Abstract
The subtelomeric domains of chromosomes are probably the most rapidly evolving structures of the human genome. The highly variable distribution of large duplicated subtelomeric segments has indicated that frequent exchanges between nonhomologous chromosomes may have been taking place during recent genome evolution. We have studied the extent and variability of such duplications using in situ hybridization techniques and a set of well-defined subtelomeric cosmid probes that identify discrete regions within the subtelomeric domain. In addition to reciprocal translocation and illegitimate recombination events that could explain the observed mosaic pattern of subtelomeric regions, it is likely that homology-based recombination mechanisms have also contributed to the spread of distal subtelomeric sequences among particular groups of nonhomologous chromosome arms. The frequency and distribution of large-scale subtelomeric polymorphisms may have direct implications for the design of chromosome-specific probes that are aimed at the identification of cryptic subtelomeric deletions. Furthermore, our results indicate that the relevance of some of the telomere closures proposed within the present Human Genome Sequence draft are restricted to specific allelic variants of unknown frequencies.
[The sequence of cosmid ICRF10 (carrying DNF92) was deposited in GenBank under accession no. Y13543.]
The telomeric regions of human chromosomes comprise essential structures ensuring the stability of the genome (Hackett et al. 2001), participating in nuclear architecture (Nagele et al. 2001) and promoting homologous chromosome pairing during meiosis (Walker and Hawley 2000). Although great attention has been recently paid to telomeres and their role in cellular senescence and carcinogenesis (Blackburn 2000), relatively little is known about the segments connecting the terminal hexameric repeats to chromosome-specific sequences in humans. The morbidity associated with cryptic subtelomeric deletions (de Vries et al. 2001) prompted the development of chromosome-specific subtelomeric probes (for review, see Knight and Flint 2000) and stimulated structural studies of subtelomeric domains (Flint et al. 1997a). However, mapping and sequencing data regarding the proterminal regions of human chromosomes remain difficult to generate and exploit owing to the complexity, nonspecificity, and size variability typical of such regions (Brown et al. 1990; Wilkie et al. 1991; Macina et al. 1994; Bailey et al. 2001; Mefford and Trask 2002).
Nevertheless, a comparative analysis of available human and yeast subtelomeric sequences allowed Flint et al. (1997a) to propose a structure common to most, if not all, human chromosome extremities. The presence of interstitial degenerate telomere repeats near the chromosome tip divides the subtelomeric domain into two structurally (and maybe functionally) different subdomains: one distal, directly connected to the hexameric telomeric repeats, and one proximal, connected to chromosome-specific sequences. The distal subdomain, up to a few kilobases long, typically presents a high density of ESTs and other short sequences that show interrupted matches to multiple distal subtelomeric regions. The proximal subdomain, which may extend over hundreds of kilobases, contains large regions of homology to a more restricted number of chromosome ends. Sequence analyses indicate that, although the distal and proximal subdomains seem to have evolved independently, frequent exchanges appear to have been taking place between nonhomologous chromosomes (Flint et al. 1997a; Mefford et al. 2001; Mefford and Trask 2002). Whereas the mechanisms leading to sequence exchange between distal subdomains remain unclear, the dissemination of proximal subtelomeric sequences among nonhomologous chromosome ends may be explained by mechanisms akin to reciprocal translocation processes, in which regions of limited homology could nevertheless be implicated (Monfouilloux et al. 1998; Vergnaud 1999).
Recently, the characterization of particular subtelomeric duplications observed in humans indicated the existence of additional structural boundaries that define discrete regions within the subtelomeric domains (Monfouilloux et al. 1998; Vergnaud 1999). Several cosmid probes issued from these regions were alternatively present or absent on a subset of chromosome extremities, thus demonstrating high variability. In particular, the most telomeric cosmid probe, carrying minisatellite DNF92 (Monfouilloux et al. 1998), was invariably or very frequently observed at some particular locations (referred to as major sites), whereas its presence was quite inconstant at others (minor sites).
The genome distribution of DNF92 is reminiscent of that described for another widespread subtelomeric sequence, f7501 (Trask et al. 1998), although major sites apparently differed between these markers. In addition, in situ hybridization analyses using P1 clones showed that sequences that extend f7501 toward the centromere have a wider subtelomeric distribution (Trask et al. 1998), which is in fact quite similar to the distribution observed for DNF92-associated sequences (Monfouilloux et al. 1998). These observations have led to the hypothesis that DNF92 and f7501 sequences occupy analogous positions within related subtelomeric structures (Vergnaud 1999).
In this work, we have conducted a systematic FISH analysis to portray the subtelomeric domains and their variations within and between individual human genomes. A set of slightly overlapping cosmid clones was designed, such that each cosmid represents distinct regions or subregions that together cover ∼200 kb. Using a cohybridization approach, we were then able to reconstitute the subtelomeric structures of specific chromosome arms.
RESULTS
The Subtelomeric Sequences DNF92 and f7501 Are Frequently Detected on the Same Chromosome Arms but Rarely Coexist
Table 1 summarizes the frequency distributions for f7501 and DNF92 (region 1, Fig. 1) in a total of 74 haploid genomes. Specific consistent signals were observed at 14 chromosome extremities. Of these, only the 3qter and 17qter loci were invariable, always carrying f7501 and DNF92, respectively, in agreement with previous observations (Monfouilloux et al. 1998; Trask et al. 1998). The other chromosome ends were polymorphic, bearing either one of the markers or none (Fig. 1B). Two particular cases were observed: the locus 1pter, where f7501 is never detected, and the locus 6qter, where one of the two markers is always present. As anticipated, major sites are different for the two markers (Table 1). Moreover, the simultaneous presence of DNF92 and f7501 sequences on the same chromosome extremity was very rarely observed, being limited to minor sites 11pter and 19pter (Table 1). In total, DNF92 was detected at 442 chromosome ends, whereas f7501 was present at 279 ends, that is, 5.97 and 3.77 times per haploid genome, respectively. This situation is in clear contrast with that observed in other higher primates in which both markers have unique locations (Monfouilloux et al. 1998; Trask et al. 1998).
Table 1.
Observed Locations and Frequencies for Probes ICRF10 (Carrying DNF92) and f7501 in 31 Caucasian and 6 Pygmy Individuals
| Location | Caucasian (62) | Pygmy (12) | Total (74) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DNF92 | f7501 | No signal | DNF92 | f7501 | No signal | DNF92 | f7501 | No signal | |
| 1pter | 55 | 0 | 7 | 10 | 0 | 2 | 65 | 0 | 9 |
| 3qter | 0 | 62 | 0 | 0 | 12 | 0 | 0 | 74 | 0 |
| 5qter | 60 | 1 | 1 | 12 | 0 | 0 | 72 | 1 | 1 |
| 6pter | 14 | 4 | 44 | 1 | 2 | 9 | 15 | 6 | 53 |
| 6qter | 61 | 1 | 0 | 12 | 0 | 0 | 73 | 1 | 0 |
| 7pter | 8 | 1 | 53 | 5 | 1 | 6 | 13 | 2 | 59 |
| 8pter | 44 | 0 | 18 | 8 | 1 | 3 | 52 | 1 | 21 |
| 9qter | 12 | 14 | 36 | 10 | 1 | 1 | 22 | 15 | 37 |
| 11pter | 27* | 28* | 10 | 8 | 2 | 2 | 35* | 30* | 12 |
| 15qter | 3 | 58 | 1 | 0 | 12 | 0 | 3 | 70 | 1 |
| 16pter | 3 | 0 | 59 | 3 | 1 | 8 | 6 | 1 | 67 |
| 16qter | 3 | 5 | 54 | 0 | 5 | 7 | 3 | 10 | 61 |
| 17qter | 62 | 0 | 0 | 12 | 0 | 0 | 74 | 0 | 0 |
| 19pter | 9* | 56* | 5 | 0 | 12 | 0 | 9* | 68* | 5 |
Takes into account colocalization observations.
Only consistent (present in all metaphases) fluorescent signals were scored. Weak, inconstant signals were occasionally observed at 2qter and 20pter/qter with DNF92, and at 9pter and 12pter with f7501 (data not shown). Boldface numerals indicate the major (most frequent sites) for both markers. No obvious differences between the ethnic groups were noticed, aside from an apparent excess of DNF92 locations at 9qter in the Pygmy population. This may be caused by sample variations because, on the other hand, f7501 was not frequently observed at 7pter in this ethnic group, contrary to a previous report (Trask et al. 1998).
Figure 1.
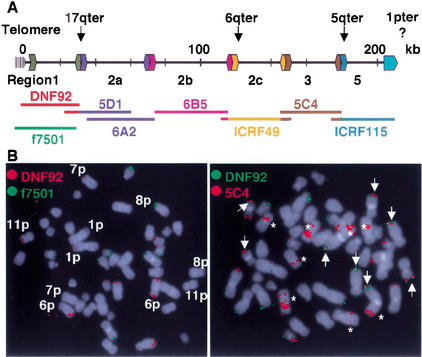
(A) Cartography of the subtelomeric domain common to Chromosome arms 1p/q, 5q, 6q, and 17q (Monfouilloux et al. 1998). The arrows indicate the limit between the subtelomeric domain and chromosome-specific sequences for three of the chromosomes. This limit has not been found for 1p/qter. The limits between regions have been previously defined (Monfouilloux et al. 1998; Vergnaud 1999), and their relative positions are indicated here. The length of the lines approximately corresponds to the size of the cosmid probes used in this study. Extremities of the same color indicate actual overlapping among clones. (B) Colocalization of probes by two-color FISH. (Left) Cohybridization obtained using region 1 probes (red and green). Chromosomes are counterstained with DAPI. Chromosome arms bearing presence/absence polymorphisms are indicated. All other locations are homozygous in this individual. (Right) Cohybridization using probes specific to regions 1 (green) and 3 (red). Arrows indicate colocalization. The 5C4 probe also hybridizes to interstitial locations (indicated with an asterisk) on Chromosomes 1, 4, 7, and 10. The results presented in Table 1 and Figure 2 only take into account signals that are distinct and consistently detected (i.e., are present in all metaphases analyzed from an individual).
Although the hypervariable character of these markers indicates some intrinsic instability, segregation studies using Southern blotting analyses and large CEPH families have previously shown that multicopy minisatellite DNF92 is stably transmitted through meiosis (Monfouilloux et al. 1998). We have confirmed and extended this observation by FISH using DNF92 and f7501 cosmid probes. In all families examined so far (including some large CEPH families, adding up to ∼100 meiotic events), both markers have shown Mendelian inheritance at all subtelomeric locations (data not shown). Also, no mitotic instability has been observed in long-term cultures of lymphoblastoid cell lines (data not shown). The term polymorphism is therefore appropriate to refer to the presence/absence variability of subtelomeric markers DNF92 and f7501.
The Reconstitution of Subtelomeric Domains of Single Chromosome Arms Reveals High Variability Within and Between Genomes
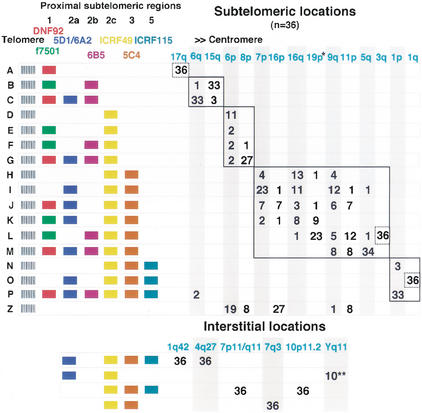
We next analyzed, by FISH, the rest of the subtelomeric domain. To do this, we labeled probes from a panel of cosmid clones representing discrete regions from the subtelomeric domains of 1p, 5q, 6q, and 17q chromosome arms (Fig. 1; Monfouilloux et al. 1998). The results of cohybridization experiments carried out in 18 individuals (including the 6 individuals of African origin) are summarized in Figure 2. The presence or absence of hybridization signals on individual chromosome extremities allowed us to distinguish 17 different subtelomeric combinations or structures (including the complete absence of fluorescent signals) distributed among 15 chromosome ends. These structures are detected in the same subset of chromosome arms already defined by DNF92 and f7501 (although these markers are not always present) plus another chromosome end, 1qter. Some of the combinations observed (namely, A, D, E, N, and O) have unique locations, the rest being polymorphically distributed among certain chromosome ends. With the exception of the invariable 17qter, 3qter, and 1qter locations, the number of variants detected per chromosome end ranged from 2 (1pter) to 6 (9qter). As reported previously (Monfouilloux et al. 1998), interstitial signals are constantly observed at specific locations with cosmids representing regions 2a, 2c, and 3 (Figs. 1B and 2). Our results now indicate that region 5 is also constantly detected at some of the same sites (Fig. 2).
Figure 2.
Cartography by FISH of the subtelomeric domains observed in 18 nonrelated individuals (36 haploid genomes). Colored boxes indicate that a consistent fluorescent signal was observed with the corresponding cosmid probe (color code for probes is as in Fig. 1), whereas their absence indicates no fluorescent signal obtained. The cohybridization of probes allowed, in all cases showing presence/absence of polymorphisms, the distinction between homologous chromosomes and the reconstitution of subtelomeric domains for individual chromosome arms. For a given combination, certain regions showed differences in fluorescence intensity between chromosome arms (not noted here), which may reflect additional size or sequence heterogeneity. The variants A to P were first arranged according to their complexity within the more proximal half of the subtelomeric domain, and then, within each group, according to the complexity of the more distal half. Variant Z corresponds to the absence of signal for all probes used (null allele). Homologous recombination events implicating the more proximal half of subtelomeric domains may have contributed to the shuffling of more distal sequences among the chromosome arms contained within the solid-line rectangles. The dotted rectangles outline invariable loci. (*) 34 observations, the remaining two correspond to f7501/DNF92 colocalizations in which a subtelomeric structure of type J/K was detected. (**) Out of 10 male individuals.
Although no obvious differences were observed in the distribution of variants between Caucasian and African individuals, such differences cannot be excluded, given the relatively small population sample. On the other hand, we did not score fluorescence intensities for each probe at every location. Nonetheless, such differences were sometimes noticeable, indicating additional levels of polymorphism (size of the corresponding region or lower degree of sequence homology).
Sequence Analyses of Genomic Fragments Comprising Subtelomeric Domains Reveal Similar Segmental Arrangements
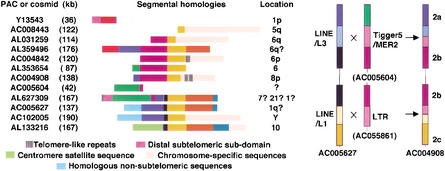
The results presented in Figure 2 indicate that the order of segments may be consistent across all chromosome ends. However, this has been formally established only for two alleles, represented by half-YACs and derived, respectively, from Chromosomes 5qter and 6qter (Monfouilloux et al. 1998). The segmental arrangement of these half-YACs corresponds, respectively, to the M and C alleles described here, which are, indeed, the configurations most frequently observed by FISH on 5qter and 6qter in our population (Fig. 2). Owing to the human genome sequencing project (Lander et al. 2001), this analysis can now be extended to other alleles. For this purpose, sequence data generated previously (Monfouilloux et al. 1998) and derived from different segments along the two half-YACs were used to search human genome sequences in the available databases. Fully (finished) sequenced PACs were thus identified and compared to each other with the help of the PipMaker program (Schwartz et al. 2000). The deduced organizations for homologous segments within these PACs (illustrated in Fig. 3, left) indicate that the order of these segments is conserved among several subtelomeric regions as well as at interstitial locations (i.e., Chromosomes 10 and Y). Overall, there is a high degree of homology (>98%) at the sequence level within the corresponding segments (data not shown), although their size may vary.
Figure 3.
(Left) Schematic representation of the segmental organization of large genomic fragments. The colored boxes (drawn to scale) represent the different subtelomeric segments according to the code in Figure 2. All sequences are identified by their accession numbers (only finished, not draft, sequences were analyzed). The proposed chromosome location is also indicated. AC004842 is assigned to 7p22 in the GenBank; however, a 6p location was found using the Genebridge4 hybrid panel (Y.-M. Borde, unpubl.). Similarly, AC004908 (no chromosomal location indicated by NCBI) has been assigned to Chromosome 8p using the Genebridge4 hybrid panel (Y.-M. Borde, unpubl.). AL627309 (previously named AC073186) has been successively assigned to Chromosomes 7, 21, and, more recently, 1, illustrating the difficulties of assigning subtelomeric genomic fragments with no obvious connection to chromosome-specific sequences. The black box represents a novel region not included in our FISH studies. (Right) Proposed illegitimate recombination events leading to the incorporation of an intervening subtelomeric region. The color code is as above with lighter colors representing the boundaries of the corresponding regions. Boxes are not drawn to scale. The sequence AC005627 carries the ancestral configuration, and AC004908 represents one of the final products after a double recombination event, implicating sequences similar to the ones found in AC005604 and AC055861. Although not found in the sequence database, the reciprocal product may correspond to the E variant observed by FISH in the population.
The comparisons between PACs also revealed additional features not detected by FISH in this study. An additional small region (black box in Fig. 3) is found between the 2a and 2c regions (blue and orange boxes) when the intervening region 2b (purple box) is absent. The analysis of the junctions between these blocks, overlapped by interspersed repeated elements, points to a history of recombination events and indicates that the blue–black–orange arrangement is ancestral. An ectopic recombination event between black–orange (as observed in AC005627) and purple–X (as in AC0055861) probably resulted in the observed organization purple–orange (observed in AC004908; Fig. 3, right). The predicted reciprocal product (black–X), not detectable by FISH, is absent from the present collection of human genome sequences and might have been lost during human evolution. A similar event between blue–black (also in AC005627) and green–purple (as in AC005604) may have produced the blue–purple combination observed again in AC004908 (Fig. 3, right). The predicted reciprocal product (green–black) is not present among the known human genome sequences but may correspond to the E allele (green–[black]–orange?) observed twice by FISH on Chromosome 6p (Fig. 2).
DISCUSSION
The results reported here clearly illustrate the extreme polymorphism that characterizes the subtelomeric domains of a subset of human chromosomes. Because the presence/absence of fluorescent signals indicates the gain/loss of segments that are several tens of kilobases long, this variability is most probably related to size polymorphisms known to affect some chromosome ends (Brown et al. 1990). For instance, the relatively rare 6qter-DNF92 variant P observed here may correspond to, and perhaps completely explain, the long 6qter allele described by Macina et al. (1995). Similarly, the four 16pter variants distinguished here by FISH techniques may be related to the length polymorphism found for this chromosome end (Wilkie et al. 1991). In this case, the frequently observed Z variant (null) apparently corresponds to the common, fully sequenced allele A, which lacks all the subtelomeric segments tested here (Flint et al. 1997b).
The molecular mechanisms by which such polymorphisms arise within the human genome or the sequence of events resulting in a particular combination of regions are still unknown. Nonetheless, this report provides the grounds for new conjectures regarding the evolution of subtelomeric domains. Our results are compatible with the idea that chromosome extremities do not evolve independently (Flint et al. 1997a; Mefford et al. 2001). As proposed before (Vergnaud 1999), balanced translocations have probably contributed to the spread of subtelomeric regions; this is clearly indicated by the detection of a P variant at 6qter, possibly originating from 1pter. At the sequence level, the presence of telomere-like repeats near some of the junctions between the subtelomeric domain and chromosome-specific sequences (PACs AC004842 and AC004908; Fig. 3, left) could indicate that these domains were grafted onto simpler subtelomeric structures (Z variants). Z variants are also compatible either with reciprocal translocation events implicating chromosome ends that carry completely unrelated subtelomeric domains or with subtelomeric domains that have been lost because of terminal deletions (eventually rescued by telomere capture mechanisms; Flint et al. 1994). The frequency of the latter events in the normal population is unknown.
Aside from P and Z, other variants tend to show a clustering distribution among chromosome arms (Fig. 2), indicating that random, reciprocal translocations that carry whole subtelomeric domains are rather rare. It may also be that the products of such translocations are rapidly eliminated from the population either by genetic drift or deleterious effects, which likely depend on the extent of chromosome-specific sequences involved.
On the other hand, mechanisms based on homologous recombination could explain the spread of distal sequences among defined groups of nonhomologous chromosome arms, outlined by rectangles in Figure 2. This hypothesis requires that the order of most segments be conserved across chromosome ends. Our results from comparisons between fully sequenced PACs comprising several subtelomeric segments to DNF92/f7501 indicate that this may be the case. Conceivably, the presence of large homologies within the more proximal half of subtelomeric domains increases, during bouquet formation in meiosis, the chance for transient interactions between otherwise nonhomologous chromosome ends (Roeder 1997). This interaction would occasionally foster recombination events, with exchange of more distal sequences. Interestingly, certain chromosome ends seem to be excluded from these events, as indicated by the absence of distal variability at some locations such as 1qter or 3qter, in spite of bearing substantial homology to other variants. A high stability is similarly observed at interstitial subtelomere-related structures, although in this case sporadic nonhomologous interactions leading to reciprocal exchanges at this level would necessarily bring about obvious genome rearrangements with inevitable deleterious consequences.
The exclusive relationship observed between DNF92 and f7501 sequences is intriguing. It has been proposed that the duplication of f7501 at subtelomeric sites throughout the genome likely predated the split of the primate clade (Trask et al. 1998) and, therefore, preceded the DNF92 duplication (observed only in the human genome; Monfouilloux et al. 1998). Our results now show that both duplications affect the same subset of chromosomes, but do not coincide, and that DNF92 is present at more chromosome ends than f7501. These observations concurrently indicate that the spread of DNF92 may have occurred to the detriment of f7501, and connote a better efficiency of the former to colonize chromosome ends and/or to become fixed in the population. Although such an opposite evolution may be the result of genetic drift, the role of as-yet unknown selective pressures cannot be formally ruled out.
The subtelomeric hybridization profiles obtained on 36 haploid genomes provide us also with some hints about the history of the DNF92 diffusion among chromosome ends. The nonobservation of variants in which DNF92 is linked to region 2b but not to 2a indicates that the duplication of DNF92 outside 17qter (its ancestral site) led first to its association with region 2a (leading from variant I to variant J). This association was later followed by the addition of 2b. The last event may correspond to an illegitimate recombination between, for instance, variants F and J, resulting in variants G and E, all four being still present in the population. In support of this hypothesis is our analysis of available sequences, which indicates that the configuration 2a–2c is ancestral and reveals the traces of ectopic recombination events that may have originated the 2a–2b–2c variant. Taken together, our results are compatible with a limited number of rearrangements being the cause of the apparently complex patterns observed. Only a detailed sequence analysis of all variants, together with sequence information from nonhuman primates, will allow a more precise phylogenetic dissection and eventually cast some light on the molecular mechanisms involved. Unfortunately, such studies are still hindered by the scarcity of fully sequenced proterminal regions.
The reasons that the polymorphisms described here affect only a particular subset of chromosome ends in the human genome are not clear. Nonetheless, the existence of other families of large subtelomeric duplications, affecting an overlapping or a completely different set of chromosomes, is indicated by the genome distribution of other, completely unrelated, subtelomeric sequences (Cross et al. 1990; Ijdo et al. 1992; Martin-Gallardo et al. 1995; Ciccodicola et al. 2000). This possibility is supported by a recent report showing that large blocks of sequences may be found polymorphically duplicated at the short arm of all human acrocentric chromosomes (Piccini et al. 2001). In any case, it would be interesting to determine how these widespread subtelomeric homologies influence the physiological mechanisms mediating homologous chromosome pairing during meiosis.
The observations reported here have also implications for the development of diagnostic tools that aim at correlating cryptic subtelomeric deletions and clinical manifestations. As noted by others (Knight et al. 2000), the nearer a probe is to the telomere, the higher its probability of being non-chromosome-specific. In fact, our data predict that this obstacle may be particularly acute on specific chromosome arms affected by extended subtelomeric length polymorphisms. Furthermore, the distance between a chromosome-specific probe and the telomere may vary substantially between individuals, a circumstance that must be taken into account when the physical location of a marker is studied (Knight et al. 2000). Conversely, because balanced translocations just affecting subtelomeric domains seem to be rare enough in the general population (as suggested above), the cartography by FISH of subtelomeric domains in individuals suspected of carrying cryptic translocations may cast a light onto difficult diagnostic situations by disclosing unusual locations or structural patterns.
Finally, at least some of the telomere closures proposed in the present draft Human Genome Sequence (Riethman et al. 2001), and especially those corresponding to chromosome extremities shown here to be polymorphic, are presumably allelic variants. The subtelomeric cartography by FISH of a large number of individuals is required to estimate allelic frequencies and assess ethnic differences. Although this information will certainly contribute to our understanding of this form of genetic diversity, the full sequence characterization of different chromosome-specific subtelomeric alleles is needed to definitely sort out their phylogeny.
METHODS
DNA Probes
The isolation and characterization of cosmid probes was as described (Monfouilloux et al. 1998). Cosmids ICRF10 (carrying DNF92, GenBank accession number Y13543), ICRF49 (icrfC112N2142), and ICRF115 (icrfC112I0546Q6) were identified from a Chromosome 1-specific library (Monfouilloux et al. 1998). Cosmids 5D1 and 5C4, and cosmids 6A2 and 6B5, were obtained from 5qter and 6qter subcloned half-YACs, respectively (Monfouilloux et al. 1998). Cosmid f7501 (Trask et al. 1998) was obtained from B. Trask (University of Washington, Seattle). BACs b231F8 and h563E1 from the CEPH collection were occasionally used to identify Chromosome 8 and Chromosome 19, respectively.
Individuals
Established lymphoblastoid cell lines from unrelated individuals were selected from available collections at the CEPH, including six lymphoblastoid cell lines from two African Pygmy populations (Biaka and Mbouti). Aliquots of fresh whole blood samples from unrelated healthy donors participating in different CEPH research programs were also included. Finally, two human diploid fibroblast cell lines were obtained from ATCC. With the exception of the six African individuals, all others were of Caucasian origin. Metaphase spreads were prepared from lymphoblastoid cell lines and from PHA-stimulated PBLs following conventional methods.
FISH
Hybridizations were carried out as described (Pinkel et al. 1986), always in the presence of two different probes in order to reveal colocalization. Cosmids were labeled either with biotin or digoxigenin and revealed by Texas-red avidin (Vector) or FITC-conjugated anti-digoxin antibodies (Sigma), respectively. Cosmids ICRF10 and f7501 were tested in all individuals (n = 37). From these, 18 were tested with all possible cosmid combinations. Chromosomes were counterstained with DAPI. Signals were visualized under an epifluorescence microscope (Axioplan2, Zeiss) equipped with a computer-piloted filter wheel. Red, green, and blue fluorescent signals were independently captured through a CCD camera (Photometrics-Sensys) using the corresponding wavelength excitation filter. Merged pseudocolor images were obtained, and normalization and enhancements procedures were used to improve G-banding and specific signal detection. Most of the time, chromosomes were identified based on DAPI simulated G-banding. Occasionally, chromosome-specific probes (BACs) were cohybridized to establish chromosome locations.
Sequence Analyses
Similarity searches were done using BLAST (Altschul et al. 1990) and public human genome sequence data as accessible from the NCBI Web site. Dot-plot alignments of large sequences were performed using advanced PipMaker (Schwartz et al. 2000; accessed at http://bio.cse.psu.edu/pipmaker/). Alignments required the masking of interspersed repeats, which was obtained using RepeatMasker (A.F.A. Smit and P. Green, RepeatMasker at http://ftp.genome.washington.edu/RM/RepeatMasker.html) accessed through Infobiogen (http://www.infobiogen.fr/services/deambulum/).
WEB SITE REFERENCES
http://bio.cse.psu.edu/pipmaker/; Pipmaker program.
http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker program.
http://www.infobiogen.fr/services/deambulum/; Infobiogen analysis tools.
Acknowledgments
We thank C. de Toma, L. Cazes, and the other members of the Cell Culture Laboratory at CEPH for the excellent technical assistance. Thanks to the Centre National du Séquençage for sequencing cosmid ICRF10. Thanks also to B. Trask for providing the f7501 cosmid and to R. Berger for comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL londono@cephb.fr; FAX 33-0-15372512.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.322802.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- Brown WR, MacKinnon PJ, Villasante A, Spurr N, Buckle VJ, Dobson MJ. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- Ciccodicola A, D'Esposito M, Esposito T, Gianfrancesco F, Migliaccio C, Miano MG, Matarazzo MR, Vacca M, Franze A, Cuccurese M, et al. Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet. 2000;9:395–401. doi: 10.1093/hmg/9.3.395. [DOI] [PubMed] [Google Scholar]
- Cross S, Lindsey J, Fantes J, McKay S, McGill N, Cooke H. The structure of a subterminal repeated sequence present on many human chromosomes. Nucleic Acids Res. 1990;18:6649–6657. doi: 10.1093/nar/18.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BB, White SM, Knight SJ, Regan R, Homfray T, Young ID, Super M, McKeown C, Splitt M, Quarrell OW, et al. Clinical studies on submicroscopic subtelomeric rearrangements: A checklist. J Med Genet. 2001;38:145–150. doi: 10.1136/jmg.38.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Craddock CF, Villegas A, Bentley DP, Williams HJ, Galanello R, Cao A, Wood WG, Ayyub H, Higgs DR. Healing of broken human chromosomes by the addition of telomeric repeats. Am J Hum Genet. 1994;55:505–512. [PMC free article] [PubMed] [Google Scholar]
- Flint J, Bates GP, Clark K, Dorman A, Willingham D, Roe BA, Micklem G, Higgs DR, Louis EJ. Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum Mol Genet. 1997a;6:1305–1313. doi: 10.1093/hmg/6.8.1305. [DOI] [PubMed] [Google Scholar]
- Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett NA, King A, Higgs DR. The relationship between chromosome structure and function at a human telomeric region. Nat Genet. 1997b;15:252–257. doi: 10.1038/ng0397-252. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- Ijdo JW, Lindsay EA, Wells RA, Baldini A. Multiple variants in subtelomeric regions of normal karyotypes. Genomics. 1992;14:1019–1025. doi: 10.1016/s0888-7543(05)80125-9. [DOI] [PubMed] [Google Scholar]
- Knight SJ, Flint J. Perfect endings: A review of subtelomeric probes and their use in clinical diagnosis. J Med Genet. 2000;37:401–409. doi: 10.1136/jmg.37.6.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Lese CM, Precht KS, Kuc J, Ning Y, Lucas S, Regan R, Brenan M, Nicod A, Lawrie NM, et al. An optimized set of human telomere clones for studying telomere integrity and architecture. Am J Hum Genet. 2000;67:320–332. doi: 10.1086/302998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Macina RA, Negorev DG, Spais C, Ruthig LA, Hu XL, Riethman HC. Sequence organization of the human chromosome 2q telomere. Hum Mol Genet. 1994;3:1847–1853. doi: 10.1093/hmg/3.10.1847. [DOI] [PubMed] [Google Scholar]
- Macina RA, Morii K, Hu XL, Negorev DG, Spais C, Ruthig LA, Riethman HC. Molecular cloning and RARE cleavage mapping of human 2p, 6q, 8q, 12q, and 18q telomeres. Genome Res. 1995;5:225–232. doi: 10.1101/gr.5.3.225. [DOI] [PubMed] [Google Scholar]
- Martin-Gallardo A, Lamerdin J, Sopapan P, Friedman C, Fertitta AL, Garcia E, Carrano A, Negorev D, Macina RA, Trask BJ, et al. Molecular analysis of a novel subtelomeric repeat with polymorphic chromosomal distribution. Cytogenet Cell Genet. 1995;71:289–295. doi: 10.1159/000134129. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Linardopoulou E, Coil D, van den Engh G, Trask BJ. Comparative sequencing of a multicopy subtelomeric region containing olfactory receptor genes reveals multiple interactions between non-homologous chromosomes. Hum Mol Genet. 2001;10:2363–2372. doi: 10.1093/hmg/10.21.2363. [DOI] [PubMed] [Google Scholar]
- Monfouilloux S, Avet-Loiseau H, Amarger V, Balazs I, Pourcel C, Vergnaud G. Recent human-specific spreading of a subtelomeric domain. Genomics. 1998;51:165–176. doi: 10.1006/geno.1998.5358. [DOI] [PubMed] [Google Scholar]
- Nagele RG, Velasco AQ, Anderson WJ, McMahon DJ, Thomson Z, Fazekas J, Wind K, Lee H. Telomere associations in interphase nuclei: Possible role in maintenance of interphase chromosome topology. J Cell Sci. 2001;114:377–388. doi: 10.1242/jcs.114.2.377. [DOI] [PubMed] [Google Scholar]
- Piccini I, Ballarati L, Bassi C, Rocchi M, Marozzi A, Ginelli E, Meneveri R. The structure of duplications on human acrocentric chromosome short arms derived by the analysis of 15p. Hum Genet. 2001;108:467–477. doi: 10.1007/s004390100520. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethman HC, Xiang Z, Paul S, Morse E, Hu XL, Flint J, Chi HC, Grady DL, Moyzis RK. Integration of telomere sequences with the draft human genome sequence. Nature. 2001;409:948–951. doi: 10.1038/35057180. [DOI] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask BJ, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, et al. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet. 1998;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- Vergnaud G. Structure and evolution of human sub-telomeric regions. J Soc Biol. 1999;193:35–40. [PubMed] [Google Scholar]
- Walker MY, Hawley RS. Hanging on to your homolog: The roles of pairing, synapsis and recombination in the maintenance of homolog adhesion. Chromosoma. 2000;109:3–9. doi: 10.1007/s004120050407. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Higgs DR, Rack KA, Buckle VJ, Spurr NK, Fischel-Ghodsian N, Ceccherini I, Brown WR, Harris PC. Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell. 1991;64:595–606. doi: 10.1016/0092-8674(91)90243-r. [DOI] [PubMed] [Google Scholar]