scholar]