Abstract
The plastid genomes of several plants contain ndh genes—homologues of genes encoding subunits of the proton-pumping NADH:ubiquinone oxidoreductase, or complex I, involved in respiration in mitochondria and eubacteria. From sequence similarities with these genes, the ndh gene products have been suggested to form a large protein complex (Ndh complex); however, the structure and function of this complex remains to be established. Herein we report the isolation of the Ndh complex from the chloroplasts of the higher plant Pisum sativum. The purification procedure involved selective solubilization of the thylakoid membrane with dodecyl maltoside, followed by two anion-exchange chromatography steps and one size-exclusion chromatography step. The isolated Ndh complex has an apparent total molecular mass of approximately 550 kDa and according to SDS/PAGE consists of at least 16 subunits including NdhA, NdhI, NdhJ, NdhK, and NdhH, which were identified by N-terminal sequencing and immunoblotting. The Ndh complex showed an NADH- and deamino-NADH-specific dehydrogenase activity, characteristic of complex I, when either ferricyanide or the quinones menadione and duroquinone were used as electron acceptors. This study describes the isolation of the chloroplast analogue of the respiratory complex I and provides direct evidence for the function of the plastid Ndh complex as an NADH:plastoquinone oxidoreductase. Our results are compatible with a dual role for the Ndh complex in the chlororespiratory and cyclic photophosphorylation pathways.
Sequencing of the tobacco and liverwort plastid genomes (1, 2) led to the surprising discovery of homologues to genes encoding subunits of the proton-pumping NADH:ubiquinone oxidoreductase, or complex I, component of the mitochondrial respiratory chain. The products of these 11 ndh genes have been suggested, on the basis of sequence similarity, to form a large protein complex (Ndh complex) with NAD(P)H:plastoquinone oxidoreductase activity (1–3), but the Ndh complex had not been purified or characterized.
Structural models of complex I (3, 4) indicate that the plastid ndhA–ndhG gene products form a membrane-embedded subcomplex that is possibly involved in plastoquinone reduction and that the more hydrophilic ndhH–ndhK gene products act as a connecting fragment to an as yet undefined subcomplex involved in the binding and oxidation of reductant, which for mitochondrial and bacterial complex I is NADH (4). In Escherichia coli complex I, which contains 14 subunits and is thought to represent a minimal form of complex I (3, 5), the peripheral NADH-oxidizing domain consists of 3 proteins analogous to the bovine 24-kDa, 51-kDa (which contains binding motifs for NADH and FMN), and 75-kDa subunits (4, 5). Importantly, homologues to these subunits are not present in the plastid genome and have not been identified so far in the nuclear genome of plants (3). This has led to controversy over the substrate specificity of the enzyme. It was proposed recently that the plastid Ndh complex does not in fact oxidize NADH but, rather, products of photosynthetic electron transport—either reduced ferredoxin or NADPH (3, 6), in the latter case possibly mediated by attachment of ferredoxin-NADP+ reductase (FNR) to the Ndh complex (7).
In higher plant chloroplasts, several Ndh proteins have been detected in the thylakoid membrane and preferentially in the nonappressed stromal lamellae fraction (8, 9). The expression of the ndh genes in chloroplasts is rather low—the level of the putative Ndh complex in pea is about 0.2% of total thylakoid membrane protein or about one complex for every 100 photosynthetic chains (10).
Homologues of the plastid ndh genes are found in cyanobacteria, which do not contain chloroplasts. The product of these genes may be analogous to the plastid Ndh complex, although a direct comparison is not possible because cyanobacteria contain both photosynthetic and respiratory electron transfer pathways in the same membrane (11). The cyanobacterial complex is also ill-defined with only an inactive subcomplex consisting of the NdhHIJK subunits so far isolated from Synechocystis sp. PCC 6803 (12). Studies with ndh mutants in cyanobacteria have shown a possible role for the complex both in cyclic and respiratory electron transport (13, 14).
The function of the Ndh complex in higher plants in vivo is not established, although it has been suggested to be either a part of a putative respiratory chain in chloroplasts or to serve as a component of cyclic photosynthetic electron transport (3, 6, 13, 14).
To characterize the chloroplast Ndh complex in terms of substrate specificity, size, and subunit composition, we describe herein the isolation of the complex from pea thylakoid membranes.
MATERIALS AND METHODS
Materials.
Pea plants (Pisum sativum cv. Little Marvel) were grown (16-h day/8-h night cycle, 24°C) for 10–11 days after germination and harvested after reaching a stage of young but not fully developed leaves. n-Dodecyl β-d-maltoside (DM) was purchased from Calbiochem, column chromatography materials were from Pharmacia, and other chemicals were from Sigma.
Preparation of Plant Material.
Chloroplasts were isolated as in ref. 8 with modifications as indicated to exclude mitochondrial contamination. All steps, including subsequent purification of the Ndh complex, were performed at 4°C or on ice, under very dim light and within 2 days of the start. Pea leaves were ground in a Waring blender in STE buffer (0.33 M sorbitol/50 mM Tris⋅HCl/2 mM EDTA, pH 7.5), and the homogenate was filtered through muslin and cotton wool. Intact chloroplasts were collected by low-speed centrifugation of the filtrate (1,500 × g; 3,000 rpm for 3 min in A6.9 Kontron rotor, rav = 12 cm). This washing step was repeated two more times in STE buffer, and then chloroplasts were subjected to osmotic shock in MNM buffer (20 mM Mes/15 mM NaCl/5 mM MgCl2, pH 6.0) and centrifuged at 35,000 × g (20,000 rpm, JA20 Beckman rotor, rav = 9 cm) for 20 min. The pellet, containing thylakoid membranes, was washed again under the same conditions. The membranes were then resuspended to 2 mg of chlorophyll per ml in MNM buffer containing 10% glycerol and 1 mM benzamidine (TM preparation).
Highly purified intact chloroplasts were obtained by centrifugation of the chloroplasts, obtained after the first wash (1,500 × g) through self-generating Percoll gradients (15). The green band at the bottom of the gradient, containing only intact chloroplasts, was collected, washed in Percoll-free buffer, and reapplied to another Percoll gradient. In this way, intact chloroplasts were reisolated two more times on Percoll gradients, then subjected to osmotic shock in MNM buffer, and further fractionated on sucrose gradients (16) to obtain thylakoid membranes (TP preparation). Mitochondria were prepared from pea leaves by centrifugation through Percoll gradients essentially as described (17) and were free of chlorophyll contamination. TM and TP preparations of thylakoid membranes were further fractionated into granal and stromal lamellae by using a sonication procedure as described (8).
Purification of the Ndh Complex from Thylakoid Membranes.
Thylakoid membranes (TM preparation, about 80 ml at 2 mg of chlorophyll per ml) were allowed to stack in MNM buffer for at least 30 min and then solubilized by adding DM [10% (wt/vol) stock in MNM buffer] dropwise to 1% (final concentration) with constant stirring. After a 30-min incubation, unsolubilized material was removed by centrifugation at 35,000 × g (JA20 rotor) for 30 min, and the supernatant was applied to Q-Sepharose HP column (1.6 × 12 cm), equilibrated at 2 ml/min with MEG buffer (20 mM Mes/2 mM EDTA/10% glycerol/20 mM NaCl/1 mM benzamidine/0.05% DM, pH 6.0). Bound proteins were eluted with a 140–300 mM (including NaCl present in MEG buffer) linear NaCl gradient applied in 480 ml of MEG buffer. The NdhK, I, and J proteins (as determined by immunoblotting) and the second peak of NADH:FeCN (ferricyanide) activity were eluted at about 240 mM NaCl. Two or three peak fractions (20 ml each) were combined, diluted 1:2 with MEG buffer (without NaCl), and applied to a Mono Q HR5/5 FPLC column equilibrated at 0.5 ml/min with MEG buffer. After application of 140–280 mM linear NaCl gradient in 140 ml of MEG buffer, all the NADH:FeCN activity and Ndh proteins were eluted at about 220 mM NaCl. One or two peak fractions (10 ml each) were concentrated about 20-fold by using a Centricon-100 concentrator (Amicon). This sample was then applied to Sephacryl S-300 HR (1.6 × 45 cm) size-exclusion column equilibrated at 0.5 ml/min with MEG buffer. Collected fractions (2.5 ml each) were concentrated 10-fold to the same degree by using Centricon-10 concentrators for analysis by SDS/PAGE. Molecular mass standards, thyroglobulin (669 kDa), ferritin (440 kDa), and aldolase (158 kDa), were chromatographed separately under identical conditions.
Gel Electrophoresis, Immunodetection, and N-Terminal Sequencing.
SDS/PAGE was performed in 14% polyacrylamide gels according to Laemmli (18). Silver staining of protein was performed according to ref. 19. For immunodetection, protein was transferred to nitrocellulose membranes. Affinity-purified antibodies against tobacco NdhI and NdhJ and wheat NdhK were obtained as described (8, 20). Antibodies against spinach FNR, ferredoxin, maize mitochondrial cytochrome oxidase subunit 1, and cytochrome bc1 complex were gifts from K.-H. Suss, L. J. Rogers, and C. J. Leaver, respectively. Immunodetection was performed by using a ECL immunoblotting kit (Amersham) according to manufacturer’s instructions. For quantitative immunoblotting, a dilution series of known amounts of recombinant NdhI, J, and K proteins were also blotted with samples of interest. For estimation of the amount of Ndh complex, the presence of one copy each of these Ndh proteins in the complex was assumed. Protein concentrations were determined either by the bicinchoninic acid method (21) or by a dot-blot Coomassie blue-staining assay using BSA as a standard. For N-terminal sequencing, proteins after SDS/PAGE were electroblotted onto Pro-Blott (Applied Biosystems) poly(vinylidene difluoride) membranes, and bands of interest were excised after Coomassie blue staining and then subjected to automatic Edman degradation on a Perkin–Elmer/Applied Biosystems model 477A sequencer.
Enzyme Assays.
The NAD(P)H:FeCN oxidoreductase activity was assayed by spectrophotometrically measuring the reduction of FeCN at 420 nm (extinction coefficient = 1.03 mM−1⋅cm−1). Assays were performed at 25°C in 1 ml of assay buffer (50 mM Tris⋅HCl/0.5 mM EDTA/0.1% DM, pH 7.5) containing 0.1 mM NADH or NADPH and 0.5 mM FeCN. Oxidation of NADH in presence of quinones was monitored at 340 nm (extinction coefficient = 6.22 mM−1⋅cm−1) in assay buffer containing 0.1 mM NADH and 0.2 mM menadione or 0.2 mM duroquinone. All specific activities are expressed as μmol of NAD(P)H oxidized per min per mg of protein. The detergent present in the assay buffer was sufficient for solubilization of chloroplasts or thylakoid membranes, added to the equivalent of 5 μg of chlorophyll per ml. All reactions were started by the addition of sample and the rates were corrected for the low background nonenzymatic rate.
All reported results were reproduced at least three times.
RESULTS
Thylakoid Membranes from Pea Chloroplasts Possess NAD(P)H Dehydrogenase Activities.
Attempts to assign an NAD(P)H dehydrogenase activity to the plastid Ndh complex are complicated by the possible contamination by plant mitochondrial complex I. It was therefore important to isolate pure thylakoid membranes and to quantify the degree of mitochondrial contamination.
Two different preparations of pea thylakoid membranes were analyzed. The first, designated TP, was obtained by sucrose density centrifugation of osmotically shocked intact chloroplasts that had been repurified a total of three times through Percoll gradients. For the generation of preparative amounts of thylakoid membranes, a second preparation, designated TM, was used. In this method, intact chloroplasts are first obtained from three consecutive low-speed centrifugation washing steps. After osmotic shock, thylakoid membranes were washed twice to ensure removal of soluble proteins.
Intact chloroplasts prepared by either method showed similar NADH DH activities, measured (after solubilization in the assay buffer) as NADH:FeCN oxidoreductase activities of about 0.05 μmol per min per mg of protein. In both cases, 60–70% of this activity remained bound to thylakoid membrane at pH 6.0 (Table 1). The NADPH:FeCN activity of intact chloroplasts was 60–80% of NADH:FeCN activity, with about half of it remaining bound to thylakoid membranes at pH 6.0. When thylakoids were washed at pH 8.0, instead of pH 6.0, only about 10% of the NADH:FeCN activity, but still about half of NADPH:FeCN activity, remained membrane-bound (data not shown). This apparent instability may be similar to dissociation of the E. coli complex I at higher pH (5). The NADH:FeCN activity of mitochondrial membranes purified from pea leaves was ∼3.0 μmol per min per mg of protein, about 50-fold higher than that associated with pea thylakoids, but consistent with the low level of plastid Ndh complex—about 0.2% of total thylakoid membrane protein (Table 1).
Table 1.
Purification of the Ndh complex from pea thylakoid membranes
| Step | Total protein, mg | Total NADH: (NADPH:) FeCN activity, μmol/min | Specific NADH: (NADPH:) FeCN activity, μmol per min per mg | Amount of Ndh complex (estimated by immunoblot), mg | Yield (immunoblot-based), % | Purification (immunoblot-based), fold |
|---|---|---|---|---|---|---|
| Chloroplasts | 2,300 | 103 (78) | 0.045 (0.034) | 2.3 | — | — |
| Thylakoid membranes | 1,100 | 72 (43) | 0.065 (0.039) | 2.3 | 100 | 1 |
| Dodecylmaltoside (1%) extract | 450 | 67 (42) | 0.15 (0.093) | 2.3 | 100 | 2.4 |
| Q-Sepharose HP anion exchange | 21 | 8.1 (0.47) | 0.39 (0.022) | 1.3 | 56 | 29 |
| Mono Q anion exchange | 1.1 | 1.6 (0.05) | 1.45 (0.045) | 0.60 | 26 | 260 |
| Sephacryl S-300 size exclusion (fractions 4 and 5) | 0.15 | 0.11 (0.003) | 0.73 (0.02) | 0.14 | 6.1 | 450 |
Estimation of the amount of Ndh complex at each step was done by quantitative immunobloting with anti-NdhK, I and J antibodies.
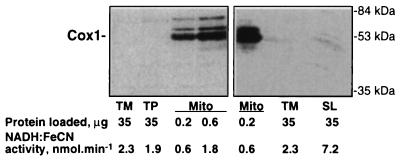
To check for any possible mitochondrial contamination in the thylakoid preparations, we used as probes antibodies specific for plant mitochondrial membrane proteins—either cytochrome oxidase subunit 1 (Fig. 1) or the cytochrome bc1 complex (data not shown). No contamination was normally detected in either the TM or TP preparation with both antibodies. Although some mitochondrial contamination (or a nonspecific cross-reaction) could be sometimes seen in highly exposed blots as in Fig. 1 Right, the amount detected was very low. We can estimate from these blots that contamination of our thylakoid membranes preparations by mitochondrial membrane protein is less than 0.05% (<0.02 μg in 35 μg). The contamination of the observed NADH:FeCN activity by activity originating from mitochondria is less than 2.5% and is likely to be lower as judged from comparison (Fig. 1 Right) with the stromal lamellae fraction that is essentially similar in protein content to the solubilized extract used for the purification of the Ndh complex.
Figure 1.
Estimation of mitochondrial contamination in preparations of thylakoid membranes. The TM and TP preparations of pea thylakoid membranes and mitochondrial membranes (Mito) were separated by SDS/PAGE and immunoblotted with antibodies specific for cytochrome oxidase subunit 1 (Cox1). The bottom line indicates NADH:FeCN activity corresponding to the amounts of protein loaded on the gel. A stromal lamellae (SL) fraction of thylakoid membranes, which is enriched in NADH:FeCN activity and Ndh proteins content (8, 10) is shown for comparison.
Purification of the Ndh Complex.
Young pea leaves were used as a starting material. Because the Ndh complex kept its integrity longer at pH 6.0 (see above), all purification steps were conducted at this pH. Incubation of stacked thylakoid membranes with 1% DM led to preferential solubilization of the nonappressed stromal lamellae membranes (data not shown). More than 90% of NAD(P)H:FeCN activity and all the NdhK, I, and J proteins were solubilized under these conditions (Table 1).
Purification of the Ndh complex was monitored through immunoblotting experiments (Table 1). After each chromatography step, about half of the applied NADH:FeCN activity was lost, with even more (about 80%) lost after the size-exclusion column, possibly due to the delipidation of the enzyme (22). To verify that activity is not fractionated away, all eluted fractions, including the 1 M NaCl wash, were assayed (Figs. 2 and 3). In Table 1, we used the amount of Ndh complex, estimated by quantitative immunoblotting, as a basis for evaluating the degree of purification at each step.
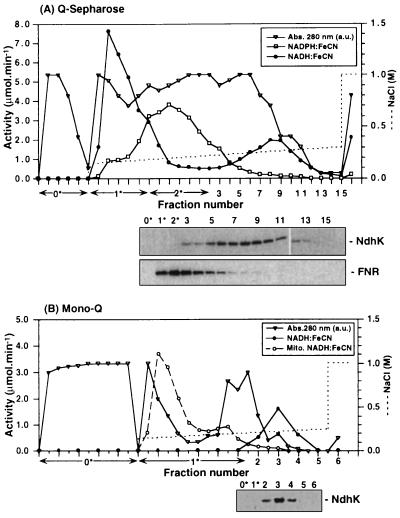
Figure 2.
Anion-exchange chromatography. (A Upper) Q-Sepharose HP step. A sample of selectively solubilized thylakoid membranes was applied to the column and eluted with a linear NaCl gradient. All fractions were assayed for NADH:FeCN (•) and NADPH:FeCN (□) activity. Protein concentration was monitored by absorbance at 280 nm (▵). Some of the fractions, known not to contain Ndh proteins from preliminary experiments, were pooled prior to SDS/PAGE. (Lower) Immunoblots of the fractions probed with antibodies specific for NdhK and FNR. (B Upper) Mono Q HR 5/5 step. Peak fractions from Q-Sepharose step (fractions 8 and 9) were diluted, applied to the column, and eluted with a linear NaCl gradient. Fractions were assayed for activity and probed with anti-NdhK antibody (Lower). The NADPH:FeCN activity of all the fractions was less than 3% of the NADH:FeCN activity. The elution profile of the activity [Mito.NADH:FeCN (○)] from solubilized mitochondrial membranes (20 μmol/min of activity applied) under identical conditions of chromatography is shown for comparison.
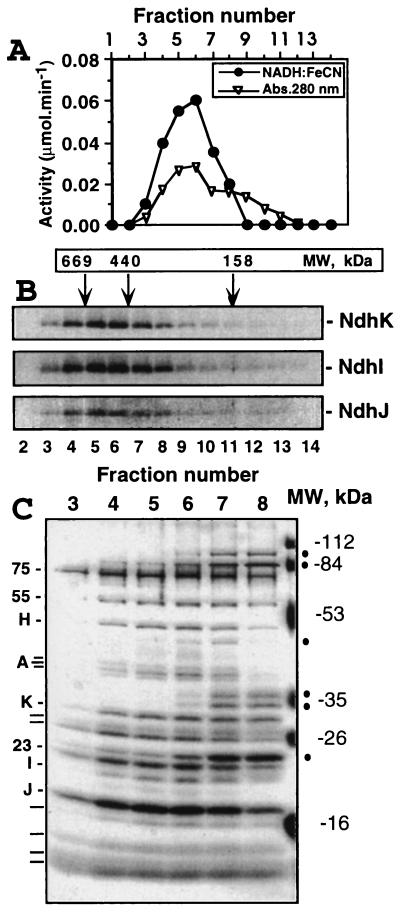
Figure 3.
Size-exclusion chromatography of the Ndh complex. The peak fraction after the Mono Q step (fraction 3) was concentrated and applied to the Sephacryl S-300 HR column. (A) Assay of all fractions containing protein (estimated by absorbance at 280 nm) for NADH:FeCN activity. (B) Immunoblot analysis. The same fractions separated by SDS/PAGE and probed with antibodies specific for NdhK, I, and J. Positions corresponding to the elution of the molecular mass standards are indicated by arrows. (C) Silver-stained SDS/PAGE gel. The coeluting subunits of the Ndh complex, some of which were identified (see text), are indicated on the left. Several impurities of smaller total molecular mass are marked by dots on the right. Positions of molecular mass markers are also indicated on the right.
In the first chromatography step involving Q-Sepharose anion exchange (Fig. 2A), two peaks of NADH:FeCN activity were resolved upon application of a shallow NaCl gradient. The second peak coeluted with NdhK (Fig. 2A), but the first peak at about 140 mM NaCl did not contain NdhK. The NADPH:FeCN activity was coeluted with FNR (Fig. 2A) and no copurification of FNR and Ndh proteins was observed. As judged from immunoblotting experiments, ferredoxin was also eluted in the earlier part of the gradient at about 150 mM NaCl, separately from NdhK (data not shown). Immunoblotting showed that the NdhI and NdhJ subunits copurified with NdhK during this and all subsequent purification steps, confirming that these proteins are part of a larger complex (data not shown and Fig. 3).
Pooled fractions containing the Ndh proteins (Fig. 2A) were subjected to chromatography on a Mono-Q FPLC column (Fig. 2B). At this step all the NADH:FeCN activity and Ndh proteins were coeluted and separated from the majority of contaminants. The NADPH:FeCN activity of all fractions was below 3% of the NADH-specific activity.
In an additional control experiment to estimate the possible degree of mitochondrial contamination, we analyzed a sample of solubilized pea mitochondrial membranes. All NADH:FeCN activity was solubilized under conditions similar to that for thylakoids. This sample was applied to a Mono Q column and eluted under conditions identical to that for an Ndh-complex-containing sample. As can be seen from Fig. 2B, all mitochondrial NADH:FeCN activity was eluted earlier in the gradient than the activity associated with Ndh proteins. Because the elution profiles on this column were highly reproducible, we can make a conservative estimate from Fig. 2B that after the Mono Q step any remaining mitochondrial contaminating activity eluting with the Ndh complex (Fig. 2B, fractions 2–4) will be less than 2% of the initial amount (2.5%), i.e., 0.05%.
To achieve further purification and to estimate the size of the complex, we passed the concentrated Mono Q fractions through a Sephacryl S-300HR size-exclusion column. At this stage, the NdhK, I, and J proteins (Fig. 3B) and all the NADH:FeCN activity (Fig. 3A) were coeluted, peaking in fractions 5 and 6, indicating the size of the complex to be 550 ± 30 kDa, neglecting any contribution from the detergent shell. Because NdhIJK copurified with the NADH:FeCN oxidoreductase activity in all purification steps, we conclude that the Ndh complex is an NADH-specific dehydrogenase. SDS/PAGE analysis showed that about 16 proteins (indicated on the left of Fig. 3C) were coeluted with the NADH dehydrogenase activity, providing a preliminary indication that the active Ndh complex consists of at least 16 subunits. Several impurities were present, mainly eluting with a size of <400 kDa (indicated on the right of Fig. 3C). Fractions 4 and 5, however, appear to be mostly free of contamination (∼90% pure, as judged from densitometry analysis of the gel in Fig. 3C, neglecting differences in the degree of protein staining) and can be considered as an isolated Ndh complex preparation.
A control experiment, in which a solubilized TM preparation was directly fractionated on the same size-exclusion column, to exclude possible fragmentation of the Ndh complex during anion-exchange chromatography, indicated the same size (550 kDa) for the Ndh complex (data not shown). We conclude therefore that our preparation (Fig. 3) most likely represents the intact Ndh complex.
Subunit Composition of the Ndh Complex.
N-terminal sequencing of the putative subunits of the complex allowed us to identify the NdhA (apparent molecular mass, 40 kDa), NdhH (50 kDa), and NdhK (29 kDa) subunits (Fig. 4). Protein bands corresponding to NdhI (20 kDa), NdhJ (17 kDa), and NdhK could be assigned (Fig. 3C) on the basis of immunoblotting experiments (Fig. 3B). The presence of NdhA confirms that at least one of the hydrophobic Ndh proteins is present in our preparation. Interestingly, our data (Fig. 4) suggest that for some of the predicted protein sequences for plastid NdhK the initiation codon may have been wrongly assigned to a site about 22 residues upstream of the true initiation codon.
Figure 4.
Comparison between the N-terminal sequences of NdhA, NdhH, and NdhK subunits (Fig. 3C) of purified pea Ndh complex and sequences predicted from the plastid genome of other plants. The sequences of the ndh genes from pea are not known; therefore, the N-terminal data for pea (Pisum sativum) is compared with the predicted sequences of the NdhA, NdhH, and NdhK proteins from tobacco (Nicotiana tabacum) (1) and yellow lupine (Lupinus luteus) (Swiss Prot accession no. P52766). Asterisks, identical residues; dots, conservative changes. Numbers indicate position of the residue from the N terminus.
The N-terminal sequences of the 55- and 23-kDa subunits (data not shown) were not similar to any of the known plastid-encoded Ndh proteins, indicating that they are nuclear-encoded. No N-terminal sequence data was obtained for the 75- and 18-kDa bands, possibly because their N termini are blocked, and the remaining subunits were too weakly stained on poly(vinylidene difluoride) membranes for sequencing to be attempted. On the basis of size, it is tempting to speculate that the 75-, 55-, and 23-kDa subunits are analogous to the 75-, 51-, and 24-kDa subunits of bovine complex I, involved in NADH binding and oxidation (4). Identification by comparison of the N-terminal sequences was not possible because the N-terminal region of the 51- and 24-kDa subunits is not conserved; therefore, these subunits will have to be identified by other means.
Enzymatic Activity of the Ndh Complex.
The NADH:FeCN activity recovered after the last size-exclusion step was quite low—0.11 μmol/min for fractions 4 and 5 (Table 1) and about 0.25 μmol/min in total (Fig. 3). These values are, however, much higher than possible mitochondrial contaminating activity at this stage, which is less than 0.05% (Figs. 1 and 2B) of the initial activity of 72 μmol/min (Table 1), i.e., 0.036 μmol/min. Therefore, we can exclude the possibility that the observed activity may be due to the contaminating mitochondrial complex I. For the isolated Ndh complex, estimated Km values for NADH and FeCN were about 15 μM and 0.2 mM, respectively, consistent with values obtained for other complex I preparations (5, 22). The optimum pH for activity, in the pH range 5–9, was about pH 8.0, higher than the optimum pH of 6.0 observed for solubilized pea mitochondrial membranes (data not shown). The NADPH:FeCN activity of the purified Ndh complex was very low, about 3% of the NADH-specific activity, which is usual for complex I (22). Because of the significant inactivation of the Ndh complex during size-exclusion chromatography, characterization of the NADH dehydrogenase activity with respect to the quinone acceptor was performed by using the preparation obtained after the Mono Q column, which is more than 50% pure (Table 1; verified by densitometry analysis of silver-stained gels) and does not appear to contain contaminating dehydrogenase activities (Fig. 3A).
The rates with menadione and duroquinone as acceptors (Table 2) were 2–5% of the rates with FeCN, consistent with other preparations (22, 23). Importantly, deamino-NADH, which is complex I enzyme type-specific substrate (24, 25), was about as effective an electron donor as NADH (Table 2). This result was reproduced with the purified Ndh complex. Rates with quinones as acceptors are too low to be measured with thylakoid membranes, due to low abundance of the Ndh complex in the membranes and high chlorophyll absorbance at 340 nm. Therefore, assays with water-insoluble plastoquinone, which is probably the electron acceptor in vivo, assays for proton-pumping activity, and assays for rotenone sensitivity will require reconstitution of the purified enzyme into liposomes (24, 26).
Table 2.
Specific activities of the Ndh complex
| Electron donors:acceptors | Specific activity, μmol per min per mg |
|---|---|
| NADH:FeCN | 2.70 |
| NADPH:FeCN | 0.08 |
| Deamino-NADH:FeCN | 2.18 |
| NADH:menadione | 0.10 |
| Deamino-NADH:menadione | 0.07 |
| NADH:duroquinone | 0.04 |
| Deamino-NADH:duroquinone | 0.05 |
Preparation of the complex after Mono Q column was assayed. Activities are expressed per mg of Ndh complex. The experimental error was within 0.01 μmol per min per mg of protein.
DISCUSSION
In this paper we report the purification of the Ndh complex from pea thylakoids. Important aims of this work were to clarify the nature of the reductant oxidized by this complex and to characterize the composition of this type of complex I.
Control immunoblotting experiments with pea leaf mitochondria and highly purified thylakoid membranes showed unambiguously that there is indeed NADH dehydrogenase activity associated with thylakoid membranes (Fig. 1). NADH dehydrogenase activity has also been detected in barley thylakoids (27), but as yet there is no information as to whether this activity is catalyzed by the Ndh complex (28).
Because of the low amount of Ndh complex in thylakoid membranes and its instability, the key to its purification was to devise a highly selective and rapid procedure. Because our attempts and those of others (29) to isolate the complex by immunoprecipitation were unpromising (data not shown), purification was achieved by using Q-based anion-exchange chromatography at pH 6.0, which was found to be the most effective from a variety of techniques analyzed. Of the three higher plant species examined, only thylakoid membranes from pea were suitable as a starting material, because the spinach Ndh complex was not separated sufficiently from other proteins on the Q and other columns and the tobacco enzyme appeared to be less stable even at pH 6.0 (data not shown).
About 30% of the pea thylakoid membrane-bound NADH:FeCN activity was associated with the Ndh complex (Fig. 2A, second peak), whereas the rest appeared to be catalyzed by a different enzyme (Fig. 2A, first peak), although we cannot exclude the possibility that some dissociated peripheral subunit(s) of Ndh complex is involved. All the NADPH:FeCN activity bound to thylakoid membranes seems to be due to the diaphorase activity of FNR (Fig. 2A), which is known to be partially membrane-bound (6). In contrast to recent suggestions (3, 7), FNR does not appear to be associated with the detergent-solubilized Ndh complex (Fig. 2A).
The complex appears to consist of about 16 subunits, 5 of which are probably nuclear-encoded. Of the 8 unassigned bands (Fig. 3C Left), 6 are expected to be the hydrophobic plastid-encoded NdhB–G subunits. It was difficult to assign these proteins because of the known aberrant mobility of highly hydrophobic proteins during SDS/PAGE (5). There is still uncertainty as to the true number of subunits because of potential problems in resolving and staining some of the protein components. The predicted molecular mass for the sum of the 11 known NdhA–K subunits is about 380 kDa, and with the 75-, 55-, and 23-kDa subunits, this will give a total of about 530 kDa, close to the measured molecular mass for the complex. Therefore, according to our preliminary analysis, the Ndh complex appears to be more similar in composition to the minimal version of complex I from E. coli [∼550 kDa, 14 subunits (5)] than to the larger mitochondrial complex I, for example, from potato [∼900 kDa, about 32 subunits (24)].
Our data show that the Ndh complex is an NADH- but not an NADPH-specific dehydrogenase (Figs. 2 and 3 and Table 2). As could be expected for the complete Ndh complex (4), quinones can be used as acceptors (Table 2). The NADH dehydrogenase activities of the Ndh complex are rather low compared with most of other complex I preparations (5, 23, 24), with the exception of Neurospora crassa [NADH:quinone activity of 0.15 μmol per min per mg of protein (26)]. One of the reasons for lower activity in our case could be the necessity to use prolonged anion-exchange chromatography (Fig. 2), which could lead to extensive delipidation of the complex. Indeed, our preliminary experiments showed that addition of phospholipids to the assay buffer led to about 50% increase in NADH:quinone activity of the purified complex. If we assume that about a third (see Fig. 2A) of the NADH:FeCN activity in pea thylakoid membranes is due to the Ndh complex, then its specific NADH:FeCN activity in the membrane will be about 10 μmol per min per mg of protein, not too dissimilar to other complex I preparations, e.g., from red beet mitochondria [29 μmol per min per mg of protein (23)].
Our identification of the Ndh complex as an NADH-specific complex I analogue has important implications for the understanding of the function of the enzyme in vivo. We have shown (20) in studies with ndh mutants of tobacco that the Ndh complex in higher plants mediates donation of electrons from an unidentified stromal reductant to the plastoquinone pool. On the basis of the results presented herein, we conclude that the stromal reductant is NADH and that the Ndh complex functions as an NADH:plastoquinone oxidoreductase.
As an NADH-specific enzyme, the Ndh complex could have a function both in cyclic electron transport or in a putative chlororespiratory pathway (Fig. 5). In cyclic mode, part of the NADPH produced by photosystem I activity could be used to reduce NAD+ to NADH via a putative transhydrogenase or by a substrate cycle involving, for example, NAD+- and NADP+-specific malate dehydrogenases. In vivo pool sizes of NAD(H) and NADP(H) inside the chloroplast have been reported to be similar, both in the light and in the dark (30). A role for the Ndh complex in cyclic photophosphorylation is also supported by its location in the stromal lamellae close to photosystem I (8, 9, 31). In addition, an independent ferredoxin-mediated cyclic pathway involving possibly the PsaE subunit of photosystem I (Fig. 5) is likely to be present in the chloroplast (6).
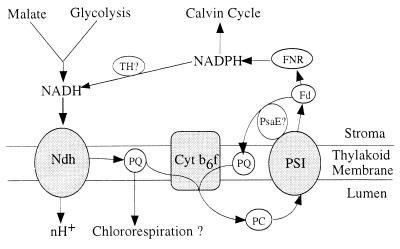
Figure 5.
Possible role of the Ndh complex. Scheme shows the Ndh complex as a component both of cyclic electron flow around photosystem I (PSI) and of a putative chlororespiratory pathway. Fd, ferredoxin; PC, plastocyanin; PQ, plastoquinone; FNR, ferredoxin:NADP+ reductase; nH+, an unknown number of protons pumped. Reduction of NAD+ by NADPH may be catalyzed either directly through a putative transhydrogenase (TH) or indirectly through substrate cycles. Independent cyclic pathway possibly catalyzed by PsaE subunit of PSI is also indicated.
In the dark a possible role for the Ndh complex could be to use NADH [produced, for example, by plastidial glycolysis (32) or accumulated in the light] as a part of a putative chlororespiratory pathway (Fig. 5), including also plastoquinone and a putative oxidase (13, 33–35). This report, which provides the direct evidence that the Ndh complex is indeed an NADH dehydrogenase, will be helpful in further studies on the function of this enzyme in vivo.
Acknowledgments
We gratefully acknowledge Dr. A.Carne (Institute of Cancer Research, London, U.K.) for help with N-terminal sequencing. We thank Prof. C. J. Leaver (University of Oxford, U.K.), Prof. K.-H. Süss (Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany), and Prof. L. J. Rogers (University of Aberystwyth, U.K.) for gifts of antiserum against maize mitochondrial cytochrome oxidase subunit 1 and cytochrome bc1 complex, spinach FNR, and spinach ferredoxin, respectively. This work was supported by grants from The Royal Society and the Biotechnology and Biological Sciences Research Council.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: DM, n-dodecyl β-d-maltoside; FNR, ferredoxin-NADP+ reductase.
References
- 1.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torozawa K, Meng B Y, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kata A, Tohdoh N, Shimada H, Sugiura M. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohyama K, Fukuzawa H, Koichi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, Aota S -I, Inokuchi H, Ozeki H. Nature (London) 1986;322:572–574. [Google Scholar]
- 3.Friedrich T, Steinmüller K, Weiss H. FEBS Lett. 1995;367:107–111. doi: 10.1016/0014-5793(95)00548-n. [DOI] [PubMed] [Google Scholar]
- 4.Walker J E. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 5.Leif H, Sled V D, Ohnishi T, Weiss H, Friedrich T. Eur J Biochem. 1995;230:538–548. doi: 10.1111/j.1432-1033.1995.tb20594.x. [DOI] [PubMed] [Google Scholar]
- 6.Bendall D S, Manasse R S. Biochim Biophys Acta. 1995;1229:23–38. [Google Scholar]
- 7.Guedeney G, Corneille S, Cuiné S, Peltier G. FEBS Lett. 1996;378:277–280. doi: 10.1016/0014-5793(95)01473-x. [DOI] [PubMed] [Google Scholar]
- 8.Nixon P J, Gounaris K, Coomber S A, Hunter C N, Dyer T A, Barber J. J Biol Chem. 1989;264:14129–14135. [PubMed] [Google Scholar]
- 9.Berger S, Ellersiek U, Westhoff P, Steinmüller K. Planta. 1993;190:25–31. [Google Scholar]
- 10.Sazanov L A, Burrows P, Nixon P J. Biochem Soc Trans. 1996;24:739–743. doi: 10.1042/bst0240739. [DOI] [PubMed] [Google Scholar]
- 11.Scherer S. Trends Biochem Sci. 1990;15:458–462. doi: 10.1016/0968-0004(90)90296-n. [DOI] [PubMed] [Google Scholar]
- 12.Berger S, Ellersiek U, Kinzelt D, Steinmüller K. FEBS Lett. 1993;326:246–250. doi: 10.1016/0014-5793(93)81800-f. [DOI] [PubMed] [Google Scholar]
- 13.Mi H, Endo T, Schreiber U, Ogawa T, Asada K. Plant Cell Physiol. 1992;33:1233–1237. [Google Scholar]
- 14.Mi H, Endo T, Ogawa T, Asada K. Plant Cell Physiol. 1995;36:661–668. [Google Scholar]
- 15.Douce R, Joyard J. In: Methods in Chloroplast Molecular Biology. Edelman M, Hallick R B, Chua N-H, editors. Amsterdam: Elsevier Biomedical; 1982. pp. 244–245. [Google Scholar]
- 16.Douce R, Holtz R B, Benson A A. J Biol Chem. 1973;248:7215–7222. [PubMed] [Google Scholar]
- 17.Douce R, Bourguignon J, Brouquisse R, Neuburger M. Methods Enzymol. 1987;148:403–417. [Google Scholar]
- 18.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Blum H, Beier H, Gross H J. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 20.Burrows, P. A., Sazanov, L. A., Svab, Z., Maliga, P. & Nixon, P. J. (1998) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 21.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Anal Biochem. 1985;156:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 22.Leterme S, Boutry M. Plant Physiol. 1993;102:435–443. doi: 10.1104/pp.102.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmusson A G, Mendel-Hartvig J, Moller I M, Wiskich J T. Physiol Plant. 1994;90:607–615. [Google Scholar]
- 24.Herz U, Schroder W, Liddell A, Leaver C J, Brennicke A, Grohmann L. J Biol Chem. 1994;269:2263–2269. [PubMed] [Google Scholar]
- 25.Matsushita K, Ohnishi T, Kaback H R. Biochemistry. 1987;30:7732–7737. doi: 10.1021/bi00398a029. [DOI] [PubMed] [Google Scholar]
- 26.Ise W, Haiker H, Weiss H. EMBO J. 1985;4:2075–2080. doi: 10.1002/j.1460-2075.1985.tb03894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuello J, Quiles M J, Albacete M E, Sabater B. Plant Cell Physiol. 1995;36:265–271. [Google Scholar]
- 28.Quiles M J, Albacete M E, Sabater B, Cuello J. Plant Cell Physiol. 1996;37:1134–1142. [Google Scholar]
- 29.Funk E, Steinmüller K. In: Photosynthesis: from Light to Biosphere. Mathis P, editor. II. Boston: Kluwer; 1995. pp. 701–704. [Google Scholar]
- 30.Hampp R, Goller M, Fullgraf H. Plant Physiol. 1984;75:1017–1021. doi: 10.1104/pp.75.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubicki A, Funk E, Westhoff P, Steinmüller K. Planta. 1996;199:276–281. [Google Scholar]
- 32.Plaxton W C. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:185–214. doi: 10.1146/annurev.arplant.47.1.185. [DOI] [PubMed] [Google Scholar]
- 33.Bennoun P. Proc Natl Acad Sci USA. 1982;79:4352–4356. doi: 10.1073/pnas.79.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lajko F, Kadioglu, Borbely G, Garab G. Photosynthetica. 1997;33:217–226. [Google Scholar]
- 35.Harris G C, Heber U. Plant Physiol. 1993;101:1169–1173. doi: 10.1104/pp.101.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]