Abstract
Microarrays containing 195,000 in situ synthesized oligonucleotide features have been created using a benchtop, maskless photolithographic instrument. This instrument, the Maskless Array Synthesizer (MAS), uses a digital light processor (DLP) developed by Texas Instruments. The DLP creates the patterns of UV light used in the light-directed synthesis of oligonucleotides. This digital mask eliminates the need for expensive and time-consuming chromium masks. In this report, we describe experiments in which we tested this maskless technology for DNA synthesis on glass surfaces. Parameters examined included deprotection rates, repetitive yields, and oligonucleotide length. Custom gene expression arrays were manufactured and hybridized to Drosophila melanogaster and mouse samples. Quantitative PCR was used to validate the gene expression data from the mouse arrays.
[The sequence data from this study have been submitted to GEO under accession nos. GPL208, GSM2409, GSM2410, GSM2411, GSM2412, GSM2413, GSM2414, GSE81, GSE82.]
The engineering technology that underpins the DNA microarray field is undergoing a period of rapid innovation and expansion. Currently, there are two predominant methods for creating DNA microarrays: physical deposition of presynthesized DNA onto a solid substrate (spotted arrays) or in situ synthesis of oligonucleotides on a solid substrate (Southern et al. 1992; Pease et al. 1994; Schena et al. 1995). DNA arrays manufactured using physical deposition of presynthesized material require labor-intensive preparation and record keeping of the DNA samples, resulting in a prohibitive amount of time and cost in the development of a new custom array. In situ synthesized oligonucleotide arrays using the photolithographic method pioneered by Fodor et al. (1991) require only that the DNA sequence of interest be known, but the cost and time to manufacture the photolithographic masks make it uneconomical for the manufacture of small production runs of custom DNA arrays. Ink-jet deposition and electrochemical in situ synthesis have the ability to produce custom DNA arrays (Dill et al. 1999; Hughes et al. 2001), but these methods produce arrays with more than an order of magnitude fewer features compared with that achieved with the photolithographic method (Schena 1999). Earlier, we described the initial design features of an instrument, the Maskless Array Synthesizer (MAS), that modifies the photolithographic process by using a digital light processor (DLP) from Texas Instruments to create “virtual” masks, making photolithography a much more flexible and user-friendly technology (Singh-Gasson et al. 1999). The MAS synthesizes a microarray with 195,000 24mer features in an area of 13.1 × 17.4 mm2 in <3 h. Probes up to 90 bases long can be synthesized with the MAS, which enables the researcher to adapt the MAS arrays to a wide variety of applications. In this report, we present results that demonstrate the chemical synthesis quality of MAS and the utility of the arrays for gene expression analysis.
RESULTS
Photolabile Chemistry
We have developed a second-generation MAS using DLPs that have been optimized for reflecting UV light. The DLPs obtained from Texas Instruments, contain >786,000 individually addressable mirrors, and these create the patterns of light that drive the light-directed DNA synthesis. We extensively tested a commercially available photolabile DNA synthesis blocking reagent, 2-nitrophenyl propoxycarbonyl (NPPOC), (Hasan et al. 1997) to determine its deprotection rates, its chemical yield during array synthesis, and the maximum length of oligonucleotide probes that could be synthesized using standard glass microscope slides as the solid support.
To test the deprotection rates, we used a method similar to that described previously (McGall et al. 1997). Briefly, a hydroxyalkyl-silanated slide was placed in the DNA synthesis chamber, and the NPPOC-blocked nucleotide was attached using standard automated phosphoramidite chemistry procedures. To optimize the amount of light required for deprotection with this new reagent, we exposed a set of features to different amounts of energy. After the features were exposed, a biotin phosphoramidite was coupled to all the features. After staining with streptavidin-cy3, the features were scanned, and an exposure time was chosen that produced saturated fluorescent features. The results of the deprotection studies indicated that 7.5 J/cm2 was sufficient to obtain complete deprotection of the NPPOC group (data not shown).
Chemical Yield
To test the stepwise chemical yield of the NPPOC chemistry, we used a method similar to that described previously (McGall et al. 1997). Briefly, a hydroxyalkyl-silanated slide was placed in the synthesis chamber, and oligonucleotides with different lengths, ranging from 1 to 12 bases, were synthesized. A biotin phosphoramidite was added to the ends of all the oligonucleotides, followed by the addition of streptavidin-cy3. Fluorescent intensity at each feature thus creates a relative measure of the free OH groups available and provides an estimate of the stepwise yield. These data are presented in Figure 1 and Table 1.
Figure 1.
Chemical yield. A plot of the results of the stepwise yield of the NPPOC chemistry in the MAS system. Oligonucleotides with different numbers of bases were synthesized ranging from 1 to 12 bases. A biotin phosphoramidite was coupled to the end of all the oligonucleotides. Streptavidin-cy3 was added to visualize the relative amounts of free hydroxyls in each feature. The data represent the relative numbers after subtraction of background.
Table 1.
Average Stepwise Yield for the NPPOC Amidites
| Base | Average stepwise yield |
|---|---|
| NPPOC-A(TAC) | 96% |
| NPPOC-C(ibu) | 99% |
| NPPOC-G(iPAC) | 97% |
| NPPOC-T | 98% |
Feature Size
The ability to group mirrors together to create different feature sizes is a powerful feature of the MAS. Figure 2 shows the results of an experiment in which we tested this capability. The same 24mer probes were synthesized for all of the different feature sizes and evaluated using hybridization data. The 33-μm features, created by clustering four activated mirrors surrounded by a border of inactivated mirrors, and the 16-μm features, in which a single activated mirror is surrounded by a border of inactivated mirrors, produced the most consistent results. These features are well resolved from each other with a 2.25- or 5-μm resolution scanner and produce consistently high-quality features. Because of the lack of any commercial array scanners that have <2-μm resolution, we have avoided using mirror formats that do not have a border of off mirrors to separate the features. The space between mirrors is 1 μm, and the features on images from currently available scanners are not well resolved. Based on these results, we standardized on the 33-μm features and the 16-μm features surrounded by a border of off mirrors. These formats produce microarrays with ∼85,000 or 195,000 features, respectively, when the entire printable area is used.
Figure 2.
Feature size flexibility. A 2.25-μm resolution fluorescent micrograph of a hybridization of biotin labeled cRNAs from Bacillus subtilus and Escherichia coli to a custom DNA array. This scan was obtained using a 2.25-μm resolution arrayWoRx scanner from API. The array contains 195,000 features in 14 × 17.4 mm2. From left to right, four different feature layouts were tested: 14-μm features separated by 3-μm gaps, 16-μm features in a checkerboard pattern, 16-μm features separated by 18-μm gaps, and 33-μm features separated by 18 μm.
Linker Length
To investigate the effects of linker length on hybridization efficiency, we tested the hybridization characteristics of an 18mer oligonucleotide synthesized at increasing distance from the glass surface. To this end, we fabricated an array containing 129,000 16-μm features with probes varying in length from 1 to 150 bases long. This was accomplished by synthesizing increasing lengths of oligonucleotide linker, from 1 to 132 bases in length, with the control 18mer oligonucleotide at the 5′ terminus (i.e., the solution end of the oligonucleotide, synthesized last).
We then hybridized a biotinylated oligonucleotide complementary to the control oligonucleotide on the array and visualized the hybridization with streptavidin-cy3 conjugate. Figure 3 shows the results of this experiment. The intensity rose rapidly as linker length increased. It reached a local maximum at 27mers and then decreased to ∼50% of the maximum and leveled off out to 150mers. This local maximum at 27mers corresponds to an optimum linker length of nine bases for an 18mer oligonucleotide in this experiment.
Figure 3.
Hybridization to 1mer to 150mers. Test of the 5′ sequence fidelity of oligonucleotides up to 150 bases long. A custom array was designed to compare the hybridization characteristics of an 18mer probe sequence (5′-AGGTCATTACAGCGAGAG-3′) synthesized on the 5′ end (solution phase) of different length oligonucleotides. On the X-axis is the length of the total oligonucleotide including the 18mer sequence. The array was hybridized to a biotin-labeled target (5′-biotinCTCTCGCTGTAATGACCT-3′) that was complementary to the 18mer probe sequence. After hybridization, the array was stained with streptavidin-cy3.
In addition, this experiment confirmed that the sequence synthesized at the end of the oligonucleotide was of sufficient quantity and quality to hybridize. The 18mer on a 132-base linker hybridized with equal strength as the 18mer on a 60-base linker. This demonstrates the quality of DNA synthesis that can be obtained with the NPPOC chemistry.
Probe Length
There has been considerable debate in the recent literature about the optimum probe length for oligonucleotide microarrays. To date, there is no consensus on this subject. To gain insight into this question using our platform, we synthesized custom microarrays with a variety of different probe lengths.
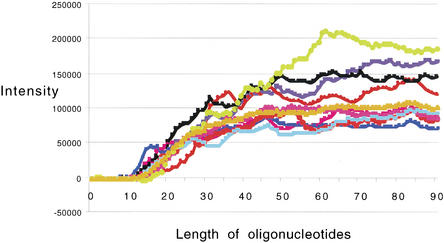
To test the influence of length on hybridization, oligonucleotides ranging from one to 90 bases were synthesized. These oligonucleotides were specific through their entire length to one of 20 different genes from Drosophila. These oligos were selected based on a set of 24mer oligonucleotides that were empirically proven to hybridize well to RNAs in Drosophila samples. The 1mer oligonucleotides started with the first 3′ base of the proven oligonucleotide sequence, and the 90mer was composed of the 90 bases starting at the 1mer position and moving 90 bases in the 5′ direction. Controls were included to study the behavior of mismatches for different lengths of oligonucleotides. We created mismatches in the control sequences by switching every tenth base to its complement; thus, a 19mer would have a single base mismatch at the tenth position, and a 20mer would have two mismatches (one at the 10th position and one at the 20th position). The 90mer had nine mismatches. For the purposes of simplicity, a relatively short five-base linker was used for all sequences.
Analysis of the 90mer data revealed a significant relationship between probe length and hybridization efficiency. Figure 4 shows the plots of intensity versus probe length. All 20 probe sets displayed similar length-dependence. For clarity, data from only 10 of the 20 genes are shown. As shown, intensity rose above background when the probes reached 17mers and climbed rapidly to up to 50mers and then leveled off. The mismatch control sequences showed a similar curve, but with lower intensities (data not shown).
Figure 4.
Hybridization to 1mer to 90mers. Plot of relative intensity from a hybridization to a custom array containing probes to 20 Drosophila genes. The perfect match value (with mismatch data subtracted) is shown. Each gene had a set of probes that ranged from 1 to 90 bases long. The array was hybridized to a biotin-labeled cRNA sample from Drosophila. After hybridization, the array was stained with streptavidin-cy3.
Mouse Expression Arrays
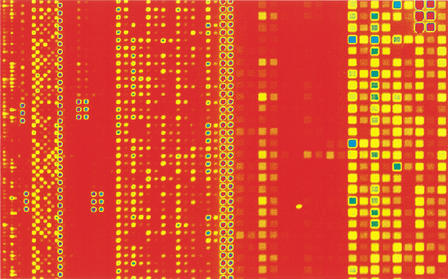
To investigate the use of this maskless technology in gene expression studies with complex genomes, we designed a mouse expression array containing 130,044 features representing 3240 genes, with 20 probe pairs per gene. Each probe was 24 bases long and had a mismatch control. We synthesized six of these mouse arrays and hybridized a biotin-labeled mouse liver mRNA sample to three of them and a biotin-labeled mouse spleen mRNA sample to the other three arrays. Figure 5 shows subsections from two of the arrays. As is evident in the images, very different patterns of gene expression were observed in the two tissue types.
Figure 5.
Scanner images of sections from two custom mouse arrays. (A) A custom mouse array containing 3240 genes with 20 probes per gene with mismatch controls was made and hybridized with biotin-labeled cRNA from a mouse liver sample. The features on this array were 16 μm. The array contained a total of 130,044 features. (B) Another mouse array of the same design hybridized to a mouse spleen sample.
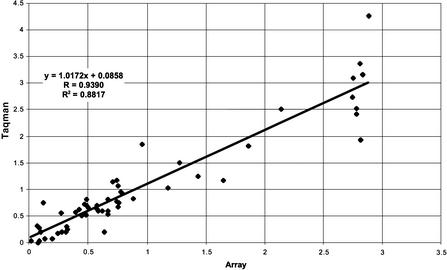
To test the accuracy of gene expression measurements using NimbleGen arrays, we compared the differential gene expression data shown in Figure 5 with quantitative PCR values. Sixty genes were selected for quantitative PCR validation based on their distribution in the array gene expression profile, such that the validated dataset would span the entire range of expression ratios. The selection was otherwise unbiased and random. PCR primers were designed to these 60 genes, and their expression levels were measured in the two samples. Of these 60 genes, we were unable to obtain valid quantitative PCR data from five. Quantitative PCR and array data of the remaining 55 are represented in Table 2. Reasons for PCR failure included generation of multiple products, inability to reach log phase, or improper primer design. As shown, an extremely high correlation was observed between array data and quantitative PCR data. Figure 6 shows a log ratio versus log ratio plot of the same data presented in Table 2.
Table 2.
Fold Change Predictions from NimbleGen Arrays and Quantitative PCR
| Accession | Array | PCR |
|---|---|---|
| NM009286 | 5984 | 268 |
| NM023125 | 5961 | 342 |
| NM018819 | 5247 | 15 |
| AK004839 | 2870 | 87 |
| NM007443 | 2554 | 2360 |
| NM008878 | 822 | 1296 |
| Y11356 | 724 | 1462 |
| NM008877 | 610 | 18458 |
| NM009349 | 510 | 554 |
| NM007409 | 110 | 328 |
| NM007494 | 79 | 67 |
| NM016661 | 28 | 18 |
| NM022310 | 10 | 32 |
| NM008705 | 7.4 | 6.8 |
| NM009945 | 6.2 | 9 |
| NM013541 | 5.8 | 4.7 |
| BC008241 | 5.6 | 5.6 |
| NM025628 | 4.9 | 4 |
| NM007747 | 4.8 | 6.6 |
| NM009941 | 4.7 | 4.3 |
| AK012602 | 4 | 3.4 |
| NM010239 | 4 | 1.6 |
| NM007506 | 3.5 | 8.7 |
| NM009735 | 3.2 | 3.3 |
| NM009429 | 2.6 | 4.2 |
| NM009080 | 2.5 | 3.6 |
| NM008160 | 2.3 | 4 |
| NM031868 | 2.2 | −1.6 |
| NM007572 | 2.1 | 2 |
| NM018853 | 2 | 1.8 |
| NM025586 | 1.9 | 1.5 |
| NM011664 | 1.8 | −1.6 |
| NM008503 | 1.6 | −1.2 |
| NM009081 | 1.3 | 1 |
| NM009082 | 1.3 | 1.1 |
| NM016750 | 1.3 | −1.6 |
| BC003861 | 1.2 | −1.9 |
| NM013647 | 1.2 | −1.1 |
| NM013765 | 1.2 | −2.1 |
| NM011042 | −1.1 | −5.7 |
| NM009091 | −1.2 | −1.2 |
| NM011701 | −2.3 | −4.6 |
| AK008273 | −2.6 | −5.1 |
| NM011693 | −2.6 | −3.8 |
| AK021200 | −3 | −6.5 |
| BC005803 | −3.3 | −5.3 |
| M12056 | −3.3 | −5 |
| NM008048 | −3.3 | −3.2 |
| BC006026 | −3.9 | −6.1 |
| NM008339 | −4.7 | −12 |
| NM008774 | −5.3 | −4 |
| NM023182 | −8 | −71 |
| AK007392 | −13 | −11 |
| NM009008 | −13 | −15 |
| NM008594 | −510 | −14 |
Values in the Array and PCR columns represent fold changes for these genes as measured using the respective method.
Figure 6.
Log ratio plot comparing fold change data from arrays and Taqman. The fold changes for 55 mouse genes in mouse liver and mouse spleen samples were calculated from array and Taqman data, and the log ratio of the fold changes was plotted.
We estimated the repeatability of the expression ratios by calculating the coefficient of variance of the fold changes for the genes. We hypothesized that the coefficients of variation (CVs) would be higher for genes that had expression levels near background, so we looked at the CVs for the fold changes for all genes and then used a sliding cutoff to remove genes with low expression levels. The data from this study are presented in Table 3.
Table 3.
Coefficient of Variation (CV) of Fold Changes from Mouse Array Data
| Threshold | Mean CV of fold changes | CV standard deviation |
|---|---|---|
| 1 | 44.6% | 0.243 |
| 25 | 34.6% | 0.136 |
| 50 | 31.7% | 0.111 |
| 75 | 30.7% | 0.104 |
| 100 | 29.9% | 0.095 |
| 125 | 29.3% | 0.092 |
| 150 | 29.1% | 0.088 |
| 175 | 29.2% | 0.089 |
| 200 | 28.9% | 0.089 |
| 225 | 29.1% | 0.091 |
| 250 | 29.3% | 0.093 |
Threshold is the average difference value that all the samples had to have for the ratios to be calculated for that gene.
DISCUSSION
These results demonstrate that maskless photolithographic technology produces high-quality oligonucleotide arrays that provide reliable gene expression data. The flexible nature of this technology and its rapid turnaround time allow researchers to quickly test new experimental designs and applications. Potential applications for this capability include antisense optimization, novel SNP identification strategies, rapid insertion and deletion mapping, pathogen detection, yeast three hybrid to find RNA 3′ UTR protein binding sites, splice variant studies, enhancer mapping in yeast using chromosomal immunoprecipitation, and host/pathogen dual genome arrays, to name a few. As this technology becomes more widely available, the number of new applications will increase as researchers use its unique flexibility to answer their specific scientific questions.
A critical aspect of in situ DNA synthesis is the quality of oligonucleotides created. The yields demonstrated in this paper are the highest yet reported for a complex functional microarray. The yields achievable with the NPPOC chemistry allow for the synthesis of high fidelity probes and allow a researcher to synthesize very long probes (90mers). However, selecting the optimum length probe for an application is not trivial. Steric hindrance, linker length, charge, hydrophobicity, probe quality, and other factors influence the behavior of probes on a microarray. The results of the hybridization to the 18mer on the 5′ end of various length probes indicate there is an optimum distance from the surface for a probe. Other researchers have reported similar results (Shchepinov et al. 1997). Other factors, such as steric hindrance, will vary depending on the application, and the complexity of the system will make theoretical predictions of the optimum conditions difficult. Thus, it is advantageous to have a flexible system that can test a wide variety of conditions simultaneously to determine the optimum probe and linker length for the application under development. The large number of features available to the researcher allows a wide variety of controls to be placed on the arrays. These controls can be designed to facilitate normalization between arrays and to increase the statistical robustness of the experimental design.
Validation of the accuracy of the results reported by the MAS arrays using 16-μm features was provided by the independent method of quantitative PCR. These results demonstrate the ability of the MAS arrays to reliably discern changes in gene expression, and they validate the MAS method for gene expression profiling.
One of the surprising outcomes from the completion of the Drosophila, mouse, and human genomes was the relatively low number of predicted genes. However, because this number relies on imperfect computational algorithms that are in turn based on imperfect experimental information, one must view it with caution. For example, the recent discovery of a large number of “microRNAs,” that is, small RNA molecules that do not encode proteins, underscores the necessity that the number of genes encoded by a genome be experimentally determined (Wasserman et al. 2001). The ability to make custom high-density oligonucleotide arrays provides a new opportunity for researchers to empirically test the prediction algorithms and scan genomes for expressed regions.
METHODS
Silanization
Erie Gold seal microscope slides (Fisher), arranged in odd-numbered slots of a stainless-steel slide rack, undergo a 10-min, room-temperature incubation in 10% (w/v) sodium hydroxide. The slides are then rinsed (for 2.5 min in each bath) in two dishes of deionized water. After rinses, the slides are transferred to a 2% bis(2 hydroxyethyl)-aminopropyltriethoxysilane solution (v/v; United Chemical Technologies) with shaking for 1 h. After the silane coating, slides are rinsed in 95% ethanol for 5 min. Slides are dipped immediately into ether and air-dried. Once slides are completely dry, they are baked for 15 min at 100°C. Immediately after baking, slides are stored desiccated at −20°C.
Array Synthesis
Standard DNA synthesis reagents (Glen Research, Proligo, Amersham Pharmacia, or Applied Biosystems) were used on Expedite DNA synthesizers (Applied Biosystems). The photolabile phosophoramidites (NPPOC-d-adenosine [N6-tac] β-cyanoethylphosphoramidite, NPPOC-d-cytidine [N4-isobutyryl] β-cyanoethylphosphoramidite NPPOC-d-guanosine [N2-ipac] β-cyanoethylphosphoramidite, NPPOC-d-thymidine-β-cyanoethylphosphoramidite) were from Proligo. The MAS units (NimbleGen Systems) were connected to the expedites to manufacture the custom arrays. Arrays were designed with JazzSuite software (NimbleGen Systems). After synthesis on the MAS was completed, the base-protecting groups were removed in a solution of ethylenediamine/ethanol (1:1 v/v; Aldrich) for 2 h. The arrays were rinsed with water, dried, and stored desiccated until use.
Sample Isolation
Mouse liver mRNA was isolated from adult Swiss Webster liver tissue obtained from Pel-Freez Biologicals. mRNA isolation was performed using oligo-dT hybrid capture on magnetic streptavidin beads using a commercially available kit (PolyATract System 1000, Promega), essentially according to the manufacturer's protocol. Drosophila RNA was isolated from a mixed population of embryos using Qiagen Rneasy columns as per the manufacturer's instructions. For the experiments described in Figure 2, cRNA corresponding to 10 Escherichia coli or Bacillus subtilis genes were hybridized to the array. These samples were derived from the following 10 bacterial gene sources: Aqpz, AppA, BioB, BioC, BioD (E. coli), and Lys, Phe, Thr, Trp, and Dap (B. subtilis). The clones were purchased from American Type Culture Collection, grown in TB media, and plasmid-isolated using Qiagen Maxiprep plasmid prep kits according to the manufacturer's instructions.
Sample Labeling
Total RNA or poly A+ RNA was converted to double-stranded cDNA using GIBCO BRL's SuperScript Choice System and an oligo dT primer containing the T7 RNA polymerase promoter (5′-GGCCAGTGAATTGTAATACGACTCACTATAGG GAGGCGG-T24-3′). Briefly, 15 μg total RNA or 3 μg polyA+ RNA was incubated with 1× first-strand buffer, 10 mM DTT, 500 μM dNTPs, and 5 pM/μL primer for 60 min at room temperature. Second-strand synthesis was accomplished by incubation with 200 μM dNTPs, 0.07 U/μL DNA ligase, 0.27 U/μL DNA polymerase I, 0.013 U/μL RNase, 1× second-strand buffer, and 10 U T4 DNA polymerase for 2 h. Double-stranded cDNA was purified using phenol-chloroform extraction and Eppendorf Phase-Lock Gel tubes, ethanol precipitated, washed with 80% ethanol, and resuspended in 3 μL water. In vitro transcription (IVT) was used to produce biotin-labeled cRNA from the cDNA using the Ambion MEGAscript T7 kit. Briefly, 1 μg double-stranded cDNA was incubated with 7.5 mM ATP and GTP, 5.6 mM UTP and CTP, and 1.9 mM bio-11-CTP and bio-16 UTP (Sigma-Aldrich), 1× transcription buffer, and 1× T7 enzyme mix for 5 h at 37°C. For the experiment outlined in Figure 2, 100 ng of each of the bacterial plasmids was used as template in the IVT reaction to generate the corresponding cRNA for hybridization. cRNA was purified using Qiagen RNeasy RNA purification columns according to the manufacturer's instructions. RNA yield was determined by absorbance at 260 nM. Before hybridization, cRNA was fragmented to an average size of 50 to 200 bp by incubation in 100 mM potassium acetate, 30 mM magnesium acetate, and 40 mM tris-acetate for 35 min at 94°C. Fragmentation was checked by gel electrophoresis in 1% agarose.
Hybridization and Washing
Microarrays were hybridized with 12 μg cRNA in 300 μL, in the presence of 50 mM MES, 0.5 M NaCl, 10 mM EDTA, and 0.005% (v/v) Tween-20 for 16 h at 45° C. Before application to the array, samples were heated to 95°C for 5 min, heated to 45°C for 5 min, and spun at 14,000g for 5 min. Hybridization was performed in disposable adhesive hybridization chambers from Grace BioLabs in a hybridization oven with agitation. After hybridization, arrays were washed in nonstringent (NS) buffer (6× SSPE, 0.01% [v/v] Tween-20) for 5 min at room temperature, followed by washing in stringent buffer (100 mM MES, 0.1 M NaCl, 0.01% Tween-20) for 30 min at 45°C. After washing, arrays were stained with streptavidin-cy3 conjugate from Amersham Pharmacia for 25 min at room temperature, followed by a 5-min wash in NS buffer, a 30-sec rinse in 1× NimbleGen final rinse buffer, and a blow-dry step using high-pressure grade-5 Argon (Badger Welding).
Yield Measurements
Custom arrays were designed to study the repetitive yield of four photolabile phosphoramidites: NPPOC-d-adenosine (N6-tac) β-cyanoethylphosphoramidite, NPPOC-d-cytidine (N4-isobutyryl) β-cyanoethylphosphoramidite, NPPOC-d-guanosine (N2-ipac) β-cyanoethylphosphoramidite, NPPOC-d-thymidine-β-cyanoethylphosphoramidite (Proligo). The arrays had features containing oligonucleotides ranging from 1 to 12 bases long. After synthesizing the features, biotin phosphoramidite (Glen Research) was coupled to all of the features. After deprotection, the arrays were placed in the streptavidin-cy3 (Amersham Pharmacia) buffer for 10 min, washed with NS wash buffer, and rinsed for 30 sec with 1× NimbleGen final rinse buffer.
Data Analysis
Prior to data extraction, images were rotated and doubled in size (without interpolation) using ImageJ software (http://rsb.info.nih.gov/ij/). Features were extracted using GenePix 3.0 software (Axon Instruments, Inc.), using a fixed feature size. The local background correction from the GenePix software was not applied to raw signal intensities.
Quantitative PCR
Quantitative PCR was performed using a TaqMan 5700 sequence detection system from Applied Biosystems. Primers were designed for 60 mouse genes that were represented on the microarray, as well as mouse α-tubulin and β-actin, using Primer Express 2.0 (Applied Biosystems). Reactions were prepared using the SYBR Green PCR Core Reagents kits (Applied Biosystems) to the kit specifications. A standard curve was prepared using primers to mouse α-tubulin and 0, 0.5, 1, 2, 5, and 10 ng of mouse liver cDNA (prepared as above); 2 ng of mouse liver and spleen cDNA were used with primers to β-actin (loading control) and each of the 60 experimental genes. Each reaction was replicated four times. Of the 60 experimental genes, five produced either multiple bands or the amplification reaction failed to reach log-phase. These reactions were dropped from the analysis. The relative expression change between liver and spleen was calculated for the remaining 55 genes and adjusted for the β-actin loading control.
WEB SITE REFERENCE
http://rsb.info.nih.gov/ij/; ImageJ software.
Acknowledgments
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rgreen@nimblegen.com; FAX (608) 218-7601.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.362402.
REFERENCES
- Dill K, Stanker LH, Young CR. Detection of salmonella in poultry using a silicon chip-based biosensor. J Biochem Biophys Meth. 1999;41:61–67. doi: 10.1016/s0165-022x(99)00027-5. [DOI] [PubMed] [Google Scholar]
- Fodor SPA, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Hasan A, Stengele KP, Giegrich H, Cornwell P, Isham KR, Sachleben RA, Pfleiderer W, Foote RS. Photolabile protecting groups for nucleosides: Synthesis and photodeprotection rates. Tetrahedron. 1997;53:4247–4264. [Google Scholar]
- Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman S, Schleter JM, Meyer MR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotech. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- McGall GH, Barone AD, Diggelmann M, Fodor SPA, Gentalen E, Ngo N. The efficiency of light-directed synthesis of DNA arrays on glass substrates J. Am Chem Soc. 1997;119:5081–5090. [Google Scholar]
- Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SPA. Light-generated oligonucleotide arrays for rapid DNA-sequence analysis. Proc Natl Acad Sci. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M. DNA microarrays: A practical approach. Oxford, UK: Oxford University Press; 1999. [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Shchepinov MS, Case-Green SC, Southern EM. Steric factors influencing hybridization of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Gasson S, Green RD, Yue YJ, Nelson C, Blattner F, Sussman MR, Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array Nat. Biotech. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- Southern EM, Maskos U, Elder JK. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: Evaluation using experimental models. Genomics. 1992;13:1008–1017. doi: 10.1016/0888-7543(92)90014-j. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes & Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]