Abstract
Various portions of the region surrounding the site where two ancestral chromosomes fused to form human chromosome 2 are duplicated elsewhere in the human genome, primarily in subtelomeric and pericentromeric locations. At least 24 potentially functional genes and 16 pseudogenes reside in the 614-kb of sequence surrounding the fusion site and paralogous segments on other chromosomes. By comparing the sequences of genomic copies and transcripts, we show that at least 18 of the genes in these paralogous regions are transcriptionally active. Among these genes are new members of the cobalamin synthetase W domain (CBWD) and forkhead domain FOXD4 gene families. Copies of RPL23A and SNRPA1 on chromosome 2 are retrotransposed-processed pseudogenes that were included in segmental duplications; we find 53 RPL23A pseudogenes in the human genome and map the functional copy of SNRPA1 to 15qter. The draft sequence of the human genome also provides new information on the location and intron–exon structure of functional copies of other 2q-fusion genes (PGM5, retina-specific F379, helicase CHLR1, and acrosin). This study illustrates that the duplication and rearrangement of subtelomeric and pericentromeric regions have functional relevance to human biology; these processes can change gene dosage and/or generate genes with new functions.
[Supplemental material is available online at http://www.genome.org. Sequence data reported in this paper have been deposited in GenBank and assigned the following accession nos.: AF452722, AF452723, and AF452724.]
Two ancestral chromosomes fused head-to-head to form human chromosome 2 (Yunis and Prakash 1982). This gross karyotypic change probably contributed to the reproductive barrier between the early humans who carried this new structure and closely related hominids. Sequences that once resided near the telomeres of the two fusion partners are now interstitially located in band 2q13–2q14.1 (2qFus, for short) (Ijdo et al. 1991), but portions of these regions had already duplicated and spread to/from subtelomeric and pericentromeric regions before the fusion. More recent exchanges propagated some blocks of sequence to additional sites and/or homogenized the sequences of subtelomeric segments on different chromosomes. These past events are now apparent in the cross-hybridization patterns of 2qFus- and subtelomere-derived probes (Ijdo et al. 1991; Trask et al. 1993, 1998; Hoglund et al. 1995; Martin-Gallardo et al. 1995; Ning et al. 1996; Ciccodicola et al. 2000; Park et al. 2000; Mefford et al. 2001; Martin et al. 2002; Fan et al. 2002) and the extensive sequence similarity among paralogous sites (Bailey et al. 2002; Martin et al. 2002; Fan et al. 2002).
Duplications can spawn new genes. In many cases, the duplicates will degenerate into pseudogenes; in other cases, the parent and daughter copies evolve to take on distinct functions (Nei et al. 1997; Lynch and Conery 2000). We were therefore interested in examining the gene content of the intra- and interchromosomal duplications involving 2qFus-related sequence. Because these duplications took place during hominid evolution, resulting variation in gene number and function might contribute to phenotypic differences among primates. In addition, we were interested in comparing the fates of genes sequestered at the interstitial fusion site with their counterparts in subtelomeric and pericentromeric locations, in light of the different evolutionary processes and possible constraints on expression that might act on these different genomic compartments.
The Sanger Centre annotated seven genes or gene homologies in its GenBank entry of BAC RP11–395L14 (http://www.sanger.ac.uk/HGP/), which derives from 2qFus (Martin et al. 2002; Fan et al. 2002). Martin et al. (2002) reported that several of these genes occur in more than one copy in the human genome. Prior to our study, however, detailed analyses of only two functional genes within the multicopy interstitial “subtelomeric” region at 2qFus had been reported in the literature. These are RABL2A (RAS oncogene family-like 2A) (Wong et al. 1999) and a retina-specific transcript called F379 (Mah et al. 2001). RABL2A's close relative, RABL2B, is located at the subtelomeric region of 22q13.3 (22qter); both are ubiquitously expressed (Wong et al. 1999). Mah and colleagues (2001) found evidence for at least eight copies of F379, mostly near telomeres, and demonstrated that multiple copies are transcriptionally active.
In this paper, we analyze the gene content of 614 kb surrounding the fusion site and paralogous segments in other genomic locations. Of the 11 genes within this region, nine are duplicated at least once elsewhere in the genome. Our analyses of publicly available sequence uncover four members of CBWD and FOXD4-like gene families dispersed at 2qFus, 9pter, 9p11.2, and 9q13. At least two members of each family are transcriptionally active, and others are potentially functional. We have re-evaluated and revised the chromosomal assignments of several members of the new F379 gene family. We have also characterized the genomic structure of the single-copy TIC and PAX8 genes, which map within 400 kb of the fusion site, and functional copies of CHLR1, SNRPA1, and PGM5 genes. The 2qFus paralogs of these genes are pseudogenes. In addition, the 2qFus fusion site contains one of at least 53 RPL23A-processed pseudogenes in the human genome; some arose via retrotransposition of processed mRNA, others were propagated via segmental duplications.
RESULTS
Gene Content of the Fusion Region and Paralogous Segments
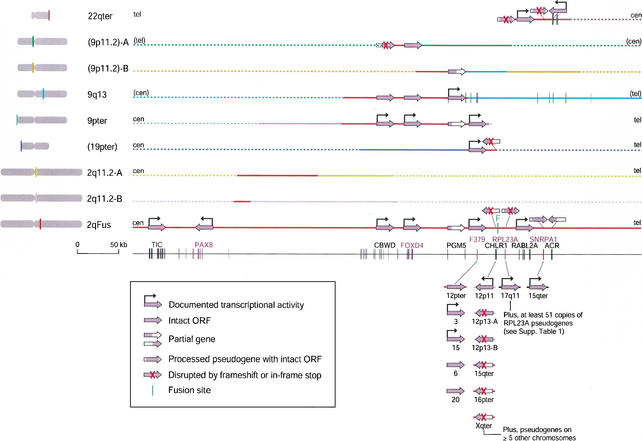
Figure 1 summarizes the gene content of the 614-kb region surrounding the fusion site, which is marked by two head-to-head arrays of degenerate telomere repeats. Over 360 kb of the region is duplicated elsewhere in the genome (Martin et al. 2002; Fan et al. 2002). Only the sequences on either end of the contiguous sequence are unique to 2qFus. The region surrounding the telomere-repeat arrays is a medley of small blocks of sequence that are duplicated on seven or more chromosomes, primarily near telomeres (Fan et al. 2002). Surrounding this complex region are much larger duplicated segments shared by 2qFus and one to three other genomic locations. These larger paralogous blocks range in size from 20 kb to over 168 kb, and they are 96% to 99% identical to 2qFus sequence (Martin et al. 2002; Fan et al. 2002). The accompanying paper provides information on the assembly of these paralogous segments, their chromosomal localization, and their evolutionary history (Fan et al. 2002).
Figure 1.
Gene content of sequences at the 2q13–q14.1 fusion site and paralogous regions. Each line corresponds to sequence from a different chromosomal region. For the 68-kb region immediately surrounding the fusion site, only sequences with homology to genes are shown. Locations in parentheses are tentative (see below) and based on FISH, hybrid panel analysis, and the human genome assembly (Fan et al. 2002). The blocks on 9p11.2 may derive from 9q13. The large arrows indicate the state of each gene in each chromosomal location (e.g., full-length, partial, or disrupted ORF; see legend in figure) and point in the predicted direction of transcription relative to the genomic sequence. They are not drawn to scale. Bent black arrows indicate the genes for which transcriptional activity has been documented by us or others (see text). The exons of the 11 genes or pseudogenes identified in the 2qFus sequence are drawn to scale in the bottom line. As the 2qFus copies of PGM5 and ACR are partial genes, the exons for these genes are drawn to scale on the lines for 9q13 and 22qter, respectively. Details of sequence identity and chromosomal mapping data of homologous sequence blocks are provided in Fan et al. 2002. Solid lines indicate the extent of sequence coverage of available clones in the regions of interest as of March 1, 2002. Red solid lines indicate the regions with >95% average identity to 2qFus sequence. Red dotted lines indicate adjoining regions with no available sequence, but that were shown by PCR to be homologous to 2qFus (Fan et al. 2002). Different colors are used to indicate divergent sequence, with solid lines indicating the extent of contiguous sequence coverage, and dotted lines indicating either unavailable sequence or sequence in (non)overlapping clones that lack homology to any of the other segments shown. Orientations indicated in parentheses are tentative. In the bottom section, short lines extending from the gene arrows indicate flanking homology to 2qFus (red) or among each other (gray); the full extent of homology is not indicated. Gene arrows without protruding lines are sequences of PCR products (Mah et al. 2001). The GenBank accessions for sequences shown are 22qter (AC002055, AC002056), 9p11.2-A (AL512605), and 9p11.2-B (AL445925) (tentative; one or both may derive from 9q13, see Fan et al. 2002), 9q13 (AL161457, AL353608, AL353616), 9pter (AL356244, AL449043), 2q11.2-A (AC008268), 2q11.2-B (AC009238), 19pter (AL627309, tentative; Fan et al. 2002), 12pter (AC026369), 12p11 (AC008013), 12p13-A (AC009533), 12p13-B (AC092821), 15qter (AF282022/Z96310 and AC023024 (SNRPA1)), 16pter (Z84812), Xqter (AJ271736/M57752), 17q11 (AF001689), and 2qFus (AC016683, AC016745, AC017074, AL078621). All are finished sequence except AC092821.
At least 11 known genes have homology to sequences within the 614-kb region surrounding the fusion site (Fig. 1). Only two genes, TIC and PAX8, are single-copy. The other nine are duplicated in at least one other location in the genome. Our analyses of the genomic organization and transcriptional activity of the multicopy genes are described in the following sections. The RAS oncogene family genes RABL2A (2qFus) and RABL2B (22qter), which were described and shown to be expressed by Wong et al. (1999), and the acrosin (ACR) gene, which was shown earlier to be intact on 22qter and truncated in 2qFus (Bailey et al. 2002; Martin et al. 2002), are not discussed further here. Tables of the sequences at the intron–exon boundaries of the multiexon genes are included in Supplementary Table 3, available online at http://www.genome.org. Gene coordinates are given with respect to the 614-kb 2qFus contiguous sequence (available as a FASTA file at http://www.fhcrc.org/labs/trask/subtelomeres/index.html), as the current practice of trimming bacterial artificial chromosome (BAC) sequences to minimize overlap redundancy in final GenBank entries renders ambiguous coordinates given with respect to the actual full-length BAC sequences.
Cobalamin Synthetase W Domain (CBWD) Genes
Each of the segmental duplications on chromosome 2qFus, 9pter, 9q13, and 9p11.2 contains a CBWD gene. The paralogs on 2qFus and 9pter were identified previously from genomic sequence (Martin et al. 2002), but neither was evaluated for transcriptional activity. Cobalamin synthetase W (COBW) is an intronless gene isolated from Pseudomonas denitrificans, which encodes a 354-amino-acid cobalamin (vitamin B12)-synthesis protein (Crouzet et al. 1991). Three of the human CBWD paralogs (CBWD1, CBWD2, CBWD3 in 9pter, 2qFus, and 9q13, respectively) encompass 59 kb (nucleotides 275125–332999 in 2qFus) and encode an 1188-bp open reading frame (ORF) composed of 15 exons (Figs. 1 and 2). Of the 395 amino acids in the predicted human protein sequences, ∼50% are similar and ∼27% are identical to the Pseudomonas protein (M62866). Available sequence for the 9p11.2-A block covers only exons 7–15, but this portion contains an in-frame stop codon (named CBWD4P, for COBW domain-containing four pseudogenes) (Fig. 2).
Figure 2.
Comparison of cDNA and DNA sequences of human CBWD genes. CBWD1, CBWD2, CBWD3, and CBWD4P represent cobalamin synthetase W domain-containing sequences from 9pter, 2qFus, 9q13, and 9p11.2 (or another block in 9q13, see Fig. 1), respectively. Sequences of four transcripts are compared with the genomic sequences. Only the coding portions are shown. AF257330 is a cDNA sequence from GenBank (Shi et al. 2001). AF452722 is a sequence we obtained by PCR amplification of human brain Marathon-Ready cDNAs. A cDNA sequence from GenBank, BC005996 (Mammalian gene collection, http://mgc.nci.nih.gov) and an EST contiguous sequence assembled from EST sequences AL540640, AL545680, and AL571299 match the 9pter sequence exactly and are therefore not shown. Blue residues are identical to AF257330; black residues differ from it. Dashed lines indicate incomplete sequence information. Arrows indicate exon boundaries. Predicted amino acid sequences are indicated below the nucleotide sequences. Accession numbers for genomic copies as in Figure 1.
As expected from the recent origin of these paralogous duplications (Fan et al. 2002), the mouse genome contains only one COBW-like gene; it maps to mouse chromosome 19 within a region that contains orthologs to both 9q13 and 9pter.
No two of the human CBWD paralogs are identical (Fig. 2). The maximal difference is between 9pter and 9q13 (1.2% and 2% at nucleotide and amino-acid levels, respectively). The sequence conservation of the three intact human CBWD genes indicates that they are under selective pressure for function. Overall, the coding exons are 0.4% –0.7% more conserved than the 5′ and 3′ flanking noncoding portions of the duplicated segments.
At least two CBWD paralogs, on 9pter and 2qFus, are transcriptionally active. The kidney cDNA BC005996 and a 1275-bp EST contiguous sequence assembled from three placental ESTs (see Fig. 2 legend) are identical to CBWD1, indicating that the 9pter locus is transcriptionally active. AF212253, an 1131-nucleotide transcript isolated from the adrenal gland, appears to be an alternatively spliced form lacking exons 9 and 10 transcribed from 9pter (only 3 nucleotide differences). We assembled another cDNA sequence from eight PCR products that we amplified from human fetal brain cDNA (AF452722). This sequence spans the ORF, and its best match is the 2qFus paralog (4 nucleotide and 3 amino-acid differences in 1188 nucleotides/395 amino acids). Assuming that these are allelic polymorphisms, we conclude that the 2qFus copy, CBWD2, is also transcribed. Another full-length, 1680-nucleotide cDNA derived from human fetal brain, AF257330 (Shi et al. 2001), most closely matches copies on 9pter and 2qFus at the nucleotide level (6 nucleotide differences each), but is more similar to 9pter than 2qFus at the amino-acid level (2 vs. 4 amino-acid differences). It is not clear if these differences are SNPs among 9pter alleles or if they represent yet another paralog in the genome.
FOXD4-Like Genes
One FOXD4-like gene resides in each of the four 2qFus-paralogous segments (Figs. 1 and 3). These genes are members of a large family of transcription factors with highly conserved 100-amino-acid DNA-binding forkhead domains (Lai et al. 1991). Forkhead proteins regulate embryonic development (Ruiz i Altaba et al. 1993) and act as oncogenes when overexpressed (Li and Vogt 1993). In 1994, Pierrou et al. identified a 318-bp partial human cDNA sequence (U13223) with high homology to the forkhead domain and named it FREAC5 for forkhead related activator 5 (Pierrou et al. 1994). This gene was recently renamed FOXD4 (Kaestner et al. 2000). Larsson et al. (1995) assigned this gene to the pericentromeric region of chromosome 9 by PCR analysis of a hybrid panel and FISH, although FISH signals were also seen on chromosome 2. Our sequence analyses indicate that the U13223 cDNA instead derives from 9pter, and that three other potentially functional FOXD4 genes reside in 2qFus (nucleotides 336513–337739), 9q13, and 9p11.2. These three genes are assigned the names FOXD4L1, FOXD4L3, and FOXD4L2, respectively. The similarity among the human FOXD4 genes (97.7% to 98.9% identity at the nucleotide level) correlates with the overall homology among the four segmental duplications that generated them (Fan et al. 2002).
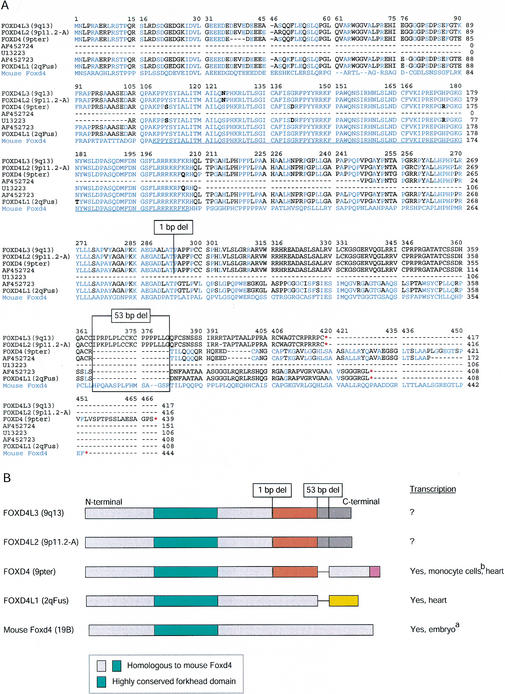
Figure 3.
(A) Comparison of predicted amino-acid sequence of cDNA and paralogous genomic copies of forkhead FOXD4-like genes. FOXD4, FOXD4L1, FOXD4L3, and FOXD4L2 represent human forkhead domain-containing genes from 9pter, 2qFus, 9q13, and 9p11.2, respectively, derived from BAC sequences. Foxd4 is an orthologous mouse forkhead protein (fkh-2, X86368). All genes are single-exon. AF452723 and AF452724 are cDNA sequences we generated by PCR from human heart Marathon-Ready cDNAs. U13223 is a published 318-bp partial human FOXD4 (FREAC5) cDNA (Pierrou et al. 1994). Blue residues are identical to the mouse Foxd4 protein; black residues differ from it. The underlined region indicates 100-amino-acid conserved forkhead DNA-binding domain. The locations of the 1-bp and 53-bp deletions relative to mouse Foxd4 are indicated; each causes a frame shift. Red stars indicate both stop codons. An alignment of the corresponding nucleotide sequences and locations of primers used in our analysis are in Supplemental Figure A (available online at http://www.genome.org). (B) Summary of the putative protein structures of human FOXD4-like subfamily. Colors indicate homologous regions at the amino-acid level. Light blue blocks regions are ≥49% identical to mouse Foxd4. Green blocks are conserved forkhead domains (≥96% amino-acid identity). References for transcriptional activity are a (Kaestner et al. 1995); b (Pierrou et al. 1994); others, present study. The sequence containing FOXD4L2 is tentatively assigned to 9p11.2 based on FISH results (Fan et al. 2002), but may derive from 9q13.
All four FOXD4 paralogs are predicted to encode single-exon ORFs of ≥408 amino acids with conserved amino termini, including the highly conserved forkhead domain (Fig. 3). A TATA box and a transcription start site lie 189 and 160 nucleotides, respectively, upstream of the predicted translation start site in all four genes (data not shown). A poly-A signal sequence, AATAAA, can be found 544–697 nucleotides downstream of the stop codon in each gene (data not shown). Despite these similarities, two frameshift-causing mutations make the carboxy-terminal portions (after amino acid 292) of the predicted human proteins grossly different (Fig. 3; Supplemental Fig. A, available online at http://www.genome.org). A 1-bp deletion after amino acid 292 changes the reading frame of the copies in 9pter, 9q13, and 9p11.2 relative to that in 2qFus. Further on, a 53-bp deletion after amino acid 363 in 2qFus and 349 in 9pter alters their reading frame relative to the other two human versions. We resequenced these genes from overlapping PCR products amplified from BACs representing the four FOXD4 loci and a chromosome-2 hybrid line and confirmed the accuracy of the publicly available sequence (data not shown). Using primers flanking the 53-bp deletion, we also amplified two PCR products differing by ∼53 bp from a chromosome-9 hybrid line, thereby confirming the presence of both long and short FOXD4-like forms on this chromosome (data not shown).
The mouse genome appears to contain only one Foxd4 gene, for which a transcript has been identified from embryonic tissue (X86368, previously called fkh-2) (Kaestner et al. 1995). Like its human orthologs, the mouse Foxd4 protein is encoded by a single-exon gene. It maps to mouse chromosome 19B. Overall, the mouse gene is ∼79% to ∼82% identical to the human genes at the nucleotide level, but the level of identity varies across the gene (Fig. 3; Supplemental Fig. A, available online at http://www.genome.org). The human paralogs are remarkably similar to the mouse gene within the 297-bp/99-amino-acid forkhead domain (92.3–93.3% identity at the nucleotide level and 96–99% identity at the amino-acid level). In contrast, the human genes have only ∼65% nucleotide identity and ∼49% amino-acid identity to the mouse gene in the amino-terminal region preceding of the forkhead domain because of a combination of single nucleotide changes and insertions or deletions (10 or 11 involving from 1–12 bp, 8 of which cause frameshifts). The mouse and human genes show ∼70% nucleotide identity and 52–56% amino-acid identity in the region between the forkhead domain and amino acid 292. After this point, only the proteins encoded by 2qFus and 9pter are homologous to the mouse protein, and then only partly so. The 2qFus gene lacks the 1-bp deletion after amino acid 292 and is therefore homologous to the mouse protein up to the 53-bp deletion. As FOXD4 in 9pter has both the 1-bp and 53-bp deletions, the carboxl terminus of its predicted protein product is read in the same frame as the mouse product. Except for these two partial homologies to the predicted mouse protein, the carboxyl termini of the human paralogs are not similar to the carboxyl termini of any known forkhead-domain protein.
The nucleotide changes that have occurred in the forkhead domain since divergence of the mouse and human orthologs are highly biased for synonymous changes (34–39 synonymous changes/100 synonymous sites vs. 1–3 nonsynonymous changes/100 nonsynonymous sites). Ka/Ks ratios of 0.022–0.074, a sign of strong purifying selection, are obtained when the human sequences are compared with the mouse sequence in the forkhead domain. We observe the signature of milder purifying selection in the less conserved 84-amino-acid region between the forkhead domain and the first frameshift-causing deletion (Ka/Ks ∼0.6, human genes relative to mouse).
So far, we have evidence that at least two of the human FOXD4 paralogs are transcriptionally active. As noted above, the 9pter paralog is identical at the nucleotide level to the partial FOXD4 (FREAC5) cDNA sequence published by Pierrou et al. (1994) (Fig. 3; Supplemental Fig. A, available online at http://www.genome.org). Because Pierrou and coworkers had shown FOXD4 to be expressed at high levels in heart and skeletal muscle and at low levels in other tissues examined by Northern blot analyses (Pierrou et al. 1994), we performed PCR analyses on cDNA from heart tissue in order to detect transcription of the 2qFus paralog. As this is a single-exon gene, genomic contamination was a concern, so we situated our FOXD4L1-specific primers so as to flank the 53-bp deletion. The genome contains at least two short and two long forms, but we detected only the short 2qFus-like form in the cDNA (only one SNP is found between the amplified cDNA, submitted as AF452723, and the 2qFus genomic sequence in 1227 nucleotides compared; Supplemental Fig. A, available online at http://www.genome.org). The SNP causes an amino-acid change in the forkhead domain at position 179 (Fig. 3), making this cDNA sequence identical to mouse Foxd4 within the forkhead domain at the amino-acid level. We also designed sets of PCR primers to specifically amplify transcripts in heart tissue derived from each of the FOXD4-like loci on chromosome 9 and confirmed the transcriptional activity of the 9pter paralog (submitted as AF452724). Transcripts of the 9p11.2 or 9q13 copies were not detected in this tissue, despite the use of a primer situated within the 53-bp region retained in these two genes and deleted from the other two. It remains possible that these genes are expressed in other tissues.
Phosphoglucomutase-Related Protein (PGM5) Genes
Segmental duplications generated three partial copies of PGM5 from the intact locus in 9q13. PGM5 (also called PGM-RP) was originally cloned as a transcript from a human uterus cDNA library and mapped to 9cen–9q13 by Moiseeva et al. (1996). Edwards and colleagues (1995) had earlier demonstrated a broad tissue expression pattern and duplication of PGM5 sequence around the centromere of chromosome 9. With the availability of the draft human genome assembly, the intact gene can now be assigned to 9q13. The gene spans 172 kb and encodes an 11-exon, 507-amino-acid ORF. The cDNA sequenced by Moiseeva et al. matches the 9q13 genomic sequence with 100% identity. Because this large gene spans two breakpoints of homology between 9q13 and other loci (Fig. 1), PGM5 copies on 9p11.2-B, 9pter, and 2qFus are truncated (9p11.2-B has 5′ UTR and first 6 exons; 9pter and 2qFus [nucleotides 379272–400554] have 5′ UTR and first 2 exons). In addition, the start codons in both the 2qFus and 9pter copies have mutated from ATG to ACG. The mouse PGM5 ortholog maps to chromosome 19, and the predicted mouse and human proteins are 97% identical.
Retina-Specific F379 Genes
The multicopy region near the fusion site contains one of at least eight copies of F379 in the human genome. F379s are retina-specific transcripts of unknown function that were identified recently by Mah and colleagues (2001). They identified a full-length 3-exon cDNA of 1188 bp, which is predicted to encode an 85-amino-acid protein. Five other transcripts with slightly different nucleotide sequences were identified in retina cDNA libraries or public EST databases, and eight F379 paralogs were identified by PCR analyses of hybrid panel DNAs (Mah et al. 2001). We confirmed the existence of F379 genes on chromosomes 2, 3, 6, 9, 12, 15, 19, and 20 by hybrid-panel PCR using primers immediately flanking the F379 gene in the 2qFus sequence (Fan et al. 2002). BAC genomic sequences are available for four of these F379 paralogs, that is, those on 2qFus (nucleotides 414983–415581), 9pter, 12pter, and 19pter, which appear to have been generated by segmental duplications among subtelomeres (Fig. 1). We confirmed the chromosomal location of each sequenced BAC by FISH (data not shown). Mah et al. (2001) also ascribe F379 sequences to chromosome 22 (AL078621), 10 (AL135795), and 21 (AC073186, now AL627309), but none of these locations is consistent with the hybrid-panel results. These chromosome assignments reflect erroneous mapping information in GenBank entries or earlier drafts of the human genome. The first two genomic clones cross the fusion site on chromosome 2. AC073186 contains no chromosome-specific DNA, but most likely represents a variant form of the subtelomeric region of 19p (Fan et al. 2002).
F379 transcripts identical to the genomic copies assigned to 2qFus, 9pter/19pter, 3, and 15 have been identified so far (in total, four distinct genes) (Mah et al. 2001) (Fig. 1). Given the frequency of subtelomeric exchanges, however, the chromosomal distribution of F379 variants in the individuals from which transcripts were obtained may differ from that in the individuals whose DNA was used for the human genome sequencing project and the prototypic individual represented in the hybrid panel (Linardopoulou et al. 2001). As expected from their subtelomeric location, F379 genes are polymorphic in number. FISH with an F379-specific probe identifies 10 or 23 copies in the genomes of two tested individuals (data not shown). F379 copies were polymorphically present in these individuals on 1pter, 5qter, 8pter, and 11pter, which are locations not identified by the hybrid-panel analysis, and 20qter. If a mouse ortholog of F379 exists, it has not yet been sequenced by public or Celera efforts.
CHLR1 Genes
A multicopy segment on the centromere-proximal side of the 2q fusion site (nucleotides 43718–438686) contains one of many incomplete copies of CHLR1 in the human genome (Fig. 1). CHLR1 is a homolog of yeast Chl1, which is a member of the DEAD/DEAH family of DNA and RNA helicases and is critical for proper chromosome transmission in mitosis (Gerring et al. 1990). Two human transcripts, CHLR1 and CHLR2, differing by 1.6% at the nucleotide and 2.6% at the predicted protein levels, were cloned previously by Amann and coworkers and mapped by FISH to chromosome 12p11 and/or 12p13 (Amann et al. 1996, 1997).
Our analyses indicate that CHLR1 sequences have been caught up in two series of segmental duplications. At least three complete copies of the gene were generated by duplication of a >30-kb segment shared by AC008013, AC009533, and AC092821, which map to chromosomes 12p11, 12p13 (A), and 12p13 (B) in the draft genome assembly, respectively (Fig. 1). AC008013 has 34 kb and ≥63 kb in common with AC009533 and AC092821, respectively (98.1% identity). These three genes each contain 26 exons spanning ∼25 kb, but only the copy assigned to 12p11 appears to encode an intact ORF. It is nearly identical to the transcript of Amann et al. (U33833) (4 nucleotide/1 amino-acid differences in 2721 nucleotide/906 amino-acid compared). The other two contain a 1-bp, frameshift-causing deletion (one in exon 19 and the other in exon 25). A rearranged portion of CHLR1 has also been propagated among many subtelomeric locations as part of another segmental duplication (Fig. 1). Available genomic sequences of these copies from 2qFus, 15qter, 16 qter, 19pter, and Xqter (Fig. 1) share ≥3.5 kb, at 97.5%–98.5% nucleotide identity, encompassing CHLR1 exons 18 and 22–25 and the 3′ UTR. In all these copies, 1286 bp containing exons 19–21 have been deleted and replaced with a 40-bp segment not present in any of the full-length copies. PCR analyses of a hybrid panel and FISH analyses (Fan et al. 2002) (Brown et al. 1990) indicate that there are at least five additional copies in the genome, on chromosomes 3, 6, 9, 20, and Y. Copy number and chromosomal location of these partial CHLR1 genes, like F379, vary among individuals (Brown et al. 1990).
Remarkably, the human genomic location containing CHLR2 has not been sequenced yet. All available genomic copies differ from this transcript (U33834) by 1.3% or more (≥29 nucleotides in 2202).
Ribosomal Protein L23A (RPL23A)-Like Processed Pseudogenes
One of >50 copies of processed RPL23A pseudogenes in the human genome resides in 2qFus (nucleotides 449035–449489). The functional, intron-containing RPL23A gene was isolated and mapped to human chromosome 17q11 previously (Fan et al. 1997). This group also estimated the existence of 30–40 other pseudogenes in the human genome (Fan et al. 1997). We can now find a total of 53 RPL23A pseudogenes in sequenced BAC and cosmid clones (Supplemental Table 1, available online at http://www.genome.org). Every human chromosome except 15 and Y appears to have been a recipient of at least one RPL23A pseudogene. Each is intronless and spans ∼471 bp. All but five have incurred mutations that disrupt the original ORFs, and all are likely to lack promoter sequences. These pseudogenes represent 42 independent retrotransposition events—cross_match analysis detects no significant homology between the clones in sequences flanking the pseudogenes. This high number of processed pseudogenes is not unusual for genes encoding ribosomal proteins and is presumably a consequence of their ubiquitous transcription at high levels (Davies et al. 1989).
Two of the retrotransposed pseudogenes were propagated further as part of segmental duplications. One pseudogene duplicated as part of a larger block (of up to 67 kb) to at least 10 locations distributed on 10 chromosomes (Supplemental Table 1, available online at http://www.genome.org). All six of the copies mapped by sequence assembly or FISH are located in subtelomeric (or 2qFus) regions. These duplicates were created over period of several million years, as they are now 95%–98.6% identical. A second RPL23A pseudogene propagated to at least three locations as part of a much smaller segmental duplication (901–924 bp) (Supplemental Table 1, available online at http://www.genome.org). These duplications are only 90%–91% identical and therefore appear to be the result of duplication events that predated hominid divergence.
Small Nuclear Ribonucleopolypeptide A1 (SNRPA1) Gene and Processed Pseudogenes
A 768-bp ORF within the 2qFus sequence homologous to 22qter (nucleotides 495164–495931) is 98% identical to X13482, a SNRPA1 cDNA (Sillekens et al. 1989) (Fig. 1). SNRPA1 is a small ribonucleoprotein constituent of the U2 snRNP particle (Sillekens et al. 1989). The 22qter paralog has two 1-bp frameshift-causing deletions. Both the 2qFus and 22qter copies are processed pseudogenes—a multi-exon SNRPA1 gene is found in RP11–299G20 (AC023024). The 1054-bp SNRPA1 cDNA spans 9 exons and 13.5 kb of sequence in this clone, with 99.9% identity (data not shown). A SNRPA1 EST contiguous sequence made up of AL522821, BI089073, AL522987, BG828833, and BE738305 is 100% identical to this genomic sequence (data not shown). We mapped RP11–299G20 to 15qter (15q26.3) by FISH and confirmed its location by PCR analysis of the hybrid panel (data not shown). We conclude that the processed pseudogenes originated by a retrotransposition from 15qter, followed by segmental duplication between chromosome 22 and the ancestor of 2qFus.
The mouse and human SNRPA1 genes define a disruption in conserved synteny: The mouse gene maps to chromosome 7 where it is surrounded by genes whose human orthologs map to 15q11–q12, not 15qter (http://www.jax.org). The human SNRPA1 gene differs from the mouse orthologs (AF230356) by a 3-bp (1-amino-acid) deletion and 66 nucleotide changes that lead to only 7-amino-acid differences (Supplemental Fig. B, available online at http://www.genome.org). Strong purifying selection has acted on the mouse and human genes: The Ka:Ks ratio is 0.04. The processed pseudogenes in 2qFus and 22qter also have low Ka:Ks ratios when compared with the mouse gene (∼0.06), indicating that insufficient time has elapsed to completely erase the signature of past purifying selection on these pseudogenes.
Single-Copy Genes TIC and PAX8
Two single-copy genes, TIC and PAX8, reside in the centromere-proximal portion of the 2qFus region. A TIC transcript from peripheral blood mononuclear cells was mapped distal of the interleukin-1 cluster and dubbed TIC, for telomeric of interleukin-1 cluster (U63127; T.P. Klenka, R. Herbst, and M.J.H. Nicklin, unpubl.). Comparison of the transcript to 2qFus genomic sequence now shows that the gene comprises 16 exons and spans 19 kb (nucleotides 19714–38672). We detect six SNPs, leading to four predicted amino-acid changes, between the cDNA and genomic sequences within the 3171-bp ORF. A second partial TIC cDNA sequence (AK023421) differs from U63127 and 2qFus genomic sequence by 6 and 5 nucleotide changes in 2028 bp compared, respectively. Because we find no other TIC-like sequences in the genome by PCR analyses of a monochromosomal hybrid panel or database mining, we conclude that these are allelic differences (or sequencing errors). As far as we are aware, no function has been ascribed to TIC, but the predicted protein has homology to yeast SEC7, which is required for membrane traffic from the Golgi apparatus (Achstetter et al. 1988), and mammalian proteins with SEC7 domains are positive regulators of ADP-ribosylation factor (ARF) GTPases (Chardin et al. 1996; Mayer et al. 2001).
The PAX8 gene spans 60 kb of the proximal single-copy region near the fusion site (nucleotides 55790–115650). PAX8 was cloned previously from a human kidney carcinoma cell line (L19606), mapped to 2q12–q14, and shown to be alternatively spliced (Kozmik et al. 1993, 1997). PAX8 codes for a paired-box homeotic protein, which is a transcription factor associated with congenital hypothyroidism, thyroid hypoplasia, and kidney development (Macchia et al. 1998; Torban and Goodyer 1998). Newly available genomic sequence indicates that the gene has 11 exons, not 10 as originally reported (Kozmik et al. 1993). We also detect one SNP, which causes no amino-acid change, in the 1353-bp ORF by comparing the cDNA and genomic sequence.
DISCUSSION
Our genomic investigations of the chromosome-2 fusion site have illuminated the complex and dynamic history of sequences in this region. Segmental duplications involving 2qFus-paralogous sequences have multiplied seven genes. Two to seven or more copies of these genes are now present in the human genome. 2qFus-related sequences have also been the target for two retrotransposed pseudogenes (of RPL23A and SNRPA1) that have functional and nonfunctional relatives elsewhere in the genome. Segmental duplications were responsible for multiplying some of these pseudogenes further.
The existence of genes in the 2qFus-paralogous segments supports the idea that duplications are an important evolutionary process for functional change (Nei et al. 1997). Of the 32 genes belonging to the gene families associated with segmental duplications of the fusion region (excluding processed pseudogenes), 14 are transcriptionally active; six more appear to be capable of encoding functional proteins and may be transcribed in tissues or at developmental stages not yet examined; and 12 are pseudogenes with partial or disrupted ORFs. The fate of these duplicated genes is therefore consistent with the birth-and-death model for gene evolution (Nei et al. 1997). The relatively high fraction of potentially functional genes could reflect the action of purifying selection on the duplicates and/or the relative young age of some of these duplications (1% –4% overall divergence; Fan et al. 2002). Both our transcriptional assays and measurements of Ka/Ks ratios, however, indicate that many of these genes encode functional proteins. Some duplicates may have maintained the original gene's function, but increased the effective dosage. In other cases, the new copies might have evolved to take on new functions.
As far as we are aware, the CBWD genes in 2qFus and paralogous blocks are the first known eukaryotic homologs of the cobalamin synthetase W gene. Martin et al. (2002) recognized that these genes were multicopy; we demonstrate here that at least two paralogs are transcriptionally active. It will be important to conduct biochemical studies to determine if they function in the synthesis of vitamin B12 as they do in Pseudomonas and whether the paralogs have adopted distinct tissue-distribution patterns and/or functions in humans.
We are especially intrigued by the striking differences among the predicted carboxyl termini of the FOXD4 paralogs, which contrast with the extremely high conservation of their forkhead domains. Gross carboxy-terminal diversity is typical for other FOXD proteins, possibly to convey cell-type specific functions (Kaestner et al. 2000). So far, we can find transcripts from heart tissue corresponding to two paralogs. We will be curious to learn whether the varied FOXD4 forms have adopted cell-type specific functions or if the carboxyl termini are unimportant for the function of these proteins.
Because forkhead genes are key regulators of embryogenesis and tumorigenesis (Kaufmann and Knochel 1996), mutations in one or more of the FOXD4 genes identified here could lead to human disease. Mutations in other forkhead genes cause specific human diseases, including glaucoma (FOXC1) (Nishimura et al. 1998), lymphodema-distichiasis syndrome (FOXC2) (Fang et al. 2000), and a speech and language disorder (FOXP2) (Lai et al. 2001). The mouse Foxd4 ortholog is a candidate gene for the mouse mdf (muscle-deficient) mutation, which is characterized by nervous tremors and degeneration of the hindlimb muscles (Blot et al. 1995; Poirier et al. 1998). Because functional orthology need not correspond to genomic orthology, any of the human FOXD4 paralogs is a candidate for a role in analogous defects in humans.
The genes we describe here are embedded in regions with extensive homology to each other. Such regions are highly susceptible to ectopic recombination events that can result in gross chromosomal rearrangements (Ji et al. 2000; Samonte and Eichler 2002; Stankiewicz and Lupski 2002). These rearrangements could disrupt or alter the dosage of any of the genes described here, leading to an abnormal phenotype. It will therefore be worthwhile to test for such alterations in the genes identified in this study as potential candidates for human diseases linked to the ancestral fusion site or paralogous blocks. Moreover, normal variation in the sequence, copy number, and/or chromosomal context of these genes, such as the subtelomerically located retina-specific F379 transcripts (Mah et al. 2001) and olfactory receptor genes (Mefford et al. 2001), may contribute to phenotypic differences among healthy individuals.
METHODS
Database Mining and Sequence Analyses
The assembly, validation, and chromosomal mapping of genomic sequences analyzed for this report are described in the accompanying manuscript (Fan et al. 2002). Homologous sequences were obtained and analyzed by BLASTN, BLASTP, and/or BLASTX (Altschul et al. 1997) (http://www.ncbi.nlm.nih.gov/BLAST/) or cross_match (http://www.genome.washington.edu/phrap_documentation.html), and percentage identities calculated as described elsewhere (Fan et al. 2002). Multiple sequence alignments were performed using ClustalW 1.8 (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html). We used Diverge to calculate Ka/Ks ratios with addition for saturation levels of mutation for pairs of genes (S. Olsen, http://www.accelrys.com/support/bio/genhelp/) (Li et al. 1985; Li 1993).
PCR Analyses of Monochromosomal Hybrid Panel and BACs
We amplified specific segments from various chromosomes isolated in monochromosomal hybrid cell lines and BAC clones by PCR (80–100 or 5–10 ng DNA/reaction, respectively) to validate sequences obtained from GenBank. The primers are available on our Web site (http://www.fhcrc.org/labs/trask/subtelomeres/index.html) and listed in Supplemental Table 2, available online at http://www.genome.org. The PCR reaction conditions are described elsewhere (Fan et al. 2002).
Fluorescence In Situ Hybridization (FISH)
The chromosomal locations of BACs and a 6-kb PCR product encompassing a F379 gene was performed as described elsewhere (Fan et al. 2002). The F379 probe was generated from P1 RMC0MP013 (Trask et al. 1998) using primers given in Supplemental Table 2 (available online at http://www.genome.org) and the Boehringer-Mannheim Expand Long PCR System.
PCR of Marathon-Ready cDNA
Human brain and heart Marathon-Ready cDNAs (Clontech, Palo Alto, CA) were assayed for expression of specific genes. The PCR reactions contained 5 μL of Marathon-Ready cDNA, 250 μM dNTPs, 10 μM gene-specific primer (Supplemental Table 2, available online at http://www.genome.org) and adapter primer, and 1 μL Advantage 2 Polymerase Mix (Clontech). Cycling conditions were 94°C for 30 sec, 30 cycles of 5 sec at 94°C and 2 min at 68°C, followed by 5 min at 70°C.
DNA Sequencing
Excess dNTPs and primers were removed from DNA produced by PCR amplification with 2 U/μL of shrimp alkaline phosphatase and 10 U/μL of exonuclease I (Amersham, Piscataway, NJ) to 5 μL of PCR product, or by purifying 20 μL of PCR product through Sephacryl 300 spin columns (Sigma). Bulk PCR products were sequenced with Ready Reaction Big-dye terminator PRISM kits with AmpliTaq FS (Perkin Elmer). Sequencing primers were the same as those used for PCR amplification.
WEB SITE REFERENCES
http://www.fhcrc.org/labs/trask/subtelomeres/index.html, Trask laboratory Web site for supplementary information.
http://www.sanger.ac.uk/HGP/, Sanger Centre.
http://genome.ucsc.edu/, UCSC Human Genome Working Draft.
http://www.ncbi.nlm.nih.gov/Blast, NCBI genome resources.
http://ftp.genome.washington.edu/cgi-bin/RepeatMasker, RepeatMasker.
http://www.genome.washington.edu/phrap_documentation.html, cross_match.
http://www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi, Primer3.
http://www.jax.org/, comparative maps of mouse and human genomes.
http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html, ClustalW.
http://www.accelrys.com/support/bio/genhelp/, Diverge.
http://mgc.nci.nih.gov, NIH Mammalian Gene Collection.
Acknowledgments
The authors thank Eleanor Williams for help in preparing the figures; Colbey Harris for administrative assistance; and members of the Trask laboratory for helpful discussions. This work was supported by a grant GM057070 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL btrask@fhcrc.org; FAX (206) 667-4023.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.338402.
REFERENCES
- Achstetter T, Franzusoff A, Field C, Schekman R. SEC7 encodes an unusual, high molecular weight protein required for membrane traffic from the yeast Golgi apparatus. J Biol Chem. 1988;263:11711–11717. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann J, Valentine M, Kidd VJ, Lahti JM. Localization of chl1-related helicase genes to human chromosome regions 12p11 and 12p13: Similarity between parts of these genes and conserved human telomeric-associated DNA. Genomics. 1996;32:260–265. doi: 10.1006/geno.1996.0113. [DOI] [PubMed] [Google Scholar]
- Amann J, Kidd VJ, Lahti JM. Characterization of putative human homologues of the yeast chromosome transmission fidelity gene, CHL1. J Biol Chem. 1997;272:3823–3832. doi: 10.1074/jbc.272.6.3823. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Viggiano L, Misceo D, Horvath JE, Archidiacono N, Schwartz S, Rocchi M, Eichler EE. Human-specific duplication and mosaic transcripts: The recent paralogous structure of chromosome 22. Am J Hum Genet. 2002;70:83–100. doi: 10.1086/338458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blot S, Poirier C, Dreyfus PA. The mouse mutation muscle deficient (mdf) is characterized by a progressive motoneuron disease. J Neuropathol Exp Neurol. 1995;54:812–825. [PubMed] [Google Scholar]
- Brown WR, MacKinnon PJ, Villasante A, Spurr N, Buckle VJ, Dobson MJ. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson C L, Chabre M. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- Ciccodicola A, D'Esposito M, Esposito T, Gianfrancesco F, Migliaccio C, Miano MG, Matarazzo MR, Vacca M, Franze A, Cuccurese M, et al. Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet. 2000;9:395–401. doi: 10.1093/hmg/9.3.395. [DOI] [PubMed] [Google Scholar]
- Crouzet J, Levy-Schil S, Cameron B, Cauchois L, Rigault S, Rouyez MC, Blanche F, Debussche L, Thibaut D. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. J Bacteriol. 1991;173:6074–6087. doi: 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, Feo S, Heard E, Fried M. A strategy to detect and isolate an intron-containing gene in the presence of multiple processed pseudogenes. Proc Natl Acad Sci. 1989;86:6691–6695. doi: 10.1073/pnas.86.17.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards YH, Putt W, Fox M, Ives JH. A novel human phosphoglucomutase (PGM5) maps to the centromeric region of chromosome 9. Genomics. 1995;30:350–353. doi: 10.1006/geno.1995.9866. [DOI] [PubMed] [Google Scholar]
- Fan W, Christensen M, Eichler E, Zhang X, Lennon G. Cloning, sequencing, gene organization, and localization of the human ribosomal protein RPL23A gene. Genomics. 1997;46:234–239. doi: 10.1006/geno.1997.5038. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Linardopoulou, E., Friedman, C., Williams, E., and Trask, B.J. 2002. Genomic Structure and Evolution of the Ancestral Chromosome Fusion Site in 2q13–2q14.1 and Paralogous Regions on Other Human Chromosomes. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Fang J, Dagenais SL, Erickson RP, Arlt MF, Glynn MW, Gorski JL, Seaver LH, Glover TW. Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am J Hum Genet. 2000;67:1382–1388. doi: 10.1086/316915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring SL, Spencer F, Hieter P. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund M, Mitelman F, Mandahl N. A human 12p-derived cosmid hybridizing to subsets of human and chimpanzee telomeres. Cytogenet Cell Genet. 1995;70:88–91. doi: 10.1159/000133998. [DOI] [PubMed] [Google Scholar]
- Ijdo J, Baldini A, Ward DC, Reeders ST, Wells RA. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc Natl Acad Sci. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Monaghan AP, Kern H, Ang SL, Weitz S, Lichter P, Schutz G. The mouse fkh-2 gene. Implications for notochord, foregut, and midbrain regionalization. J Biol Chem. 1995;270:30029–30035. doi: 10.1074/jbc.270.50.30029. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, Kurzbauer R, Dorfler P, Busslinger M. Alternative splicing of Pax-8 gene transcripts is developmentally regulated and generates isoforms with different transactivation properties. Mol Cell Biol. 1993;13:6024–6035. doi: 10.1128/mcb.13.10.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z, Czerny T, Busslinger M. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J. 1997;16:6793–6803. doi: 10.1093/emboj/16.22.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Lai E, Prezioso VR, Tao WF, Chen WS, Darnell JE., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- Larsson C, Hellqvist M, Pierrou S, White I, Enerback S, Carlsson P. Chromosomal localization of six human forkhead genes, freac-1 (FKHL5), - 3 (FKHL7), -4 (FKHL8), -5 (FKHL9), -6 (FKHL10), and -8 (FKHL12) Genomics. 1995;30:464–469. doi: 10.1006/geno.1995.1266. [DOI] [PubMed] [Google Scholar]
- Li J, Vogt PK. The retroviral oncogene qin belongs to the transcription factor family that includes the homeotic gene fork head. Proc Natl Acad Sci. 1993;90:4490–4494. doi: 10.1073/pnas.90.10.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Linardopoulou E, Mefford HC, Nguyen OT, Friedman C, van den Engh G, Farwell DG, Coltrera M, Trask BJ. Transcriptional activity of multiple copies of a subtelomerically located olfactory receptor gene that is polymorphic in number and location. Hum Mol Genet. 2001;10:2373–2383. doi: 10.1093/hmg/10.21.2373. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Macchia PE, Lapi P, Krude H, Pirro MT, Missero C, Chiovato L, Souabni A, Baserga M, Tassi V, Pinchera A, et al. PAX8 mutations associated with congenital hypothyroidism caused by thyroid dysgenesis. Nat Genet. 1998;19:83–86. doi: 10.1038/ng0598-83. [DOI] [PubMed] [Google Scholar]
- Mah N, Stoehr H, Schulz HL, White K, Weber BH. Identification of a novel retina-specific gene located in a subtelomeric region with polymorphic distribution among multiple human chromosomes. Biochim Biophys Acta. 2001;1522:167–174. doi: 10.1016/s0167-4781(01)00328-1. [DOI] [PubMed] [Google Scholar]
- Martin CL, Wong A, Gross A, Chung J, Fantes JA, Ledbetter DH. The evolutionary origin of human subtelomeric homologies—or where the ends begin. Am J Hum Genet. 2002;70:972–984. doi: 10.1086/339768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Gallardo A, Lamerdin J, Sopapan P, Friedman C, Fertitta AL, Garcia E, Carrano A, Negorev D, Macina RA, Trask BJ, et al. Molecular analysis of a novel subtelomeric repeat with polymorphic chromosomal distribution. Cytogenet Cell Genet. 1995;71:289–295. doi: 10.1159/000134129. [DOI] [PubMed] [Google Scholar]
- Mayer G, Blind M, Nagel W, Bohm T, Knorr T, Jackson CL, Kolanus W, Famulok M. Controlling small guanine-nucleotide-exchange factor function through cytoplasmic RNA intramers. Proc Natl Acad Sci. 2001;98:4961–4965. doi: 10.1073/pnas.091100698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Linardopoulou E, Coil D, van den Engh G, Trask BJ. Comparative sequencing of a multicopy subtelomeric region containing olfactory receptor genes reveals multiple interactions between non-homologous chromosomes. Hum Mol Genet. 2001;10:2363–2372. doi: 10.1093/hmg/10.21.2363. [DOI] [PubMed] [Google Scholar]
- Moiseeva EP, Belkin AM, Spurr NK, Koteliansky VE, Critchley DR. A novel dystrophin/utrophin-associated protein is an enzymatically inactive member of the phosphoglucomutase superfamily. Eur J Biochem. 1996;235:103–113. doi: 10.1111/j.1432-1033.1996.00103.x. [DOI] [PubMed] [Google Scholar]
- Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Rosenberg M, Biesecker LG, Ledbetter DH. Isolation of the human chromosome 22q telomere and its application to detection of cryptic chromosomal abnormalities. Hum Genet. 1996;97:765–769. doi: 10.1007/BF02346187. [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19:140–147. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- Park HS, Nogami M, Okumura K, Hattori M, Sakakia Y, Fujiyama A. Newly identified repeat sequences, derived from human chromosome 21qter, are also localized in the subtelomeric region of particular chromosomes and 2q13, and are conserved in the chimpanzee genome. FEBS Lett. 2000;475:167–169. doi: 10.1016/s0014-5793(00)01632-x. [DOI] [PubMed] [Google Scholar]
- Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: Binding site specificity and DNA bending. EMBO J. 1994;13:5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier C, Blot S, Fernandes M, Carle GF, Stanescu V, Stanescu R, Guenet JL. A high-resolution genetic map of mouse chromosome 19 encompassing the muscle-deficient osteochondrodystrophy (mdf-ocd) region. Mamm Genome. 1998;9:390–391. doi: 10.1007/s003359900777. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Cox C, Jessell TM, Klar A. Ectopic neural expression of a floor plate marker in frog embryos injected with the midline transcription factor Pintallavis. Proc Natl Acad Sci. 1993;90:8268–8272. doi: 10.1073/pnas.90.17.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonte RV, Eichler EE. Segmental duplications and the evolution of the primate genome. Nat Rev Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- Shi J, Cai W, Chen X, Ying K, Zhang K, Xie Y. Identification of dopamine responsive mRNAs in glial cells by suppression subtractive hybridization. Brain Res. 2001;910:29–37. doi: 10.1016/s0006-8993(01)02393-9. [DOI] [PubMed] [Google Scholar]
- Sillekens PT, Beijer RP, Habets WJ, van Verooij WJ. Molecular cloning of the cDNA for the human U2 snRNA-specific A‘ protein. Nucleic Acids Res. 1989;17:1893–1906. doi: 10.1093/nar/17.5.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Torban E, Goodyer P. What PAX genes do in the kidney. Exp Nephrol. 1998;6:7–11. doi: 10.1159/000020498. [DOI] [PubMed] [Google Scholar]
- Trask B, Fertitta A, Christensen M, Youngblom J, Bergmann A, Copeland A, de Jong P, Mohrenweiser H, Olsen A, Carrano A, et al. Fluorescence in situ hybridization mapping of human chromosome 19: Cytogenetic band location of 540 cosmids and 70 genes or DNA markers. Genomics. 1993;15:133–145. doi: 10.1006/geno.1993.1021. [DOI] [PubMed] [Google Scholar]
- Trask BJ, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, et al. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet. 1998;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- Wong AC, Shkolny D, Dorman A, Willingham D, Roe BA, McDermid HE. Two novel human RAB genes with near identical sequence each map to a telomere-associated region: The subtelomeric region of 22q13.3 and the ancestral telomere band 2q13. Genomics. 1999;59:326–334. doi: 10.1006/geno.1999.5889. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O. The origin of man: A chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]