Abstract
To facilitate association-based linkage studies we have studied the linkage disequilibrium (LD) and haplotype architecture around five genes of interest for cancer risk: ATM, BRCA1, BRCA2, RAD51, and TP53. Single nucleotide polymorphisms (SNPs) were identified and used to construct haplotypes that span 93–200 kb per locus with an average SNP density of 12 kb. These markers were genotyped in four ethnically defined populations that contained 48 each of African Americans, Asian Americans, Hispanic Americans, and European Americans. Haplotypes were inferred using an expectation maximization (EM) algorithm, and the data were analyzed using D‘, R2, Fisher’s exact P-values, and the four-gamete test for recombination. LD levels varied widely between loci from continuously high LD across 200 kb to a virtual absence of LD across a similar length of genome. LD structure also varied at each gene and between populations studied. This variation indicates that the success of linkage-based studies will require a precise description of LD at each locus and in each population to be studied. One striking consistency between genes was that at each locus a modest number of haplotypes present in each population accounted for a high fraction of the total number of chromosomes. We conclude that each locus has its own genomic profile with regard to LD, and despite this there is the widespread trend of relatively low haplotype diversity. As a result, a low marker density should be adequate to identify haplotypes that represent the common variation at a locus, thereby decreasing costs and increasing efficacy of association studies.
[Supplemental material is available online at http://www.genome.org.]
With the exploding catalog of SNPs in the human genome, there is persistent interest in exploiting these markers for linkage disequilibrium (LD)-based searches for disease-susceptibility alleles. Characterization of the structure of LD throughout the genome is a necessary companion to the successful pursuit of such studies. While the ultimate goal of having a genome-wide map of LD has not yet been met, a candidate gene/locus approach is being taken (Clark et al. 1998; Goddard et al. 2000; Kidd et al. 2000; Moffatt et al. 2000; Taillon-Miller et al. 2000; Abecasis et al. 2001; Johnson et al. 2001; Reich et al. 2001). These studies have revealed significant diversity in the amount and structure of LD, both between independent loci and between populations.
Utilization of haplotypes in association studies for identification of commonly occurring variants may have increased power over single-allele studies (Johnson et al. 2001). Recent studies of haplotype structure at several loci have noted a lack of diversity (Daly et al. 2001; Johnson et al. 2001). Minimal haplotype diversity may mean considerably fewer markers are needed to represent the common variants in a population in a haplotype-based study than have been estimated for such studies (Kruglyak 1999). However, given the diversity and complexity of LD seen across the genome, it is likely a full description of haplotype structure will be key to determining the efficacy of such an approach for a particular locus.
Here we present a comparison study of the LD and haplotype structure for five widely studied cancer-susceptibility genes: ATM, BRCA1, BRCA2, RAD51, and TP53. Unphased genotype data were generated for markers that encompassed ∼150 kb per locus. Linkage disequilibrium and haplotype diversity were assessed at each locus in four populations: African American, Asian American, Hispanic American, and European American. These data contribute to the growing picture of LD and haplotype architecture in the genome and provide evidence that haplotype-based association studies should be possible with relatively small numbers of markers.
RESULTS
SNP Allele Frequencies
The goal for SNP ascertainment in this candidate gene-based study was to generate SNPs that spanned (throughout ∼150 kb containing) each gene of interest. SNP detection was not intended to catalog all of the diversity in these genetic regions; rather, the goal was to develop informative markers that were relatively evenly spaced throughout the loci. The target SNP density was 1 SNP every 30 kb. SNPs were ascertained through two means: by resequencing of 10 chromosomes and by searching literature/databases. Resequencing was performed on PCR-amplified regions placed sporadically throughout the loci as has been previously described (Bonnen et al. 2000). Of the 13 dbSNP entries that were genotyped, six were found to be monomorphic in the 192 samples we examined (see Methods). There were 57 SNPs identified in total: 42 were detected by resequencing, 8 from literature, and 7 from dbSNP. Seven of these were dropped from the study for technical reasons but are reported in dbSNP. These 50 SNPs were genotyped in four ethnic populations.
The majority of SNPs genotyped in this study were located in introns (41/50) or outside of known genes (3/50). Six SNPs were in coding or UTR sequences. There were 34 transitions, 14 transversions, and 2 insertion/deletion mutations. Eight to fourteen SNPs per gene that span 111–200 kb per locus were genotyped in an ethnically defined population consisting of 48 each African Americans, Asian Americans, Hispanic Americans, and European Americans. SNPs that had a rarer allele frequency of ≤0.05 in any one of the four populations were excluded from allele frequency, haplotype, and linkage disequilibrium analysis.
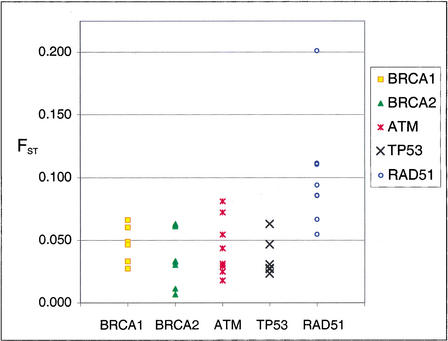
Allele frequencies of individual SNPs were found to vary between ethnic groups as has been noted in other studies (Goddard et al. 2000). The amount of allele frequency variation between ethnic groups appears to vary not only for each SNP but also by gene. The standardized variance (FST) for allele frequency across ethnic groups was measured for each SNP. The overall range of FST was from 0.007 to 0.201. The range at each gene was lower for BRCA1 (FST = 0.027–0.066), BRCA2 (FST = 0.007–0.062), TP53 (FST = 0.023–0.063), and ATM (FST = 0.018–0.081) than for RAD51 (FST = 0.055–0.201; Fig. 1). The majority of RAD51 SNPs had an FST higher than 0.081, whereas no other gene had SNPs with FST that high. Excluding RAD51, 100% of SNPs had an FST < 0.081, and 71% of SNPs had an FST < 0.05. Figure 1 illustrates that the FST for SNP allele frequency across ethnic groups at the other genes in this study tend to cluster together and the FST at RAD51 is clearly elevated.
Figure 1.
Fixation index (FST) of SNP allele frequencies across populations. SNP allele frequencies varied between ethnic groups. The standard deviation of that variation across populations was calculated for each SNP that was used in haplotype and LD analyses. The majority of RAD51 SNPs had an FST > 0.081. Excluding RAD51, 100% of SNPs had an FST < 0.081, and 71% of SNPs had an FST < 0.05.
Haplotype Frequencies
Haplotypes were constructed from genotype data using the EMHAPFRE program, which uses an expectation maximization algorithm (Excoffier and Slatkin 1995). Previous reports have demonstrated the appropriateness of the expectation maximization algorithm for inferring haplotypes from this type of data (Excoffier and Slatkin 1995; Bonnen et al. 2000; Tishkoff et al. 2000; Niu et al. 2002). The SNPs used to construct haplotypes all had rarer allele frequencies of ≥0.05 in all four populations. This criterion excluded from the analysis SNPs that had a low frequency in all groups as well as those termed population-specific (those with frequency >0.15 in one population and <0.05 in all others). These SNPs were excluded for two reasons. It has been shown that SNPs with low frequency have little power for detection of LD (Lewontin 1995; Goddard et al. 2000). Furthermore, when comparing numbers of haplotypes between ethnic populations, inclusion of SNPs that were not present in all populations introduces bias. The addition of SNPs with lower allele frequencies increases the number of lower-frequency haplotypes (data not shown), and the inclusion of population-specific SNPs leads to the addition of population-specific haplotypes (data not shown).
Comparison of the numbers of haplotypes at each locus reveals considerable differences. The total number of haplotypes at BRCA1 (10) and ATM (19) is considerably fewer than at the three other genes, RAD51 (35), TP53 (28), and BRCA2 (34; Table 1). If there were one founder haplotype and mutations yielding new alleles are the only evolutionary forces acting to create new haplotypes, n + 1 would have been the possible number of haplotypes, where n is the number of SNPs that comprise a haplotype. Following this logic, observation of >(n + 1) haplotypes would indicate the presence of other forces such as recombination, recurrent mutation, or gene conversion. BRCA1 and TP53 were each analyzed using six SNPs, giving them a theoretical minimum of 7 haplotypes. BRCA1 has 10 and TP53 has almost triple this number with 28 haplotypes.
Table 1.
A Small Number of Shared Haplotypes Account for a Large Proportion of Chromosomes
| Locus | # SNPs | kb spanned | Total # samples | Total # of haps | Theor mina | Theor maxb | # Shared haps | % Chromosomes accounted for by shared haplotypes | # of haplotypes | # of haplotypes q ≥ 0.10c | Hd | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AF | AS | EU | HI | AF | AS | EU | HI | AF | AS | EU | HI | AF | AS | EU | HI | ||||||||
| BRCA1 | 6 | 200 | n = 192 | 10 | 7 | 64 | 3 | 54 | 98 | 93 | 79 | 7 | 4 | 6 | 7 | 5 | 3 | 4 | 4 | 0.74 | 0.54 | 0.64 | 0.59 |
| ATM | 11 | 140 | n = 260 | 19 | 12 | 2048 | 5 | 85 | 90 | 100 | 94 | 12 | 9 | 5 | 12 | 6 | 4 | 4 | 4 | 0.84 | 0.66 | 0.71 | 0.72 |
| RAD51 | 9 | 150 | n = 190 | 35 | 10 | 512 | 5 | 22 | 70 | 87 | 81 | 23 | 14 | 11 | 12 | 7 | 4 | 5 | 5 | 0.88 | 0.71 | 0.77 | 0.74 |
| BRCA2 | 8 | 93 | n = 189 | 34 | 9 | 256 | 5 | 66 | 61 | 49 | 56 | 21 | 15 | 19 | 15 | 7 | 6 | 7 | 7 | 0.88 | 0.87 | 0.89 | 0.89 |
| TP53 | 6 | 149e | n = 189 | 28 | 7 | 64 | 7 | 69 | 66 | 59 | 70 | 18 | 20 | 13 | 12 | 8 | 7 | 11 | 8 | 0.90 | 0.85 | 0.91 | 0.87 |
Theor min = n + 1, where n is the number of SNPs used to construct haplotypes.
Theor max = 2n, where n is the number of SNPs used to construct haplotypes.
The number of haplotypes in that population with a frequency ≥0.10.
H is heterozygosity. H = 1 − ∑i − qi2
AF = African American, AS = Asian American, EU = European American, HI = Hispanic American.
The distance spanned by the TP53 SNPs was determined using a BAC that is predraft (less than 4X coverage), as such the distance may be considered an estimate.
Examining the haplotype heterozygosity in each ethnic group at each gene also shows differences between populations. Another measure of haplotype diversity is the expected heterozygosity based on haplotype frequencies,
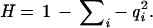 |
The African American group has the highest H in ATM, BRCA1, and RAD51, was second in TP53, and was second to last in BRCA2. Overall, African Americans demonstrated the most haplotype diversity and Asian Americans the least.
The number of shared haplotypes was relatively few when compared with the total number of haplotypes (Table 1). Haplotypes that are present in all four populations studied are termed shared haplotypes, and because they are present in all populations are considered to be the oldest haplotypes. These also tend to be the highest-frequency haplotypes. The number of shared haplotypes ranged in number from three at BRCA1 to seven at TP53, with the other loci having five each. The shared haplotypes are a small portion of the total number of haplotypes, for example, at BRCA1 there are 3 out of 10 total and 7/28 for TP53. The number of shared haplotypes and the effective number of haplotypes is similar, as would be expected from the general trend that shared haplotypes are higher frequency. However, there are populations in which a shared haplotype is at a very low frequency (sometimes <0.01). Conversely, some populations have haplotypes that are relatively high frequency and are not shared in all populations.
The most remarkable attribute of the shared haplotypes was that they account for a very high percentage of the total chromosomes studied (Table 1). For example, at ATM five out of 19 haplotypes accounted for 100% of the European American chromosomes, 94% of Hispanic, 90% of Asian American, and 85% of African American. The percentage of chromosomes accounted for by the shared haplotypes was lower in the genes that had a higher total number of haplotypes, but the fraction of total chromosomes was still quite high. For example, at BRCA2 five out of 34 haplotypes accounted for 49% of the European American chromosomes, 56% of Hispanic American, 61% of Asian American, and 66% of African American. Thus, at all loci we observe a small number of haplotypes accounting for a large proportion of chromosomes. Populations with the highest heterozygosity had the least percentage of chromosomes accounted for by the shared haplotypes. African Americans had the highest heterozygosity and the least sharing for ATM: 85%, BRCA1: 54%, and RAD51: 22%. At TP53 and BRCA2 the European Americans had the highest heterozygosity and the least percentage of chromosomes accounted for by the shared haplotypes with 59% and 49%, respectively. Although the amount of sharing varies by ethnic group and locus, it is substantial in all.
LD Analyses
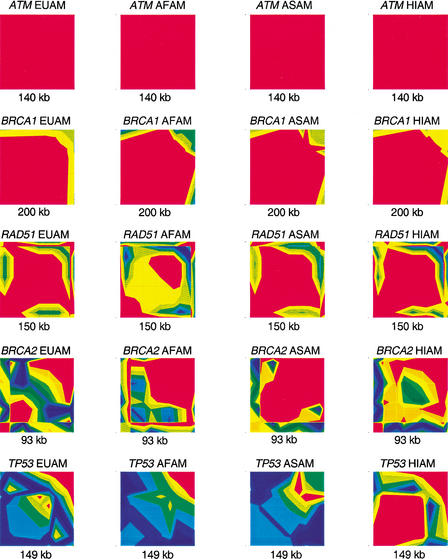
The pattern and extent of linkage disequilibrium (LD) at each genomic region differed widely. LD was measured using the statistic ‖D‘‖ and was plotted by the GOLD program to illustrate the intensity of LD along the length of the chromosome spanned by our markers (Fig. 2). This analysis reveals a spectrum in the amount of LD at the different loci with ATM and BRCA1 showing the most LD and decreasing amounts from RAD51 to BRCA2 to TP53. The 140-kb ATM region and the 200 kb spanning BRCA1 each showed a single block of LD. This extensive LD had been previously reported (Liu and Barker 1999; Bonnen et al. 2000; Thorstenson et al. 2001). RAD51 had one main block of LD and a short span of apparent recombination <10 kb, followed by what appears to be the beginning of another LD block. BRCA2 has a more complex pattern and shows significant differences between populations. TP53 shows little LD over the entire span of markers. The most extreme cases were BRCA1, in which a continuous region of strong LD extended ∼200 kb, and TP53, in which little LD was detected throughout the 140-kb region.
Figure 2.
LD intensity across loci. LD was measured for the five loci in all populations using the statistic ‖D‘‖ in a pairwise manner across markers. GOLD generates these plots through interpolation of the resulting triangular matrices. ‖D‘‖ ranges from 1 to 0, with 1 showing in red and 0 in dark blue.
The amount and pattern of LD also varied between populations at most genes (Fig. 2). At ATM and BRCA1, LD is very high and the differences between ethnic groups appear negligible. At RAD51, LD followed the same general pattern across populations but showed increased or decreased intensity at each group. In contrast, at TP53 Hispanics showed a completely different pattern of LD from the other three populations. BRCA2 has the most contrast between populations. The 3′ end shows an LD block of different lengths in each population. This is followed by a region without measurable LD that also varies in length. In some of the populations a second, smaller LD block exists in the 5′ end of the region. There is a prevailing tendency for African Americans to exhibit the least LD of all populations, which is likely owing to the age of the population leading to the accumulation of recombination and mutation. This is true for RAD51 and TP53. However, at BRCA2 European Americans showed the least LD, illustrating that the forces that act to maintain or create LD are not acting uniformly across the genome or in different populations.
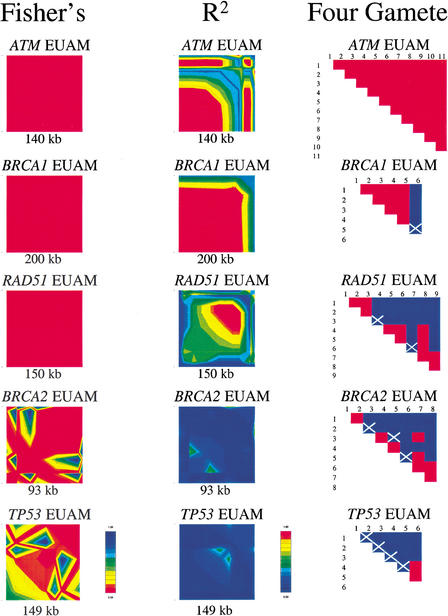
A comparison of LD patterns when determined by three different measures (‖D‘‖, r2, and Fisher’s exact test for significance) was conducted. The results are summarized in the GOLD plots for the European American population for each gene (Fig. 3). The three methods agree in showing a range in the amount of LD from BRCA1 to TP53. The overall patterns of LD are highly similar with the main difference between analyses being the intensity of LD. For example, by all three methods TP53 shows the same pattern of low LD, higher LD in the center followed by a break followed by higher LD. However, the intensity of LD indicated is higher in D‘ and Fisher than in r2. The exception here is ATM, wherein D‘ and Fisher show complete LD and r2 indicates decreased LD in the 3′ half of the region. Fisher follows D‘. r2 tends more toward 0, whereas D‘ tends more toward 1.
Figure 3.
Comparison of LD and recombination measurements at five loci. Pairwise LD was measured by Fisher’s exact P-values and r2. Each was plotted separately by GOLD with a score of 1 showing in red and 0 in dark blue. Recombination was determined using the four-gamete test with R, potential recombination sites, indicated by white Xs. Blue boxes indicate site pairs having four gametic types, which implies that recombination has occurred between these two sites. Red boxes indicate site pairs having less than four gametic types.
The results of the four-gamete test for recombination revealed similar results as the LD analysis (Fig. 3). Our interpretation of the results of the four-gamete test is to count any occurrence of a fourth gamete as evidence for recombination. However, this could also be caused by repeat mutation or gene conversion. Keeping this in mind, we use the results of the four-gamete test as an indication of recombination or disruption in LD. Comparison of the four gamete matrices and LD measurements yields close concurrence.
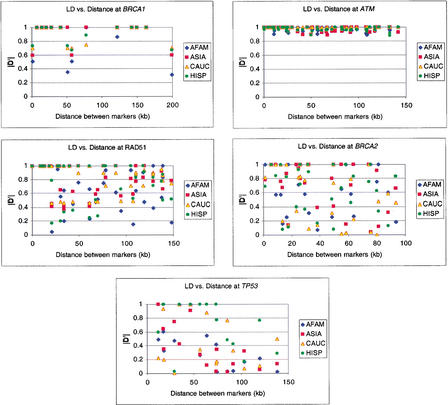
The data in this study do not show a correlation between LD and distance at these loci. Plotting LD, ‖D‘‖, versus intermarker distance results in plots with a uniform distribution of points that do not show a trend for decrease in LD corresponding with increasing distance (Fig. 4). The intensity of LD is sometimes low between markers that are closely spaced as well as those that are not and vice versa. This would support the notion of LD existing in a block-like pattern throughout the genome rather than as a continuous spectrum based on distance.
Figure 4.
LD versus intermarker distance. ‖D‘‖ values for all pairwise comparisons are plotted against the physical distance between each pair of markers. LD measurements were made for each population for each locus. African American data are plotted in blue, Asian American in red, European American in yellow, and Hispanic American in green.
DISCUSSION
We have described the most commonly occurring haplotypes for the five loci in this study. Haplotypes and linkage disequilibrium (LD) measurements were generated from SNP genotype data for ATM, BRCA1, BRCA2, RAD51, and TP53 for four populations: African Americans, Asian Americans, Hispanic Americans, and European Americans. Variation in SNP frequencies, LD pattern and intensity, and haplotype diversity was observed both between loci and populations. Despite the variation observed in this study, a trend for minimal haplotype diversity was observed at all five loci.
The conception of the configuration of LD in the genome has evolved as empirical data have accumulated. Analysis of the β-globin gene cluster was one of the initial illustrations of the complexity of the structure of LD. Regions 5′ (35 kb) and 3′ (19 kb) to the β-globin structural gene were found to have high LD with little measurable LD between these two clusters (9 kb; Chakravarti et al. 1984). The lack of LD observed at LPL (Clark et al. 1998) and TP53 (this study) is similar to the central region in the β-globin study. Empirical data from this study (BRCA1 and ATM) and others show regions of LD >100 kb (Peterson et al. 1995; Collins et al. 1999; Liu and Barker 1999; Bonnen et al. 2000; Taillon-Miller et al. 2000; Abecasis et al. 2001; Thorstenson et al. 2001). It has been suggested that the genome consists of blocks of LD (30–100 kb) interrupted by short (1–2 kb) hot spots of recombination (Daly et al. 2001; Jeffreys et al. 2001). The plots of LD versus intermarker distance support this idea (Fig. 4). Rather than a curve that would indicate a continuous degradation of LD over intermarker distance, these graphs appear as scatter plots. Data points indicate high LD between markers in an LD block and low LD in the recombination hot spots regardless of intermarker distance. Similar results have been seen by others when considering comparable distances (Johnson et al. 2001; Reich et al. 2001) and for distances as large as 1 Mb (Taillon-Miller et al. 2000). An additional feature of the LD structure in this study is that in most cases the degradation of LD is quite rapid as opposed to a gradual decline over distance. This supports the idea that there are blocks of LD interrupted by short regions of recombination with one exception. Just as there are regions of extended LD, we present data for a lengthy region without measurable LD. The TP53 locus shows no LD across as much as ∼90 kb, considerably more than the expected 1–2 kb for a recombination hot spot. A similar finding of an expansive region of little LD in two separate regions of Xq25 (129 kb and 308 kb) adds to the evidence that genome-wide LD patterns remain a complex issue that may only be resolved when a genome-wide map of LD is available (Taillon-Miller et al. 2000).
Examination of haplotype structure across these loci also supported the idea of locus-specific genomic diversity. The number of haplotypes at each locus varied widely and reflected the variation in LD patterns and intensity at the genomic locations. The seemingly substantial differences in numbers of haplotypes and LD patterns between loci underscore the importance of characterizing each locus of interest prior to association studies. This diversity also underscores the inefficiency of applying a standard marker density genome-wide for association studies or genome scans. More importantly, it points to an inability to estimate a priori the LD for a particular region—instead, LD must be characterized for each region of interest.
The variation seen between populations extends this argument to a need for characterizing haplotypes and LD in each study population as well. The amount and pattern of LD varied between populations, however, not as much as has been seen in some reports (Goddard et al. 2000; Kidd et al. 2000; Reich et al. 2001). ATM and BRCA1 showed virtually no differences between populations, perhaps because of the uniformly high LD in these regions. BRCA2 showed the least LD in European Americans, followed by Hispanic Americans, then African Americans. This illustrates an exception to the general finding that African-derived populations exhibit lower levels of LD than European-derived populations. An additional standout is Hispanic Americans at TP53, where the LD pattern is completely different from that of the other three populations. Similar to LD, the number of haplotypes and haplotype heterozygosity also varied between populations. African Americans had the highest haplotype heterozygosity at three loci: ATM, BRCA1, and RAD51. The increased number of haplotypes and haplotype diversity in African Americans correlates with the older age of the population. However, just as African Americans do not always demonstrate the least LD, this population does not always exhibit the most haplotype diversity. These discrepancies between populations support the notion that individual genomic regions may undergo different evolutionary pressures in various populations. Exploitation of such differences between populations has been suggested to have significant potential for identification of alleles contributing to common diseases (Todd et al. 1989; Reich et al. 2001).
The most salient finding of our haplotype study was that there are few haplotypes shared among all populations, and that these haplotypes account for a very high percentage of the total chromosomes (Table 1). This high degree of sharing was observed even in regions with little LD. Similarly, the total number of haplotypes at each locus is also relatively few. Considering the maximum possible as 2n (that could have been generated by free recombination), the actual number of haplotypes is closer to the theoretical minimum than maximum. We conclude that there are old mutations that can be used to mark a relatively small number of distinct haplotypic structures of chromosomes that are present in the human population at a high frequency.
The small number of haplotypes and high degree of sharing is in part due to the fact that we used more commonly occurring SNPs and no rare or population-specific SNPs. Because of the present interest in the common disease common variant (CDCV) hypothesis, we have attempted to describe the most commonly occurring haplotypes for the five loci in this study. We have not attempted to catalog all of the genetic diversity at these loci. By focusing on the most commonly occurring haplotypes, we may have missed some of the genetic diversity in these gene regions. The addition of markers, especially low-frequency markers, will partition the commonly occurring haplotypes into subgroups and add low-frequency haplotypes. In a similar manner, addition of population-specific alleles leads to population-specific haplotypes. If this were done iteratively it could lead to a situation in which each person’s haplotypes are unique. For association studies in search of a functional variant that is commonly occurring, the commonly occurring haplotypes are the most pertinent, and too many haplotypes can lead to a loss of power and information. Conversely, regions of extreme LD such as BRCA1 show few haplotypes such that a haplotype-based study for this region may suffer from a lack of discrimination. Therefore, it may be useful for some studies to refine haplotypes by breaking the commonly occurring haplotypes into subgroups through the addition of either population-specific or lower-frequency markers, especially in regions of high LD. A thorough characterization of the LD landscape at a locus in a particular population is necessary for design of effective association studies.
Focusing exclusively on regions of high LD for haplotype-based association studies may exclude informative regions. As is seen in this study of BRCA2 and TP53, shared haplotypes that are of relatively high frequency can be found spanning regions that appear virtually devoid of LD. Not only can the haplotype structure be determined across short regions without measurable LD but across extended regions of low LD as in TP53. Commonly occurring haplotypes are an important tool for a study focusing on detection of common variants, thus the haplotype structures at all five genes studied here show potential for successful detection of associations in appropriately chosen subject populations.
METHODS
Human Subjects
Genomic DNA from five unrelated European American individuals was sequenced for SNP discovery. This DNA was extracted from lymphoblast and fibroblast cell lines. SNP genotyping was carried out using genomic DNA from the Baylor College of Medicine Polymorphism Resource. This collection of ethnically defined DNAs was purified from lymphoblast cell lines established from anonymous blood donors in Houston, Texas, USA, with informed consent. Individuals reported self-described ethnicity and were subsequently divided into four ethnic groups: African American (n = 48), Asian American (n = 48), European American (n = 48), and Hispanic American (n = 48).
At least three of our samples are of composite anthropologic description. The Asian American individuals comprise an unknown number of national populations of Asia; the Hispanic Americans are genetically admixed, comprising gene pools of American Indian, European, and possibly African descent; and the African Americans are also genetically admixed, having both European and African genes in their gene pool (Chakraborty 1986). Although this may explain the high degree of haplotype sharing, because of the presence of European-derived haplotypes in at least three samples, admixture alone cannot explain the pattern of LD we observed. Furthermore, the effects of the admixture process would have been reflected at all genes, unless the initial haplotype structure were different in different parental populations before the process of admixture.
SNP Detection
Two approaches were taken for SNP detection: resequencing and database/literature searches. Resequencing was done on PCR-amplified regions placed sporadically throughout the loci and has been previously described (Bonnen et al. 2000). SNPs for ATM and RAD51 were ascertained through resequencing of five unrelated individuals. SNPs for BRCA1 and BRCA2 were obtained through dbSNP, literature searches, and resequencing. No sequencing was used for TP53 SNP detection. Any SNPs that were detected through sequencing but did not perform well under standard PCR or genotyping conditions are reported but were dropped from the study. All SNPs in this study have been entered into dbSNP and their identifiers are BRCA1: rs1054385, ss4325297, rs799923, rs799916, ss4325298, ss4328154, ss4325299, ss4328155, rs443759, rs799906; BRCA2: rs114827, rs206136, ss4325300, rs1799943, rs144848, ss4328156, ss4325301, rs206340, rs1012129; RAD51: ss4325288, ss4325289, ss4325290, rs1051482, ss4325293, ss4325294, rs752012, rs2289218, rs2289219, ss4325296, rs1801321, ss4325292, ss4325295; ATM: rs228589, rs600931, ss4328151, rs664677, rs645485, ss4328152, rs227060, rs227069, rs227074, rs664982, rs664143, rs652541, rs170548, ss4328153, rs624366, rs609261, rs172896; TP53: rs839721, rs1544725, rs1625895, rs1050528, rs727428, rs1017163, rs4227, rs1421314.
Six dbSNP entries were genotyped and not found to be polymorphic in this study population. Their dbSNP identifiers are rs1895090, rs1042526, rs916131, rs916132, rs722494, rs1059300.
Primers for DNA amplification and sequencing were designed using MacVector version 6.0.1. The genomic sequence of each gene was masked for repetitive sequences using RepeatMasker. Genomic DNA from five unrelated individuals was amplified. The 50-μL reactions included DNA (200 ng), standard 1× PCR buffer (Perkin-Elmer), dNTPs (0.1 mM), Taq (0.5 μL; Perkin-Elmer), primers (1 μM each). PCR was performed in a Perkin Elmer 9700 with an initial denaturation at 95°C for 5 min followed by 30 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec; and 72°C for 7 min.
PCR products were purified and sequenced. Preparation of DNA for sequencing included incubation of ∼60 ng of PCR product with shrimp alkaline phosphatase (2 U; Amersham) and exonuclease I (10 U; Amersham) at 37°C for 15 min, followed by enzymatic inactivation at 80°C for 15 min. Direct sequencing of each PCR product was carried out using ABI dye terminator cycle sequencing kit and run on an ABI 373A for RAD51, BRCA1, and BRCA2. The Thermo Sequenase 33P-radiolabeled terminator cycle sequencing kit (Amersham Pharmacia) was used for sequencing at ATM as previously described in Bonnen et al. (2000).
SNP Genotyping
Genotypes for each SNP were determined using allele-specific oligonucleotide (ASO) hybridizations. ASO hybridizations were executed as previously described by DeMarchi et al. (1994). Autoradiograms were read on at least two independent occasions.
PCR amplification for genotyping was combined into two multiplex PCR reactions per gene. The 50-μL reactions included DNA (250 ng), standard PCR buffer without MgCl2 (2×) (Perkin-Elmer), MgCl2 (1.8×), dNTPs (0.2 mM), and Taq (0.5 μL; Perkin-Elmer). PCR was performed in a Perkin Elmer 9700 with an initial denaturation at 95°C for 5 min followed by 30 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 2 min; and 72°C for 7 min. Primers include some of those originally designed for sequencing and some newly designed to alter the size of the amplicons. Products were separated by at least 20 bp in length so that they could be resolved from one another on a 2.5% agarose gel. Multiplex PCRs were checked to have amplified all products by running 6 μL of product on a 2.5% agarose gel.
One SNP, rs1625895, was genotyped through restriction fragment length polymorphism digest of PCR-amplified DNA with the enzyme MspI (Roche). Digest fragments were resolved on a 2.5% agarose gel.
See Supplemental Material for multiplex PCR primer sequences and ASO probe sequences (available online at http://www.genome.org). All oligonucleotides used to assay ATM SNPs were reported previously in Bonnen et al. (2000).
Estimation of Haplotypes and Frequencies
Haplotypes and their frequencies were estimated from unphased genotype data by the computer program EMHAPFRE (Excoffier and Slatkin 1995). EMHAPFRE uses an expectation-maximization algorithm that determines the maximum likelihood frequencies of multilocus haplotypes in diploid populations. Only individuals who were scored for the complete set of SNPs for a gene were included in the data analysis.
Statistical Methods
Haplotype heterozygosity was calculated from
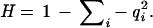 |
To test for recombination, we used the four-gamete test and the Hudson and Kaplan recombination statistic R (Hudson and Kaplan 1985). For a given haplotype AB, mutation may result in Ab or aB. Haplotype ab arises only in the case of recombination or repeat mutation. The four-gamete test was executed on unphased genotype data in a pair-wise fashion across all SNP loci. Based on the resulting matrix of the four-gamete test, R estimates the location and number of recombination events that have taken place in the sample.
LD was computed by performing pair-wise comparisons for all SNP loci. P-values from Fisher's exact test were used to determine significance levels. SNPs having a rarer allele frequency ≤0.05 were excluded from LD analyses. LD statistic D is a pair-wise comparison of gametic frequencies such that D = p11p22–p12p21. r2 is calculated from D2/(p1p2q1q2) (Hill and Robertson 1968). D‘, relative disequilibrium, is D‘ = D/‖D‖max, where ‖D‖max = max(p1p2,q1q2) if D < 0 and ‖D‖max = min(q1p2,p1q2) if D > 0 (Lewontin 1964).
All recombination and LD statistics were generated using the software program DnaSP 3.00 by J. Rozas and R. Rozas, Universitat de Barcelona. LD plots were generated using the GOLD software (Abecasis and Cookson 2000).
WEB SITE REFERENCES
http://www.bio.ub.es/∼julio/DnaSP.html; DnaSP.
http://www.ncbi.nlm.nih.gov/SNP/; dbSNP.
http://www.sph.umich.edu/statgen/abecasis/GOLD/docs/graphic.html; GOLD software.
Acknowledgments
This work was supported in part by a grant from the National Cancer Institute of the United States National Institutes of Health (CA75432).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nelson@bcm.tmc.edu; FAX (713) 798-5386.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.483802. Article published online before print in November 2002.
REFERENCES
- Abecasis GR, Cookson WO. GOLD—Graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Noguchi E, Heinzmann A, Traherne JA, Bhattacharyya S, Leaves NI, Anderson GG, Zhang Y, Lench NJ, Carey A, et al. Extent and distribution of linkage disequilibrium in three genomic regions. Am J Hum Genet. 2001;68:191–197. doi: 10.1086/316944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnen PE, Story MD, Ashorn CL, Buchholz TA, Weil MM, Nelson DL. Haplotypes at ATM identify coding-sequence variation and indicate a region of extensive linkage disequilibrium. Am J Hum Genet. 2000;67:1437–1451. doi: 10.1086/316908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R. Gene admixture in human populations: Models and predictions. Yearbook Phys Anthropol. 1986;29:1–43. [Google Scholar]
- Chakravarti A, Buetow KH, Antonarakis SE, Waber PG, Boehm CD, Kazazian HH. Nonuniform recombination within the human β-globin gene cluster. Am J Hum Genet. 1984;36:1239–1258. [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengard J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, et al. Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet. 1998;63:595–612. doi: 10.1086/301977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Lonjou C, Morton NE. Genetic epidemiology of single-nucleotide polymorphisms. Proc Natl Acad Sci. 1999;96:15173–15177. doi: 10.1073/pnas.96.26.15173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- DeMarchi JM, Richards CS, Fenwick RG, Pace R, Beaudet AL. A robotics-assisted procedure for large scale cystic fibrosis mutation analysis. Hum Mutat. 1994;4:281–290. doi: 10.1002/humu.1380040409. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Goddard KA, Hopkins PJ, Hall JM, Witte JS. Linkage disequilibrium and allele-frequency distributions for 114 single-nucleotide polymorphisms in five populations. Am J Hum Genet. 2000;66:216–234. doi: 10.1086/302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, et al. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- Kidd JR, Pakstis AJ, Zhao H, Lu RB, Okonofua FE, Odunsi A, Grigorenko E, Tamir BB, Friedlaender J, Schulz LO, et al. Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet. 2000;66:1882–1899. doi: 10.1086/302952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The detection of linkage disequilibrium in molecular sequence data. Genetics. 1995;140:377–388. doi: 10.1093/genetics/140.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Barker DF. Evidence for effective suppression of recombination in the chromosome 17q21 segment spanning RNU2–BRCA1. Am J Hum Genet. 1999;64:1427–1439. doi: 10.1086/302358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Traherne JA, Abecasis GR, Cookson WO. Single nucleotide polymorphism and linkage disequilibrium within the TCR α/δ locus. Hum Mol Genet. 2000;9:1011–1019. doi: 10.1093/hmg/9.7.1011. [DOI] [PubMed] [Google Scholar]
- Niu T, Qin ZS, Xu X, Liu JS. Bayesian haplotype inference for multiple linked single-nucleotide polymorphisms. Am J Hum Genet. 2002;70:157–169. doi: 10.1086/338446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AC, Di Rienzo A, Lehesjoki AE, de la Chapelle A, Slatkin M, Freimer NB. The distribution of linkage disequilibrium over anonymous genome regions. Hum Mol Genet. 1995;4:887–894. doi: 10.1093/hmg/4.5.887. [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Taillon-Miller P, Bauer-Sardina I, Saccone NL, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice JP, Kwok PY. Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat Genet. 2000;25:324–328. doi: 10.1038/77100. [DOI] [PubMed] [Google Scholar]
- Thorstenson YR, Shen P, Tusher VG, Wayne TL, Davis RW, Chu G, Oefner PJ. Global analysis of ATM polymorphism reveals significant functional constraint. Am J Hum Genet. 2001;69:396–412. doi: 10.1086/321296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Pakstis AJ, Ruano G, Kidd KK. The accuracy of statistical methods for estimation of haplotype frequencies: An example from the CD4 locus. Am J Hum Genet. 2000;67:518–522. doi: 10.1086/303000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Mijovic C, Fletcher J, Jenkins D, Bradwell AR, Barnett AH. Identification of susceptibility loci for insulin-dependent diabetes mellitus by trans-racial gene mapping. Nature. 1989;338:587–589. doi: 10.1038/338587a0. [DOI] [PubMed] [Google Scholar]