Abstract
Human chromosome 2 was formed by the head-to-head fusion of two ancestral chromosomes that remained separate in other primates. Sequences that once resided near the ends of the ancestral chromosomes are now interstitially located in 2q13–2q14.1. Portions of these sequences had duplicated to other locations prior to the fusion. Here we present analyses of the genomic structure and evolutionary history of >600 kb surrounding the fusion site and closely related sequences on other human chromosomes. Sequence blocks that closely flank the inverted arrays of degenerate telomere repeats marking the fusion site are duplicated at many, primarily subtelomeric, locations. In addition, large portions of a 168-kb centromere-proximal block are duplicated at 9pter, 9p11.2, and 9q13, with 98%–99% average sequence identity. A 67-kb block on the distal side of the fusion site is highly homologous to sequences at 22qter. A third ∼100-kb segment is 96% identical to a region in 2q11.2. By integrating data on the extent and similarity of these paralogous blocks, including the presence of phylogenetically informative repetitive elements, with observations of their chromosomal distribution in nonhuman primates, we infer the order of the duplications that led to their current arrangement. Several of these duplicated blocks may be associated with breakpoints of inversions that occurred during primate evolution and of recurrent chromosome rearrangements in humans.
[Supplemental material is available online at http://www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: T. Newman, C. Harris, and J. Young.]
Humans have 46 chromosomes, whereas chimpanzee, gorilla, and orangutan have 48. This major karyotypic difference was caused by the fusion of two ancestral chromosomes to form human chromosome 2 and subsequent inactivation of one of the two original centromeres (Yunis and Prakash 1982). As a result of this fusion, sequences that once resided near the ends of the ancestral chromosomes are now located in the middle of chromosome 2, near the borders of bands 2q13 and 2q14.1. For brevity, we refer henceforth to the region surrounding the fusion as 2qFus. Two head-to-head arrays of degenerate telomere repeats are found at this site; their head-to-head orientation indicates that chromosome 2 resulted from a telomere-to-telomere fusion (Ijdo et al. 1991). Furthermore, cross-hybridization between 2qFus and various subtelomeric regions has been observed by fluorescence in situ hybridization (FISH) (Ijdo et al. 1991; Trask et al. 1993; Hoglund et al. 1995; Martin-Gallardo et al. 1995; Ning et al. 1996; Lese et al. 1999; Ciccodicola et al. 2000; Park et al. 2000; Bailey et al. 2002; Martin et al. 2002). Thus, the fusion must have occurred after subtelomeric sequences present at the ends of the ancestral fusion partners had already duplicated to/from at least one other chromosome end.
The subtelomeric regions of human chromosomes are particularly dynamic relative to most of the human genome. Sequences have recurrently exchanged, recombined, and duplicated among the ends of nonhomologous chromosomes (for review, see Mefford and Trask 2002). Thus, the entrapment of subtelomeric regions at the more sequestered interstitial fusion site provides a potential opportunity to compare the composition of two ancestral subtelomeres to their counterparts that have persisted at, and propagated among, subtelomeric locations.
Martin et al. (2002) recently presented a clone contig encompassing the fusion and showed homology with several interstitial sites, in addition to subtelomeric sites. Here, we provide more detail on the structure of the DNA surrounding the fusion site and these paralogous relationships. We quantify the extent and degree of homology between this region and paralogous segments elsewhere in the human genome, including sites not described previously. Using these data and observations of the chromosomal location of these sequences in nonhuman primates, we infer the history of some of the duplications and rearrangements that have occurred during recent primate evolution. The extensive homology among 2qFus-related regions of the genome may have mediated—or been the result of—some of the rearrangements that distinguish the karyotypes of higher primates and that may now interact to cause chromosome rearrangements in humans.
RESULTS
Chromosomal Distribution of Sequences from the 2q13–2q14.1 Fusion Region
Bacterial Artificial Chromosome (BAC) RP11–395L14 (AL078621) contains 789 bp of degenerate telomere repeats organized in two head-to-head arrays and overlaps the ancestral fusion site (Fig. 1A) (Martin et al. 2002). Using this BAC as the seed, we independently assembled a 614-kb contig surrounding the fusion site using publicly available BAC sequences (Fig. 1A). The finished BACs in the contig are 99.9%–100% identical in their regions of overlap. Our 2qFus contig is consistent with the automated assembly of this region performed by University of California in Santa Cruz (UCSC) and National Center for Biotechnology Information (NCBI) (http://genome.ucsc.edu/ and http://www.ncbi.nlm.nih.gov) and recent analyses by Martin et al. (2002).
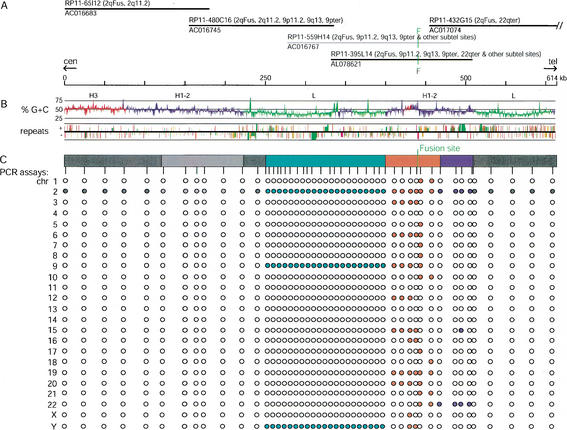
Figure 1.
Chromosomal distribution, GC content, and repeat content of 614-kb DNA sequence surrounding the 2q13–2q14.1 fusion site (2qFus). (A) Each black line indicates the sequence coverage of finished Bacterial Artificial Chromosome (BAC) clones assembled into the 2qFus contig on the basis of their 99.9%–100% identity in regions of overlap. BAC RP11–559H14 is unfinished (gray line); it overlaps RP11–480C16 and –395L15 with 99.7% identity, thereby confirming their overlap. Clone names are followed by chromosomal locations determined by fluorescence in situ hybridization; accession numbers are given below the lines. RP11–432G15 extends 35 kb off the right side of the map. Note that final GenBank entries for some of the BACs have been trimmed to remove overlap among clones exceeding 2 kb. The green vertical line marks the location of the inverted degenerate telomere repeats at the fusion site. The fusion site is immediately flanked by a telomere-associated repeat, TAR1, a repeat that is commonly found in close association with terminal telomere arrays and sometimes found near interstitial degenerate telomere repeats. (B) The G + C trace shows a graph of %GC content with window sizes of 500 bp for local content. Red, blue, and green regions represent H3, H1–H2, and L isochores, respectively, as defined by GESTALT using a 30-kb sliding window. The location, strand, age, and type of interspersed repetitive elements are shown in the third trace, also generated with GESTALT (Long Interspersed Elements [LINEs], green; Alus, red; Mammalian-wide Interspersed Repeats [MIRs], purple; all other repeats including retroviruses, Long Terminal Repeats [LTRs], Mammalian Apparent LTR-retrotransposons [MaLRs], etc., brown). The age of each element is indicated by the height of the feature; taller features represent evolutionarily more recent insertions. (C) Chromosomal distributions of sequence homologous to the 2qFus region as determined by PCR assays on a monochromosomal hybrid panel. Filled and open circles denote positive and negative PCR assays, respectively. The precise locations of the PCR assay are indicated by the vertical tick marks. The colored bar delineates regions with different chromosomal distributions. The light gray block within gray block indicates sequence duplicated within chromosome 2 (see Fig. 3).
We used a combination of three approaches in order to confirm the assignment of BACs forming the 614-kb contig to chromosome 2qFus and to investigate the chromosomal distribution of paralogous sequences.
PCR Analyses of Monochromosomal Hybrid Panel
First, we designed 48 PCR primer pairs, which amplify DNA free of known repeats, across the contig and performed PCR assays on DNA from a panel of hybrid cell lines, each containing a different human chromosome against a rodent background (Fig. 1C). As expected, all primer pairs amplified products from chromosome 2. However, only a portion of the assembled sequence—a total of ∼350 kb on the ends of the contig—is unique to chromosome 2.
Sequences closely flanking the telomere-repeat arrays (red zone, Fig. 1C) amplified from seven or more chromosomes, with one assay amplifying a product from 13 different chromosomes, including chromosome 22. A 40-kb block common to only chromosomes 2 and 22 and defined by four PCR assays (blue zone) adjoins the region of multichromosomal segments. One assay within this block is also positive for chromosome 15, due to the retrotransposition of a processed pseudogene of SNRPA1 from the intron-containing copy on chromosome 15 prior to the segmental duplication that gave rise to the larger block of homology between 2qFus and 22qter (Fan et al. 2002). On the opposite side of the telomere-repeat arrays is a 150-kb block (green zone) defined by 21 PCR assays that are common to chromosomes 2, 9, and Y in the hybrid panel.
Fluorescence In Situ Hybridization (FISH)
Second, in order to more precisely define the chromosomal location of sequences homologous to this region, we performed FISH analyses using the five BAC clones comprising the 2qFus contig. The results are summarized in Figure 1A and shown schematically for three of the BACs in Figure 2. Sequences in RP11–395L14, which contains the fusion site, hybridize to five prominent sites, 2qFus, 9q13, 9p11.2, 9pter (9p24), and 22qter (22q13.3), as well as to several other chromosomal ends with lower intensity. RP11–480C16 produces FISH signals at 2qFus, 2q11.2, 9p11.2, 9q13, and 9pter, as observed recently by Martin et al. (Martin et al. 2002). RP11–65I12 produces signals at 2qFus and 2q11.2 (not shown). On the other side of the fusion site, RP11–432G15 produces FISH signals at 2qFus and 22qter. Although the hybrid-panel analyses had implicated these chromosomes, FISH demonstrates that there are at least three sites of homology on chromosome 9 and two sites on chromosome 2.
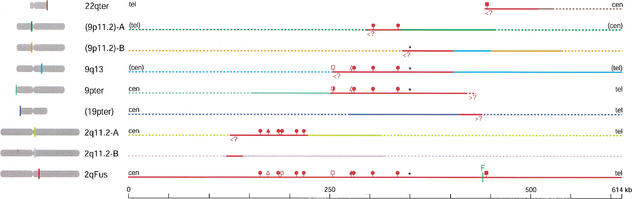
Figure 2.
Summary of fluorescence in situ hybridization (FISH) analyses of hominid chromosomes using BACs RP11–480C16 (green), RP11–395L14 (blue), and RP11–432G15 (red) derived from the ancestral fusion site in human 2q13–q14.1. Data for chromosomes 2, 9, and 22, which carry the major blocks of paralogy in human, are shown in the top panels so that the banding patterns are not obscured by the FISH signals. Other chromosomes are shown in the bottom panel. A total of at least six metaphase spreads were examined for each probe in each species (one individual each). Hybridization signals seen at each location were scored from digitized images on an intensity scale of 1–4 on each probe in each species. The cumulative scores were normalized to that of the location with highest cumulative score in each experiment. The area of each dot is proportional to this normalized score. Dots are aligned with the midpoint of observed FISH signals for each location. The asterisk indicates where hybridization was seen on one homolog only. Ideograms are redrawn from Yunis and Prakash (1982). Accession numbers for the three clones are given in Fig. 1. Human 2p- and 2q-specific clones, RP11–90H11 and RP11–47E6, respectively, were used to verify the identity of hominid chromosomes orthologous to 2p and 2q (i.e., chimpanzee 12 and 13, respectively) (not shown).
Surprisingly, FISH signals were not observed on chromosome Y in five tested individuals, despite the fact that the PCR analyses of the Y-containing hybrid indicated the presence of ∼100 kb of paralogy to the two clones used as probes. FISH also failed to detect this homologous sequence in metaphase chromosomes prepared directly from the hybrid line. A visibly intact Y was the only human material detected in this hybrid cell line when 20 cells were analyzed by FISH with a human-specific repetitive-sequence probe or by reverse painting (not shown). However, these techniques could miss a small fragment of non-Y human material, especially if present in a small subpopulation of cells. Because the sequences of PCR products generated from the Y hybrid are 99.8% identical to sequences derived from the 9q13 paralog (see following), over a total of 3.7 kb sampled (not shown), we conclude that the “Y” homology is actually a 9q13 contaminant in the hybrid line.
Database Mining and Sequence Alignment
Third, we conducted a BlastN search of all finished and draft sequences publicly available as of February 28, 2002 in order to identify sequences paralogous to the 614-kb region of 2qFus. Results are summarized in Table 1 and Figure 3. Some, but not all, of these paralogous segments were detected in earlier whole-genome scans for duplications (Bailey et al. 2001; Martin et al. 2002).
Table 1.
Extent and Degree of Sequence Similarity Among Paralogous Blocks Related to 2qFus
| 2q11.2-A 34G16 AC008268 | 2q11.2-B 468G5 AC009238 | 9pter 174M15/143M1 AL356244/AL449043 | 9q13 561O23/88I18/274B18 AL353608/AL161457/AL353616 | (9p11.2)-A 15J10 AL512605 | (9p11.2)-B 403A15 AL445925 | (19pter) 34P13 AL627309 | 22qter n1g3/n94h12 AC002055/AC002056 | |
|---|---|---|---|---|---|---|---|---|
| 2qFus | ≥97.5 kb 95.8% [355, 2740] | 20.5kb 96.0% [84, 278] | ≥167.8 kb 98.9% [257, 6822] | ≥149.4 kb 98.2% [270, 12821] | ≥42.3 kb 98.5% [63, 541] | ≥62.5 kb 98.1% [240, 12524] | ≥29.5 kb 98.8% [41, 104] | ≥67.3 kb 98.6% [83, 3715] |
| 2q11.2-A | ≥16.1 kb 99.0% [22, 76] | |||||||
| 9pter | ≥150.5 kb 98.2% [321, 12908] | ≥42.1 kb 98.4% [71, 413] | ≥64.2 kb 98.1% [224, 12100] | ≥8.5 kb 99.6% [6, 11] | ||||
| 9q13 | ≥42.4 kb 99.0% [75, 380] | ≥109.8 kb 98.8% [193, 1286] | ||||||
| 9p11.2-A | ? |
Extent of homology and percent identity of paralogous blocks related to 2q13–q14.1 fusion region. Only base substitutions, not insertions or deletions, are considered in the calculations of percent identity (see Methods section). The number of insertions or deletions and the total number of bases in these gaps are given in brackets. The extent of homology is defined by the sequence indicated at the beginning of each row and is a minimal estimate in all but one case, since available sequence terminates within the regions of homology. All clones are from the RP11 BAC library except the chromosome-22 cosmids, n1g3 and n94h12.
Figure 3.
Summary of regions of homology with portions of the 614-kb sequence surrounding the fusion site on 2q13–2q14.1 that were identified by BlastN searches of finished genomic sequence publicly available as of February 28, 2002. Table 1 gives the clone names and accession numbers for the paralogous segments. For simplicity, only one of the sequences with homology with only the 68-kb region immediately surrounding the fusion site is shown. Red solid lines indicate the regions with >95% average identity to the 2qFus sequence. These lines are drawn with reference to the 2qFus sequence; the actual lengths of the paralogous segments may be slightly longer or shorter than those drawn because of distributed insertions and deletions. Red dotted lines indicate adjoining regions with no available sequence, but that were shown by PCR to be homologous to 2qFus (see Fig. 1). Different colors are used to indicate divergent sequence, with solid lines indicating the extent of contiguous sequence coverage, and dotted lines indicating either unavailable sequence or neighboring sequence that lacks homology with any of the other segments shown. Blocks 9p11.2-A and -B map to the pericentromeric region of 9 by fluorescence in situ hybridization and hybrid panel analyses and are tentatively assigned to 9p11.2. Orientation indicated in parentheses is tentative; the presence of alpha-satellite-like repeats in the right-most portion of the 9p11.2-A sequence indicates that it runs telomere to centromere as drawn. (<?) Indicates that 2qFus homology could extend farther in direction of arrowhead. The small red symbols on stalks indicate the most recent Alu insertions in the 2qFus region and/or its paralogs. The most recently active families, AluYb8 and AluYa5, are indicated by squares and triangles, respectively, and circles indicate the older class of AluY elements. Filled and open symbols indicate that the particular element is present or absent, respectively. The partially filled symbol in 9pter indicates that this AluYb8 element is not present in all 9pter alleles (see text). The asterisks mark the position of a highly variable SATR1 repeat common to at least four of the paralogous segments; it is 15,109 bp long in 9q13, 15,021 bp in 9p11.2-B, 4919 bp in 9pter, and 3881 bp in 2qFus.
At least two paralogous segments reside in 2q11.2. One, called 2q11.2-A, is found in the finished sequence of RP11–34G16; it encompasses ≥98 kb and is 95.8% identical to the centromere-proximal portion of 2qFus. Our FISH analyses of this clone confirm its 2q11.2 location as indicated by the NCBI and UCSC assemblies: Signals are observed at both 2q11.2 and 2qFus (Supplementary Fig. IA, available online at http://www.genome.org). This intrachromosomal identity explains why clones RP11–65I12 and −480C16 from 2qFus give FISH signals on 2q11.2: over 55 kb and 45 kb of their inserts, respectively, match this paralogous segment in 2q11.2 at >95% identity. A second 20.5-kb block of 2qFus homology (96.0% identity), called 2q11.2-B, is in the finished sequence of RP11–468G5. FISH confirms the 2q11.2 location of this paralogy: This BAC gives a strong FISH signal in 2q11.2 and a weak signal at 2qFus (not shown). Although they are 99.0% identical over ∼16 kb, clones RP11–34G16 and −468G5 represent distinct paralogous blocks in 2q11.2. Sequences neighboring the paralogy are very different (Fig. 3), and a dissimilarity of 1% is greater than expected from the combination of allelic variation and sequencing errors. The two 2q11.2 blocks are not resolvable by FISH in metaphase chromosomes, however.
The paralogy between 9pter (9p24) and 2qFus was reported to reside in RP11–174M15 and RP11–143M1 by Martin et al. (2002), but was not analyzed in detail. Our analyses show that the paralogy extends at least 168 kb with 98.9% overall identity. The clones overlap by 49 kb with 100% identity, and FISH to 9pter (most intensely), 9p11.2, 9q13, and 2qFus. In addition, RP11–143M1, the more distal clone, hybridizes to multiple chromosomal ends (Supplementary Fig. IB, available online at http://www.genome.org).
Paralogy in 9q13 can be found in overlapping finished sequences from RP11–561O23, RP11–88I18, and RP11–274B18. The first two clones overlap by ∼35kb with 99.7% identity, and the latter two by ∼25 kb with 99.9% identity (before overlaps were trimmed to 2 kb in the latest GenBank entries), and thus are very likely to derive from the same locus. Our FISH analyses of RP11–561O23 confirm its assignment to 9q13 by NCBI and UCSC. It generates FISH signals on 9q13, 9p11.2, 2qFus, and 9pter, and 9q13 is the brightest site (Supplementary Fig. IC, available online at http://www.genome.org). This 9q13 paralogy to 2qFus spans at least 149 kb with overall identity of 98.2%.
We identify two additional blocks of 2qFus paralogy that derive from 9p11.2 or 9q13. One segment, which we call (9p11.2)-A, spans >42 kb within RP11–15J10 and is 98.5% identical to 2qFus within the region that is paralogous to 9q13. A second block of 2qFus paralogy, called (9p11.2)-B, is in RP11–403A15. This clone contains ∼63 kb homology at 98.1% identity to 2qFus, and ∼110 kb homology at 98.8% identity to the 9q13 sequence (red and blue lines in Fig. 3). RP11–15J10 and −403A15 hybridize by FISH most intensely to 9p11.2 and 9q13, and less intensely to 2qFus, 9pter, and many pericentromeric sites (Supplementary Fig. ID for RP11–15J10, available online at http://www.genome.org). Although these clones are assigned to 9q13 in the NCBI and UCSC maps, there is no strong justification for this assignment over 9p11.2. These sequences are not connected to other 9q13 BACs by overlapping sequence or end-sequenced BACs, and they contain no radiation-hybrid or linkage markers that are unambiguously assigned to one side of the chromosome-9 centromere or the other. RP11–403A15 and −15J10 have no sequence in common, but must lie sufficiently close to each other that they are not resolvable by metaphase FISH.
As expected from our hybrid panel data and FISH observations made by us and others (references earlier), the multicopy regions immediately flanking the fusion site match many publicly available BAC and cosmid sequences. Clones with homology with these multicopy regions belong to at least 15 different contigs representing different chromosomal ends (not shown). We show only one of the longest available homologies, that in RP11–34P13 (AC073186/AL627309), which is 98.8% identical to 2qFus over ≥29.5 kb and 99.6% identical to the 9pter sequence. This clone contains no chromosome-specific DNA and has been variously assigned to chromosome 1, 7, 18, and 21 in GenBank entries and draft assemblies over the last 2 years. It does not derive from chromosome 1, 18, or 21, since it does not cross-hybridize by FISH to these chromosomes in any of several individuals analyzed (not shown). It is likely to be a variant chromosome allele of 19, as it shares extensive homology with the 19pter allele sequenced by the Department of Energy Joint Genome Institute (http://www.jgi.doe.gov) and contains sequence variants of the olfactory receptor gene most often found on chromosome 19 in 22 individuals sampled from different ethnic groups (180 chromosomes) (Mefford et al. 2001).
Over 67 kb of sequence distal to the fusion site is 98.6% identical to the q-terminus of chromosome 22. This homology ends just ∼1.4 kb from the array of degenerate telomere repeats and was described previously (Ning et al. 1996; Eichler et al. 1997; Martin et al. 2002). The extent of this homology explains why the 2qFus-clone RP11–395L14 was initially given a GenBank assignment of chromosome 22 when the Sanger Centre (http://www.sanger.ac.uk/HGP) sequenced it.
The putative 150-kb region of paralogy on chromosome Y is not present in the sequence available for chromosome Y in GenBank or Celera's human genome assembly, consistent with the idea that the Y-hybrid results are due to a 9q13 contaminant.
In summary, no more than 254 kb of the 614-kb 2qFus contig is single copy in the genome (≤22 kb and 104 kb on the two sides and 28 kb between the regions of homology with 2q11.2-A and 9pter). The remainder of the sequence is duplicated in at least one other location. So far, we have detected 16 locations with at least 5 kb of homology with 2qFus by at least two of three methods (a reproducible FISH signal, more than one positive PCR assay, or >5 kb of >95% sequence match in a chromosomally assigned genomic sequence). Of these locations, 11 are subtelomeric, and 3 are pericentromeric. An additional 14 sites of homology (of which 11 are subtelomeric) were detected with only a single method. The failure to detect these 14 sites with more than one method is likely due to incomplete sequence coverage, insensitivity of FISH, low density of PCR assays, mismatches to primer sequences, and/or normal polymorphism among the chromosomes analyzed in the three methods.
FISH Analyses of Nonhuman Primates
We confirmed the centromere–telomere orientation of the 2qFus contig by FISH analyses of constituent clones on chimpanzee chromosomes (Fig. 2). Chimpanzee chromosomes 12 and 13 are homologous to human 2p and 2q, respectively (Yunis and Prakash 1982; Wienberg et al. 1994). RP11–480C16, from one end of the 2qFus contig, hybridizes to chimpanzee 12, indicating that it maps to the centromere-proximal side of the fusion site. RP11–432G15, from the other end of the 2qFus contig, hybridizes to chimpanzee 13, indicating that it lies on the centromere-distal side of the fusion site in human. As expected, RP11–395L14, which contains the fusion site, generates signals on both chimpanzee chromosomes 12 and 13.
FISH analyses also reveal changes in location and copy number of paralogous segments that have occurred during hominid evolution. In both human and chimpanzee, RP11–432G15 (red symbols in Fig. 2) hybridizes only to the regions corresponding to 2qFus and 22qter. These two sites are also detected in gorilla and orangutan, indicating that the transfer of material between these locations predated hominid divergence. However, sequences homologous to this clone are distributed on at least 38 additional telomeres and two interstitial sites in gorilla. Hybridization is detected at 14 of the same locations in orangutan. Given the generally accepted hominid lineage (Chen and Li 2001), either orangutan and gorilla independently acquired copies of portions of the RP11–432G15 sequence at these locations, or homologous sequence was deposited at these sites before hominids diverged and then was lost in the ancestor of human and chimpanzee. One interstitial and 26 subtelomeric integration sites are unique to gorilla, indicating that a burst of duplications also occurred along the gorilla-specific branch. One subtelomeric and five interstitial sites are unique to orangutan.
Sequences in RP11–480C16, which hybridize by FISH to five sites in human (two on 2, three on 9), are present in four of the orthologous sites in chimpanzee and three in gorilla and orangutan (green symbols in Fig. 2). Chimpanzee, gorilla, and orangutan all lack cross-hybridizing sequences at the 9p11.2-equivalent location, and gorilla and orangutan are missing an additional signal corresponding to 9pter or 9q13. Because of an inversion with breakpoints in these bands that differentiates human chromosome 9 from its counterpart in gorilla and orangutan, it is not clear whether the remaining conserved location corresponds to 9pter or 9q13, but other evidence (see below) indicates that 9q13 holds the ancestral copy. Five additional subtelomeric locations have detectable homology with RP11–480C16 in chimpanzee.
As in human, blocks immediately flanking the fusion site and contained in RP11–395L14 are multicopy in the chimpanzee, gorilla, and orangutan genomes, and the copies are primarily subtelomerically located (blue symbols, Fig. 2). Because this BAC encompasses blocks whose chromosomal positions were assayed by the two BACs discussed in the preceding two paragraphs, we expected to see marked species differences in the distribution of its FISH signals. Indeed, of 30 subtelomeric locations detected in either human or chimpanzee with reasonable efficiency, 15 are common to both species, and 15 are seen in only one of the two species. Signals were also observed in two additional interstitial locations in chimpanzee. Of the ∼50 locations detected with RP11–395L14 in any of the four tested hominid species, only seven are common to all four species, ∼13 are species-specific locations, and the rest are common to different combinations of two or three species.
Almost all gorilla chromosome ends and half of chimpanzee ends are capped with AT-rich, DAPI-bright bands. These caps are not present on human or orangutan chromosomes. 2qFus homologous sequences are invariably found centromere proximal of these caps when both are present.
Genomic Structure
Base-Pair Composition
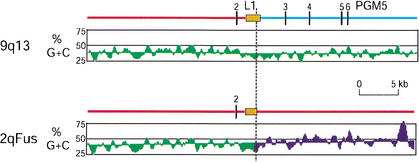
The GC content of the 2q fusion region averages 44%, but it fluctuates markedly across the 614-kb sequence (Fig. 1B). The GESTALT program (Glusman and Lancet 2000) divides the region into five isochores, from centromere to telomere: H3, H1–2, L, H1–2, and L, as defined by Bernardi and colleagues (Bernardi 1995). Each of the isochore boundaries except one corresponds to a boundary between blocks duplicated on different sets of chromosomes (compare Fig. 1B with 1C), consistent with the evolution of the 2qFus region as a patchwork of pieces copied from other genomic locations. For example, the breakpoint in homology between 9q13 and 2qFus (and 9pter) is marked in 2qFus/9pter by an L-to-H1–2 isochore transition, whereas it lies in the middle of an L isochore in 9q13 (Fig. 4). Similarly, the breakpoint between 2q11.2-A and 2qFus creates an isochore transition in 2qFus (H1–2 to L), whereas it lies amid an H1–2 isochore in 2q11.2-A (not shown).
Figure 4.
Disruption of isochore, L1 repetitive element, and PGM5 gene by the breakpoint of duplication between 9q13 and 2qFus. (Green) L isochore; (blue), H1–2 isochore. The regions shown correspond to nucleotides 381651 to 423067 in the 2qFus contig and, for 9q13, from nt 139050 in RP11–561023 to nt 9713 in RP11–88I18. The breakpoint of homology (dotted line) was determined by cross_match (http://www.genome.washington.edu/phrap_documentation.html) analysis, the L1PBa repeat was identified by RepeatMasker, and the GC-content and isochore classification were determined by GESTALT. The vertical bars are exons of the PGM5 gene (see Fan et al., 2002 for more details).
Interspersed Repeats
The density and nature of repetitive elements also vary across the 614-kb 2qFus sequence (Fig. 1B). Overall, interspersed repeats occupy 40% of the sequence, with Short Interspersed Elements (SINEs) and Long Interspersed Elements (LINEs) accounting for 12% and 15% of the sequence, respectively. Recent repeat activity helps to date some of the duplication events involving the 2qFus sequence. The full-length AluY, AluYa5, and AluYb8 insertions into the 2qFus-paralogous blocks are indicated in Figure 3. These are the youngest classes of Alu elements found in the region. The AluYa5 and AluYb8 subfamilies have been transpositionally active very recently: 99% of the insertions of these elements are human specific, and ∼25% exhibit presence/absence polymorphism in humans (Carroll et al. 2001). In contrast, the Y class was active earlier during hominid evolution; insertions of Y elements are rarely polymorphic in humans, because the Y class is ∼1000 times less active now than the Ya5 and Yb8 subfamilies (Roy-Engel et al. 2001).
Four Alu insertions in the region are informative for inferring phylogeny (Fig. 3). (1) We find an AluY element in 2q11.2-A that is not present at the corresponding site in the 2qFus paralogous block. Other AluY elements are common to both blocks. Thus, the duplication that spawned these two blocks must have occurred during AluY activity. (2 and 3) Within the region shared by 2qFus, 9q13, and 9pter, we find an AluYb8 inserted only in some alleles of 9pter and an AluYa5 element inserted uniquely in 2qFus (sequence from 9p11.2 for this portion of the paralogous region is unavailable). The 2qFus and 9pter copies are otherwise very similar (Table 1), but the duplication that generated these two copies must have occurred before these Alu elements inserted. The AluYb8 element is present in two of three sequenced clones that overlap this region of 9pter (in RP11–59O6 [AL158832] and RP13–39F9 [AL591968], but lacking from RP11–174M15 [AL356244]). These clones represent allelic variants of 9pter because their overlaps are contiguous and ≥99.7% identical. (4) Another AluYb8 element is common to both 2qFus and 22qter in their region of homology. Its presence in both blocks indicates that sequence was transferred between 22qter and 2qFus (or its unfused predecessor) after the AluYb8 element was inserted. The implications of these observations are discussed below.
Three repetitive elements cross breakpoints of homology and therefore provide clues to the ancestral and derived states of the duplicative transfers. (1) An L1PBa element crosses the red-to-light blue breakpoint in 9q13, but is truncated in the 9pter and 2qFus sequences; consistent with the creation of an isochore transition in 9pter/2qFus (Figs. 1 and 4). (2) An AluJb element is truncated in 19pter at the dark blue-to-red breakpoint of homology with 2qFus, but crosses the breakpoint in 2qFus. The breakpoint also creates an L-to-H-2 isochore transition in 19pter, but leaves the H1–2 isochore intact in 9pter/2qFus (not shown). (3) An L1ME element is truncated by the duplication from 2qFus to 2q11.2 (red-to-light green breakpoint); it crosses the breakpoint in 2qFus. This case is the only one encountered in which the direction of transfer indicated by the repeat-element is opposite that inferred from the isochore-transition pattern.
Sequence Variation Across the 2q13–2q14.1 Fusion Site
The head-to-head arrays of repeats at the fusion site in RP11–395L14 have degenerated significantly (14%) from the near perfect arrays of (TTAGGG)n found at telomeres. Comparison of the fusion site in RP11–395L14 with an 1873-bp sequence from a different individual (M73018) (Ijdo et al. 1991) reveals a high degree of variation in the length and sequence of the head-to-head arrays of degenerate telomere repeats (not shown). Overall, the two sequences show only 90% sequence identity. More differences are observed within the degenerate telomere arrays (88% identity) than in the sequences immediately flanking them (97.6% identity; 94.9% when each of the bases in insertions and deletions, which range in size from 1 to 8 bp, are counted as mismatches). Only 48% of the 127 repeats in RP11–395L14 and 46% of the 158 repeats in M73018 are perfect TTAGGG or TTGGGG units. Deviation from the canonical telomeric repeat appears to be randomly distributed across the fusion site in both alleles (not shown).
Two short arrays of degenerate telomere repeats, in addition to the arrays marking the fusion site, are found within 2qFus. They are 181 bp and 248 bp long, and 17 kb and 21 kb distal of the fusion site, respectively. Interstitial arrays of degenerate telomere arrays are common in the human genome, particularly in subtelomeric regions (Riethman et al. 2001). Like the array at the fusion site, these arrays are highly diverged from the prototypic telomeric repeats (70% and 86% identical to [TTAGGG]n, respectively). A SATR1 (satellite) repeat cluster within the block common to 2qFus, 9pter, 9q13, and 9p11.2-B (asterisks in Fig. 3) also shows high variability in length, especially when compared with the overall high identity of these blocks.
DISCUSSION
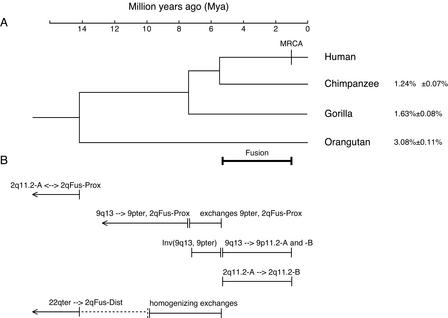
The gross characteristics of the chromosomal fusion that gave rise to human chromosome 2 were apparent 20 years ago, when Yunis and Prakash aligned the high-resolution banding patterns of human, chimpanzee, gorilla, and orangutan chromosomes (Yunis and Prakash 1982). The identities of the fusion partners were confirmed 10 years later when human chromosome-2 specific DNA was observed to “paint” chimpanzee chromosomes 12 and 13 (Jauch et al. 1992; Wienberg et al. 1992). Because the fused chromosome is unique to humans and is fixed, the fusion must have occurred after the human–chimpanzee split, but before modern humans spread around the world, that is, between ∼6 and ∼1 million years ago (Mya; Chen and Li 2001; Yu et al. 2001) (Fig. 5). This gross karyotypic change may have helped to reinforce reproductive barriers between early Homo sapiens and other species, as the F1 offspring would have had reduced fertility because of the risk of unbalanced segregation of chromosomes during meiosis.
Figure 5.
Estimated timing of duplications, inversions, and relocations of 2qFus-paralogous blocks based on sequence identity measures and fluorescence in situ hybridization analyses of hominids. (A) Phylogeny of hominids. The branch lengths and estimated speciation dates are based on sequence analyses of 53 autosomal intergenic nonrepetitive DNA segments analyzed by Chen and Li (2001). The speciation dates are drawn at the midpoint of estimated ranges: human–chimpanzee, 4.6–6.2 Mya; human–gorilla, 6.2–8.4 Mya; human–orangutan, 12–16 Mya. MRCA marks the estimate of the time of the most recent common ancestor of all modern humans (Yu et al. 2001). The average ± SD percent sequence substitution (Jukes-Cantor model, excluding indels) between human and each of the three other hominids is given at the right (Chen and Li 2001). (B) Estimated timing of events involving 2qFus-paralogous blocks. The blocks are identified by their current positions in humans (i.e., 2qFus-Dist and 2qFus-Prox are the regions forming the distal and proximal sides of the 2q fusion site, respectively; both were at chromosome tips when the duplicative transfers occurred). Ancestral and derived states, when indicated, are inferred from breaks that disrupt genes, specific repetitive elements, or isochore patterns; the copy carrying the full-length gene/element and/or lacking an isochore transition at the breakpoint is assumed to represent the ancestral version.
Molecular Characteristics of the Fusion
When observed at the sequence level, the ancestral chromosomes appear to have undergone a straightforward fusion. The sequence of RP11–395L14, like the cosmid partially sequenced by Ijdo et al. (1991), shows two head-to-head arrays of degenerate telomere repeats at the 2q fusion site, with no other sequence between the arrays. This observation indicated that the two ancestral chromosomes had joined end-to-end within the terminal telomeric repeats, with subsequent inactivation of one of the two centromeres. Kasai et al. (2000) showed using FISH that the chromosomes underwent no gross alteration in structure: The relative order of 38 cosmids derived from 2q12–2q14 was the same on human chromosome 2 and the short arms of chimpanzee chromosomes 12 and 13. Although the sequence is not yet available from the terminal regions of chimpanzee chromosomes 12p and 13p with which to compare to human 2q13–2q14.1, the human sequence is very similar to two extant human subtelomeres (9pter and 22qter) (Fig. 3, Table 1). Very little, if any, distal material is unaccounted for in the two comparisons. Although neither 9pter nor 22qter has been sequenced into the telomeric arrays, the available sequences for these chromosomes match 2qFus to within 21 kb and 1.4 kb of the array at the fusion site, respectively, and PCR assays indicate that homology with 9pter extends to at least 8.4 kb from the array.
If the fusion occurred within the telomeric repeat arrays less than ∼6 Mya, why are the arrays at the fusion site so degenerate? The arrays are 14% diverged from canonical telomere repeats (not shown), whereas noncoding sequence has diverged <1.5% in the ∼6 Mya since chimpanzee and humans diverged (Chen and Li 2001) (Fig. 5). There are three possible explanations: (1) Given the many instances of degenerate telomeric arrays within the subtelomeric regions of human chromosomes (Riethman et al. 2001), the chromosomes joined at interstitial arrays near, but not actually at, their ends. In this case, material from the very ends of the fusion partners would have been discarded. (2) The arrays were originally true terminal arrays that degenerated rapidly after the fusion. This high rate of change is plausible, given the remarkably high allelic variation observed at the fusion site. The arrays in the BAC and the sequence obtained by Ijdo et al. (1991) differ by 12%, which is high even if some differences are ascribed to experimental error. (3) Some array degeneracy could be a consequence of sequencing errors. We have not been able to PCR successfully across the fusion site, which would be required to assess the contribution of sequencing errors to this measure of fusion-site sequence polymorphism. However, explanation 2 is supported by the high variability among allelic copies of other interstitial telomeric repeats and associated regions sequenced by Mondello et al. (2000) (AF236886 and AF236885). Considering the high mutability of interstitial telomere repeat arrays, the fusion partners could have joined either within terminal or subterminal arrays to form chromosome 2.
Segmental Duplications
By using PCR analyses of a hybrid panel, genomic sequence alignment, and FISH, we demonstrate that ≥360 kb of the region surrounding the fusion site is duplicated at least once elsewhere in the genome. These paralogous segments are distributed primarily in subtelomeric and pericentromeric locations, consistent with the distribution of segmental duplications found in a recent whole genome survey (Bailey et al. 2001) and earlier FISH analyses (Ijdo et al. 1991; Trask et al. 1993; Hoglund et al. 1995; Martin-Gallardo et al. 1995; Ning et al. 1996; Lese et al. 1999; Ciccodicola et al. 2000; Park et al. 2000; Bailey et al. 2002; Martin et al. 2002). Subtelomeric homology spans ∼258 kb. The long blocks shared by 9pter or 22qter on the proximal and distal side of the fusion site, respectively, account for the bulk of this homology. In addition, highly dispersed, multicopy blocks comprise the 68 kb directly surrounding the fusion site. These blocks are relatively short and show 93%–99% identity to various subtelomeres. This complex pattern of homology among present-day subtelomeres and the fusion site indicates that various DNA segments had duplicated among subtelomeric regions, including those of the fusion partners, before the fusion took place (see following).
Very large segments of 2qFus also have homology with nontelomeric sites. These interstitial paralogs are less similar to the 2qFus sequence than are the subtelomeric paralogs (Table 1) and presumably result from earlier duplication events (see following). The fusion region and 2q11.2 share at least 100 kb as the result of large intrachromosomal duplications. Intrachromosomal duplications have also generated at least three large interstitial blocks of homology on chromosome 9, in addition to 9pter.
Many other cross-hybridizing sites were observed in the genomes of nonhuman primates (Fig. 2), reflecting the evolutionary mobility of sequences homologous to the region surrounding the fusion site.
The size and high similarity of these duplications have been problematic for the automated assembly of human genome sequence across these regions. For example, RP11–15J10 has migrated from 2q11.2, 9q13, 9p23, 9q21, and 9q12 in various versions of the genome assembly. It contains Sequence-tagged Sites (STSs) that have been assigned to chromosomes 2, 9, 7, and X, but none is single copy in the genome. Based on our FISH and hybrid-panel results, this clone most likely derives from 9p11.2. RP11–143M1 has migrated from 9q13, to 9p22, to 9pter, its true location. The numbers in Table 1 provide justification for some of this confusion: 9pter and 9q13 are ∼98% identical over a span of >150 kb. We have no explanation for why several 2qFus-related clones have been assigned at one time or another to 9p22–9p23 (RP11–15J10, −403A15, −143M1 and −174M15); none produces a FISH signal there. Unfortunately, some of these localization errors have been propagated in publications (e.g., Mah et al. 2001), which augments the confusion. These examples and the deceptive Y-hybrid results we encountered illustrate the need for multiple mapping methods to address the challenges encountered in the study of segmental duplications.
The History of the Paralogous Sequences
Large Duplications and Pericentromeric Inversions
Based on our sequence comparisons, the oldest event involving 2qFus paralogous blocks was the duplicative exchange between 2q11.2-A and the progenitor of the centromere-proximal side of the fusion site (Fig. 5). These sequences have since diverged by at least 4%. The FISH results indicate that, at the time of this duplication, both regions were located on the p arm of the ancestral chromosome that was later to be a fusion partner. If there has been no ectopic recombination or gene conversion between these two regions since the original duplication, and the two copies have diverged at a rate typical for the hominid noncoding DNA (Chen and Li 2001), then this intrachromosomal duplication predates hominid divergence (16–20 Mya) (Fig. 5). Our FISH data are consistent with this timing: The block is present in both locations in all four hominids analyzed. AluY elements in the blocks are also consistent with this timing: Together, the presence of several AluY insertions common to both blocks and one AluY element in 2q11.2, but not 2qFus, dates this duplication some time during the period of the transpositional activity of this Alu class, that is, early in hominid evolution (Roy-Engel et al. 2001).
The next event was the duplicative transfer of a ≥150-kb block from what are now 9q13 and the ancestor of 9pter or the 2q-forming chromosome. Several lines of evidence indicate that the ancestral copy of this block is now in 9q13. The transfer of material from 9q13 to 2qFus/9pter disrupted an L1PBa element, the PGM5 gene (Fan et al. 2002), and an L isochore. These features cross the breakpoint in 9q13, but are truncated in 2qFus/9pter (Fig. 4). The divergence between 9q13 and these other blocks is now ∼2%. This divergence indicates that this duplication predated the gorilla–chimpanzee–human split (Fig. 5). FISH data would indicate a more recent date, because only one location is labeled in gorilla and orangutan, compared with sites corresponding to both 9q13 and 9pter in human and chimpanzee (not 9p11 and 9pter, as reported for chimpanzee by Martin et al. [2002]). The location in gorilla coincides, at cytogenetic resolution, with a breakpoint of the 9pter–9q13 inversion that occurred after human and chimpanzee branched off from gorilla. Given their sequence divergence, it is possible that the 9q13 and 9pter blocks began as a tandem duplication in the common ancestor of human, chimpanzee, and gorilla, but were visibly separated by the pericentromeric inversion that occurred later in the ancestor of human and chimpanzee (Fig. 2). Two closely juxtaposed copies would not be visible by FISH in metaphase chromosomes. We have observed a similar staging of the steps leading to two copies of a portion of chromosome 3 containing olfactory receptor genes (Brand-Arpon et al. 1999).
Two duplications of material from 9q13 to 9p11.2 were the next evolutionary events to occur involving 2qFus blocks on chromosome 9. FISH signals appear at 9p11.2 only after human and chimpanzee diverged (Fig. 2), and the 9p11.2-A and -B blocks are more similar to sequence in 9q13 than in 9pter or 2qFus (Table 1). One duplication involved a ≥42-kb segment, and the other a ≥110-kb segment. (We surmise that these blocks derive from 9p11.2, but they may represent additional copies from 9q13, as FISH signals are equally bright in 9p11.2 and 9q13.) These blocks adjoin in the 9q13 sequence, but are distinct in the regions represented by the 9p11.2-A and -B sequences (Fig. 3). The blocks could have transposed independently, or together and then been separated by the insertion of other material. Assuming that there has been no further exchange between the blocks in these two bands, the degree of their divergence (1.0% and 1.2%) also dates the duplication(s) soon after the human–chimpanzee split (Fig. 5). After human and chimpanzee diverged, the human 9q12 heterochromatic region expanded, placing the 9q13 paralogous segment much further from the centromere on the human chromosome than the chimpanzee ortholog. Chromosome 9 also underwent a second inversion along the chimpanzee branch after the chimpanzee–human split. Although one inversion breakpoint maps at cytogenetic resolution close to the 9p11.2 paralog, this sequence is unlikely to be involved in the rearrangement, because it appears at this location only on human chromosome 9. These rearrangements may explain why Martin et al. (2002) assigned this paralogous block to the site corresponding to 9p11.2 instead of 9q13 in chimpanzee (see earlier).
The two chromosomes that joined to form human chromosome 2 each underwent pericentromeric inversions in hominid evolution (Fig. 2). Two of the four breakpoints map near the telomeric regions that were involved in the fusion, but the sequences with homology with the fusion site appear to lie outside of the inverted segments.
Subtelomeric Exchanges
Our study also adds to the complex picture of interchromosomal subtelomeric duplications. Duplications among subtelomeres generated a block common to the chromosome destined to become human 2p and the ancestor of 9pter, and another block common to the chromosome destined to become human 2q and the ancestor of 22qter. Although we are not able to infer the direction of the 9pter–2qFus transfer from the available information, 22qter represents the ancestral state and 2qFus the derived state: The breakpoint is marked by an isochore transition in 2qFus, not 22qter, and the ACR gene is intact in 22qter, but truncated in 2qFus (Fan et al. 2002; Martin et al. 2002). The divergence between these pairs of blocks (1.1% and 1.4%, respectively) dates both events at or around the time of chimpanzee–human speciation. This similarity is surprising, however, given the age of the duplications indicated by FISH analyses of other hominids. The 2q clone containing the 2qFus/22qter-homology block hybridizes to both of these sites in human, chimpanzee, gorilla, and orangutan, dating the duplication before hominid divergence. However, Martin et al. (2002) observed signals only on the chromosome-22 equivalent site in orangutan and an Old World monkey when using a clone derived from chromosome 22 containing the 2qFus/22qter as their FISH probe. It is therefore possible that the signals we see on the 2q ancestor in orangutan represent an independent duplication in orangutan of a different segment of the 2qFus sequence. Even taking the more conservative view—that the duplication from 22qter to the 2qFus ancestor occurred just before the human–chimpanzee–gorilla split—the two blocks must have undergone homogenizing ectopic exchanges at least up until the fusion event to reconcile the fact that these sequences are now only 1.4% different. The fact that the blocks in 2qFus and 22qter now carry the same AluYb8 insertion is strong evidence that these blocks exchanged sequence since humans and chimpanzee diverged. Members of the AluYb8 family have been actively retrotransposing only since human–chimpanzee divergence and occur almost exclusively in the human genome (Carroll et al. 2001).
The evolutionary mobility of subtelomeric regions is further underscored by the gross differences in the location and number of subtelomeric blocks observed among hominids. Considering the extensive polymorphism and recurrent exchanges among subtelomeres (Mefford et al. 2001; Mefford and Trask 2002), it may be unreasonable to expect that a linearly branching pedigree of subtelomeric duplications can ever be deduced.
Breakpoints
Are there sequences at the breakpoints of homology blocks that might shed light on the duplication and exchange processes that have acted on these regions? Half of the breakpoints defining the pairs of major paralogous blocks in Table 1 can be pinpointed at the sequence level because sequence of both partners is available where the homology breaks down. We observe no element that is common to the available breakpoints of paralogous segments. Several occur in LINE elements, and others are in nonrepeat sequences that have no homology with each other. Four breakpoints appear to occur within a common L1PBa element, but all concern the same events—the displacement of 9q13 homology in the ancestor of 2qFus and 9pter by sequences common to multiple telomeres (see also Fan et al. 2002). The duplication that generated copies on 9pter and 2qFus occurred after this event, so that both of these locations share the same breakpoint with 9q13 and its copy in 9p11.2. Overall, the diversity among the breakpoints indicates that the duplications and exchanges occurred by mechanisms involving random double-strand breaks in DNA rather than special common sequences.
Paralogy and (Deleterious) Rearrangements
We have provided two examples in which blocks paralogous to the fusion site are potentially involved in gross chromosomal rearrangements that have occurred during hominid evolution. These large blocks of homology may also sporadically mediate gross rearrangements in humans. Bands 9p11.2 and 9q13 contain the breakpoints of common pericentromeric inversion polymorphisms in humans (Samonte et al. 1996). The highly similar blocks identified here in these bands (≥40-kb blocks of 99%) could mediate homologous recombination and cause some of these inversions. This hypothesis could be tested by comparing the sequence of the common and inverted forms of chromosome 9; the breakpoints should map within the paralogous blocks. In addition, we would expect the two interacting blocks to lie in opposite orientation on the chromosome. The tentative orientations of the blocks in 9q13 and 9p11.2-A (Fig. 3) are consistent with this expectation. These blocks may also be involved in the formation of a dicentric chromosome 9 with tandem head-to-tail duplication of the 9p11–q13 region reported by Lukusa et al. (2000). Further analyses will be needed to determine if these blocks of homology bound the duplicated segments.
The 2qFus-paralogous blocks are also good candidates for involvement in recombination events that cause other de novo rearrangements of human chromosomes. For example, a deletion of the material between 2q11 and 2qFus has been noted in a patient with acute myeloid leukemia in the Mitelman catalog of chromosome abnormalities (http://cgap.nci.nih.gov/chromosomes/CytSearchForm). The catalog also contains at least 60 cases from a wide variety of tumors in which one of the bands containing 2qFus paralogy is joined to unidentified material to form an unbalanced rearrangement.
It has also been suggested that interstitial telomeric sequences are sites of preferential chromosome breakage, amplification, and recombination (Bertoni et al. 1994; Boutouil et al. 1996; Slijepcevic et al. 1996; Simi et al. 1998; Desmaze et al. 1999). Some internal telomeric repeats map at cytogenetic resolution together with mapped fragile sites (Musio and Mariani 1999). The inverted telomeric repeat array was a candidate for the FRA2B, which is located in 2q13 (Williams and Howell 1977; Sutherland and Mattei 1987), but published data (Ijdo et al. 1992) and our own experiments (CF, YF, and BT; unpublished results) show that the FRA2B site maps proximal of the 614-kb region described here.
In the accompanying paper (Fan et al. 2002), we characterize 11 genes within the 2qFus sequence. As a consequence of the various intra- and interchromosomal duplications documented here, 9 of these genes are present in the human genome in more than one copy. Thus, in addition to their historical contributions to the gross structural changes among hominid chromosomes and possible involvement in chromosomal rearrangements in humans, duplications and rearrangements of 2qFus-paralogous blocks also have functional relevance.
METHODS
Database Mining and Sequence Analyses
The sequence of RP11–395L14 served as the entry point for this study. Homologous sequences were obtained iteratively from GenBank (Benson et al. 2002) by BlastN (http://www.ncbi.nlm.nih.gov/BLAST/). Finished sequences from different clones were assembled into the same contig only if their overlap was contiguous and ≥99.7% identical, a reasonable allowance for polymorphism and sequencing errors. We screened for interspersed repeats with RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). We used GESTALT (http://bioinformatics.weizmann.ac.il/GESTALT/) (Glusman and Lancet 2000) to characterize GC and repeat content. Pairwise alignments were performed with BLAST2, without repeat masking and with gap-initiation and gap-extension parameters adjusted to minimize breaking long matches into pieces at sites of insertion/deletions. Percent identities of paralogous blocks were calculated from the BLAST2 output as follows (Linardopoulou et al., in prep.). First, the number of nucleotide substitutions between the two sequences was counted and divided by the number of aligned bases (both numbers exclude gaps). This observed proportion (p) was entered in the Jukes-Cantor equation to estimate K, the number of nucleotide substitutions per site. The Jukes-Cantor equation takes into account that multiple substitutions might have occurred at the same site and is as follows: K = −(3/4) ln [1–(4p/3)] (Jukes and Cantor 1969). The percent identity of the aligned sequences is therefore 100%*(1−K). The number and size of gaps in alignments caused by insertions and deletions were also extracted from the BLAST2 output. The assembled 2q13–q14.1 sequences are available from our Web site (http://www.fhcrc.org/labs/trask/subtelomeres/index.html).
Monochromosomal Hybrid Panel Analyses
Forty-eight PCR assays were designed across the 614-kb assembled 2q13 sequence by Primer 3 (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3.cgi) (Supplementary Table A, also available online at http://www.genome.org). None amplified a product of the predicted size from control rodent cell lines. The PCR reactions contained 80–100 ng of DNA from the NIGMS Human Genetic Cell Repository Somatic Cell Hybrid Mapping Panel #2 (version 3, Coriell Cell Repository), 250 μM deoxyribonucleoside triphosphates (dNTPs), 0.4 μM each primer, and 1 unit Perkin Elmer AmpliTaq Gold. Cycling conditions were 95°C for 5 min, 35 cycles of 30 sec at 94°C and 1 min at 60°C, followed by 10 min at 60°C. The products were analyzed on ethidium-bromide-stained 1% agarose gels.
Fluorescence In Situ Hybridization (FISH)
Metaphase spreads were prepared from the following cells or cell lines using published procedures (Trask 1999) for FISH analyses: peripheral blood cells from various healthy human donors and human male cell line CGM1; a male chimpanzee (Pan troglodytes) cell line CRL-1857 from ATCC; a male gorilla (Gorilla gorilla) cell line CRL-1854 from ATCC; and a female orangutan (Pongo pygmaes) cell line CRL-1850 from ATCC. DNAs from BAC clones were biotinylated by nick translation and hybridized to metaphase cells fixed on slides. Methods for preparation of the slides and probe, hybridization, washing, detection with FITC, fluorescent banding, and analysis are described elsewhere (Trask 1999). We also used FISH techniques for conventional and reciprocal chromosome painting as described elsewhere (Trask et al. 1991; Trask 1999) to identify human chromosomal material contained in the Y hybrid.
WEB SITE REFERENCES
http://bioinformatics.weizmann.ac.il/GESTALT; GESTALT.
http://cgap.nci.nih.gov/chromosomes/CytSearchForm; Mitelman catalog.
http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker.
http://genome.ucsc.edu/; UCSC Human Genome Working Draft.
http://www.fhcrc.org/labs/trask/subtelomeres/index.html; Trask laboratory Web site for supplementary information.
http:www.genome.washington.edu/phrap_documentation.html; cross_match.
http://www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi; Primer3.
http://www.jgi.doe.gov; DOE Joint Genome Institute.
http://www.ncbi.nlm.nih.gov; NCBI genome resources.
http://www.sanger.ac.uk/HGP/; Sanger Centre.
Acknowledgments
We are grateful to Tera Newman for help with the figures and Mitelman catalog queries, Colbey Harris for administrative assistance, and Janet Young and other members of the Trask lab for discussion and comments on earlier drafts of the manuscript. Some information for this paper was derived through use of the Celera Discovery System and Celera Genomics' associated databases. This work was supported in part by grant GM57070 from NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL btrask@fhcrc.org; FAX (206) 667-4023.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.337602.
REFERENCES
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Viggiano L, Misceo D, Horvath JE, Archidiacono N, Schwartz S, Rocchi M, Eichler EE. Human-specific duplication and mosaic transcripts: The recent paralogous structure of chromosome 22. Am J Hum Genet. 2002;70:83–100. doi: 10.1086/338458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2002;30:17–20. doi: 10.1093/nar/30.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G. The human genome: Organization and evolutionary history. Annu Rev Genet. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- Bertoni L, Attolini C, Tessera L, Mucciolo E, Giulotto E. Telomeric and nontelomeric (TTAGGG)n sequences in gene amplification and chromosome stability. Genomics. 1994;24:53–62. doi: 10.1006/geno.1994.1581. [DOI] [PubMed] [Google Scholar]
- Boutouil M, Fetni R, Qu J, Dallaire L, Richer CL, Lemieux N. Fragile site and interstitial telomere repeat sequences at the fusion point of a de novo (Y;13) translocation. Hum Genet. 1996;98:323–327. doi: 10.1007/s004390050216. [DOI] [PubMed] [Google Scholar]
- Brand-Arpon V, Rouquier S, Massa H, de Jong PJ, Ferraz C, Ioannou PA, Demaille JG, Trask BJ, Giorgi D. A genomic region encompassing a cluster of olfactory receptor genes and a myosin light chain kinase (MYLK) gene is duplicated on human chromosome regions 3q13–q21 and 3p13. Genomics. 1999;56:98–110. doi: 10.1006/geno.1998.5690. [DOI] [PubMed] [Google Scholar]
- Carroll ML, Roy-Engel AM, Nguyen SV, Salem AH, Vogel E, Vincent B, Myers J, Ahmad Z, Nguyen L, Sammarco M, et al. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J Mol Biol. 2001;311:17–40. doi: 10.1006/jmbi.2001.4847. [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccodicola A, D‘Esposito M, Esposito T, Gianfrancesco F, Migliaccio C, Miano MG, Matarazzo MR, Vacca M, Franze A, Cuccurese M, et al. Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet. 2000;9:395–401. doi: 10.1093/hmg/9.3.395. [DOI] [PubMed] [Google Scholar]
- Desmaze C, Alberti C, Martins L, Pottier G, Sprung CN, Murnane JP, Sabatier L. The influence of interstitial telomeric sequences on chromosome instability in human cells. Cytogenet Cell Genet. 1999;86:288–295. doi: 10.1159/000015321. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, Mohrenweiser HW. Interchromosomal duplications of the adrenoleukodystrophy locus: A phenomenon of pericentromeric plasticity. Hum Mol Genet. 1997;6:991–1002. doi: 10.1093/hmg/6.7.991. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Newman, T., Linardopoulou, E., and Trask, B.J. 2002. Gene Content and Function of the Ancestral Chromosome Fusion Site in Human Chromosome 2q13–2q14.1 and Paralogous Regions. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Glusman G, Lancet D. GESTALT: A workbench for automatic integration and visualization of large-scale genomic sequence analyses. Bioinformatics. 2000;16:482–483. doi: 10.1093/bioinformatics/16.5.482. [DOI] [PubMed] [Google Scholar]
- Hoglund M, Mitelman F, Mandahl N. A human 12p-derived cosmid hybridizing to subsets of human and chimpanzee telomeres. Cytogenet Cell Genet. 1995;70:88–91. doi: 10.1159/000133998. [DOI] [PubMed] [Google Scholar]
- Ijdo J, Baldini A, Ward DC, Reeders ST, Wells RA. Origin of human chromosome 2: An ancestral telomere–telomere fusion. Proc Natl Acad Sci. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijdo JW, Baldini A, Wells RA, Ward DC, Reeders ST. FRA2B is distinct from inverted telomere repeat arrays at 2q13. Genomics. 1992;12:833–835. doi: 10.1016/0888-7543(92)90319-n. [DOI] [PubMed] [Google Scholar]
- Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, Cremer T. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc Natl Acad Sci. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, Allison JB, editors. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–123. [Google Scholar]
- Kasai F, Takahashi E, Koyama K, Terao K, Suto Y, Tokunaga K, Nakamura Y, Hirai M. Comparative FISH mapping of the ancestral fusion point of human chromosome 2. Chromosome Res. 2000;8:727–735. doi: 10.1023/a:1026745505264. [DOI] [PubMed] [Google Scholar]
- Lese CM, Fantes JA, Riethman HC, Ledbetter DH. Characterization of physical gap sizes at human telomeres. Genome Res. 1999;9:888–894. doi: 10.1101/gr.9.9.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukusa T, Devriendt K, Holvoet M, Fryns JP. Dicentric chromosome 9 due to tandem duplication of the 9p11–q13 region: Unusual chromosome 9 variant. Am J Med Genet. 2000;91:192–197. [PubMed] [Google Scholar]
- Mah N, Stoehr H, Schulz HL, White K, Weber BH. Identification of a novel retina-specific gene located in a subtelomeric region with polymorphic distribution among multiple human chromosomes. Biochim Biophys Acta. 2001;1522:167–174. doi: 10.1016/s0167-4781(01)00328-1. [DOI] [PubMed] [Google Scholar]
- Martin CL, Wong A, Gross A, Chung J, Fantes JA, Ledbetter DH. The evolutionary origin of human subtelomeric homologies—Or where the ends begin. Am J Hum Genet. 2002;70:972–984. doi: 10.1086/339768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Gallardo A, Lamerdin J, Sopapan P, Friedman C, Fertitta AL, Garcia E, Carrano A, Negorev D, Macina RA, Trask BJ, et al. Molecular analysis of a novel subtelomeric repeat with polymorphic chromosomal distribution. Cytogenet Cell Genet. 1995;71:289–295. doi: 10.1159/000134129. [DOI] [PubMed] [Google Scholar]
- Mefford H, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Linardopoulou E, Coil D, van den Engh G, Trask BJ. Comparative sequencing of a multicopy subtelomeric region containing olfactory receptor genes reveals multiple interactions between non-homologous chromosomes. Hum Mol Genet. 2001;10:2363–2372. doi: 10.1093/hmg/10.21.2363. [DOI] [PubMed] [Google Scholar]
- Mondello C, Pirzio L, Azzalin CM, Giulotto E. Instability of interstitial telomeric sequences in the human genome. Genomics. 2000;68:111–117. doi: 10.1006/geno.2000.6280. [DOI] [PubMed] [Google Scholar]
- Musio A, Mariani T. Distribution of interstitial telomere-related sequences in the human genome and their relationship with fragile sites. J Environ Pathol Toxicol Oncol. 1999;18:11–15. [PubMed] [Google Scholar]
- Ning Y, Rosenberg M, Biesecker LG, Ledbetter DH. Isolation of the human chromosome 22q telomere and its application to detection of cryptic chromosomal abnormalities. Hum Genet. 1996;97:765–769. doi: 10.1007/BF02346187. [DOI] [PubMed] [Google Scholar]
- Park HS, Nogami M, Okumura K, Hattori M, Sakakia Y, Fujiyama A. Newly identified repeat sequences, derived from human chromosome 21qter, are also localized in the subtelomeric region of particular chromosomes and 2q13, and are conserved in the chimpanzee genome. FEBS Lett. 2000;475:167–169. doi: 10.1016/s0014-5793(00)01632-x. [DOI] [PubMed] [Google Scholar]
- Riethman HC, Xiang Z, Paul S, Morse E, Hu XL, Flint J, Chi HC, Grady DL, Moyzis RK. Integration of telomere sequences with the draft human genome sequence. Nature. 2001;409:948–951. doi: 10.1038/35057180. [DOI] [PubMed] [Google Scholar]
- Roy-Engel AM, Carroll ML, Vogel E, Garber RK, Nguyen SV, Salem AH, Batzer MA, Deininger PL. Alu insertion polymorphisms for the study of human genomic diversity. Genetics. 2001;159:279–290. doi: 10.1093/genetics/159.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonte RV, Conte RA, Ramesh KH, Verma RS. Molecular cytogenetic characterization of breakpoints involving pericentric inversions of human chromosome 9. Hum Genet. 1996;98:576–580. doi: 10.1007/s004390050262. [DOI] [PubMed] [Google Scholar]
- Simi S, Attolini C, Giulotto E. Intrachromosomal telomeric repeats and stabilization of truncated chromosomes in V79 Chinese hamster cells. Mutat Res. 1998;397:229–233. doi: 10.1016/s0027-5107(97)00217-0. [DOI] [PubMed] [Google Scholar]
- Slijepcevic P, Xiao Y, Dominguez I, Natarajan AT. Spontaneous and radiation-induced chromosomal breakage at interstitial telomeric sites. Chromosoma. 1996;104:596–604. doi: 10.1007/BF00352299. [DOI] [PubMed] [Google Scholar]
- Sutherland GR, Mattei JF. Report of the committee on cytogenetic markers. Cytogenet Cell Genet. 1987;46:316–324. doi: 10.1159/000132482. [DOI] [PubMed] [Google Scholar]
- Trask B. Fluorescence. In: Birren B, Green ED, Hieter P, Klapholz S, Myers RM, Riethman H, Roskams J, editors. Situ Hybridization. In Genome analysis: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 303–413. [Google Scholar]
- Trask BJ, van den Engh G, Christensen M, Massa HF, Gray JW, Van Dilla M. Characterization of somatic cell hybrids by bivariate flow karyotyping and fluorescence in situ hybridization. Somat Cell Mol Genet. 1991;17:117–136. doi: 10.1007/BF01232970. [DOI] [PubMed] [Google Scholar]
- Trask B, Fertitta A, Christensen M, Youngblom J, Bergmann A, Copeland A, de Jong P, Mohrenweiser H, Olsen A, Carrano A, et al. Fluorescence in situ hybridization mapping of human chromosome 19: Cytogenetic band location of 540 cosmids and 70 genes or DNA markers. Genomics. 1993;15:133–145. doi: 10.1006/geno.1993.1021. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R, Jauch A, Cremer T. Homologies in human and Macaca fuscata chromosomes revealed by in situ suppression hybridization with human chromosome specific DNA libraries. Chromosoma. 1992;101:265–270. doi: 10.1007/BF00346004. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Jauch A, Ludecke HJ, Senger G, Horsthemke B, Claussen U, Cremer T, Arnold N, Lengauer C. The origin of human chromosome 2 analyzed by comparative chromosome mapping with a DNA microlibrary. Chromosome Res. 1994;2:405–410. doi: 10.1007/BF01552800. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Howell RT. A fragile secondary constriction on chromosome 2 in a severely mentally retarded patient. J Ment Defic Res. 1977;21:227–239. doi: 10.1111/j.1365-2788.1977.tb00042.x. [DOI] [PubMed] [Google Scholar]
- Yu N, Zhao Z, Fu YX, Sambuughin N, Ramsay M, Jenkins T, Leskinen E, Patthy L, Jorde LB, Kuromori T, et al. Global patterns of human DNA sequence variation in a 10-kb region on chromosome 1. Mol Biol Evol. 2001;18:214–222. doi: 10.1093/oxfordjournals.molbev.a003795. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O. The origin of man: A chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]