Abstract
Assuming that patterns of sequence variants within highly homogeneous centromeric tandem repeat arrays can tell us which molecular turnover mechanisms are presently at work, we analyzed the α-satellite tandem repeat array DXZ1 of one human X chromosome. Here we present accurate snapshots from this dark matter of the genome. We demonstrate stable and representative cloning of the array in a P1 artificial chromosome (PAC) library, use samples of higher-order repeats subcloned from five unmapped PACs (120–160 kb) to identify common variants, and show that such variants are presently in a fixed transition state. To characterize patterns of variant spread throughout homogeneous array segments, we use a novel partial restriction and pulsed-field gel electrophoresis mapping approach. We find an older large-scale (35–50 kb) duplication event supporting the evolutionarily important unequal crossing-over hypothesis, but generally find independent variant occurrence and a paucity of potential de novo mutations within segments of highest homogeneity (99.1%–99.3%). Within such segments, a highly nonrandom variant clustering within adjacent higher-order repeats was found in the absence of haplotypic repeats. Such variant clusters are hardly explained by interchromosomal, fixation-driving mechanisms and likely reflect a fast, localized, intrachromosomal sequence conversion mechanism.
[Supplemental material is available online at www.genome.org and www.pedgen.med.uni-muenchen.de. The sequence data from this study have been submitted to DDBJ, EMBL, and GenBank under accession nos. AJ509815–AJ509823, AJ509829–AJ509852, AJ509874–AJ510031. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: P. Warburton, and C. Roos.]
It has been realized for decades that the high level of homogeneity observed within repetitive sequence families is the result of a concerted evolution caused by a variety of non-Mendelian molecular drive mechanisms such as unequal crossing over, sequence conversion, and transposition (Southern 1975; Smith 1976; Horz and Zachau 1977; Dover 1982; Charlesworth et al. 1994; for review, see Elder Jr. and Turner 1995). Particularly for the centromeric tandem repeat arrays of higher eukaryotes, it would be intriguing to dissect all the underlying molecular mechanisms, because of the obvious lack of a direct sequence–function relation, which is the subject of discussion at present (Eichler 1999; Tyler-Smith and Floridia 2000; Henikoff et al. 2001; Sullivan et al. 2001). The fact that marker chromosomes lacking centromeric satellite DNA can form neocentromeres on a variety of repetitive and/or unique sequences (Choo 1997a; du Sart et al. 1997; Karpen and Allshire 1997; Barry et al. 1999; Tyler-Smith et al. 1999) implies an epigenetic control of centromere function. Without the need to maintain sequences for the sake of centromere function, mechanisms observed at centromeres might act within other regions of the genome, too.
Human α-satellite DNA comprises 2%–5% of the genome. The 0.17-kb-sized monomers are organized into monomeric, dimeric, or pentameric repeat families, on top of which a variety of modern higher-order repeat compositions have evolved (Manuelidis et al. 1978; Jorgensen et al. 1986; Waye and Willard 1986; Alexandrov et al. 1988, 1993; Thompson et al. 1989; Choo et al. 1991; Jorgensen et al. 1992; Greig et al. 1993; Choo 1997b). Whereas distant repeats and related families often share an identity of only 60%–90%, that of homogeneous arrays typically exceeds 97%. The 1–4-Mb-sized (Wevrick and Willard 1989) DXZ1 array of the human X chromosome is almost entirely composed of 2-kb-sized higher-order repeats with a dodecameric monomer organization of diverged pentamers and parts thereof (Yang et al. 1982; Willard et al. 1983; Waye and Willard 1985).
Homogeneous centromere arrays have been omitted from sequencing in the human genome project (Collins et al. 1998; Lander et al. 2001; Venter et al. 2001). Without absolute map positions, direct comparison of nucleotide variants between individuals or over the course of time is not possible, making it difficult to reconstruct the process of homogenization. In principle, the underlying molecular mechanisms fall into either one of two categories. Homogeneity is either attained by amplification of (novel) repeats, replacing more diverged ones (unequal crossing over, transposition, rolling circle amplification), or is maintained by an ongoing sequence adjustment between preexisting repeats (sequence conversion). Amplification of higher-order repeats would lead to a limited number of haplotypic variant combinations (sets of variable nucleotides within higher-order repeats would occur interdependently), and to few de novo mutations within regions of highest homogeneity. In addition, a novel variant (within an amplified haplotype) would spread throughout the population along certain gradients regarding its abundance (array size) and admixture with preexisting haplotypes, which would be accompanied by an accumulation of de novo mutations. Sequence conversion would also lead to few de novo mutations within regions of highest homogeneity, but would not (necessarily) lead to islands of haplotypic repeats (caused by short tract lengths or mixed directions of mismatch repair in intermediate heteroduplexes). Thus, variants presently in transition within a homogeneous array type and within the population would not show interdependency. However posttransition, when a variant has converted throughout the whole array, a novel haplotype results (as compared with the pretransition state). This causes problems, if closely related array types coexist within a genome but follow distinct trajectories of molecular drive. Thus, in order to analyze ongoing homogenization, it might be critical to isolate variants, which are presently in transition within homogeneous sections of one distinct array, and within the population.
Several studies showed that unequal crossing over can result in a novel higher-order repeat structure (Mashkova et al. 1998). Subsequent array expansion might lead to large homogeneous islands and coexistence of distinct array types, as known from human centromere 17 (Waye and Willard 1986; Warburton and Willard 1990, 1995). These observations, together with a long-standing model based on a computer simulation, which demonstrated that a single process of mutagenesis, unequal crossing over, and array expansion is sufficient to generate homogeneous tandem repeat arrays (Smith 1976), strongly support the role of unequal crossing over in α-satellite evolution. Although unequal pairing seems to be an inevitable feature of arrays with highly polymorphic sizes (Wevrick and Willard 1989), it has remained unknown to what extent crossing over actually takes place within homogeneous arrays. Centromere flanking markers generally show suppressed recombination rates (see Choo 1998), and a pedigree analysis of DXZ1 revealed rather stable array sizes during meioses and mitoses (Mahtani and Willard 1990). An early analysis of DXZ1 nucleotide variants used small groups of λ and plasmid-cloned higher-order repeats from unmapped regions of various X chromosomes. Sequencing underpinned the high level of homogeneity and revealed that repeats from a clone have more in common than repeats from different localizations, an observation compatible with amplification of major haplotypic higher-order repeats, as predicted by the unequal crossing-over model (Durfy and Willard 1989; Warburton and Willard 1990). In addition, it has been realized that the distinct, but closely related array types of Chromosome 17 (>95% identity, different higher-order repeat structure) harbor array-specific variation as a result of localized exchange (localized in terms of being restricted to a certain array type). At this point, without exact mapping of variants throughout larger numbers of higher-order repeats, it remained open how the process within an array would take place with respect to unequal crossing over and sequence conversion. Because some of the analyzed variants were not fixed, relatively fast, intrachromosomal exchanges along haplotypic lineages (array types) were concluded (Warburton and Willard 1995). A recent study based on genomic mapping and sequencing of a boundary of DXZ1 demonstrated a gradual decrease of identity of clearly related repeats from 97% to <85%, further confirming the unequal crossing-over model of α-satellite evolution (Schueler et al. 2001). However, the mapping of long-range restriction fragments and restriction fragment length polymorphisms in BAC clones derived from several different X chromosomes was not extended throughout a homogeneous array portion of significant size. In addition, variants, which are presently in a fixed transition state within the homogeneous array, were not subjected to the detailed mapping and sequencing analysis of this boundary.
To analyze the distribution of variants, which at present are in transition within regions of highest homogeneity within a single homogeneous array, we performed a novel combined subclone sequencing and large-scale variant mapping analysis within DXZ1 segments from a single human X chromosome. As far as known, and as supported by this work, homogeneous α-satellite DNA regions are devoid of anchorage points useful for fine mapping. The homogeneous, PAC-cloned segments analyzed here could be megabases apart from a boundary of the array, which could possess significantly diverged sequences or insertions providing unique restriction marks to integrate cloned and genomic data (Schueler et al. 2001). Therefore, the novel detailed snapshots of variant spread within segments of highest homogeneity (>99%) are solely based on stable and representative cloning of DXZ1 in PACs.
Here we report the discovery of highly nonrandom variant clustering within adjacent higher-order repeats and demonstrate that clustering occurs in the absence of haplotypic repeats. It is assumed that the most homogeneous array segments primarily present patterns of ongoing homogenization and not those of degradation of a putative, recent state of even higher homogeneity (which would not fit the ubiquitous phenomenon of concerted evolution of centromeric satellite DNA families, anyway). This is supported by the relative paucity of potential de novo mutations within the highly homogeneous segments analyzed in this study. Because the variants analyzed here are presently in a fixed transition state, they have had sufficient evolutionary time to be subjected to all ongoing mechanisms driving the concerted evolution, and to develop typical patterns according to the most prominent mechanisms involved. The observed regular clustering of variants within adjacent higher-order repeats (not to be confused with unmapped variants localized to array types; see Warburton and Willard 1995) implies a process confined to a narrow region of a single chromosome. Several rounds of this intrachromosomal clustering mechanism must obviously take place, before fixation driving interchromosome spread has a chance to disintegrate the clusters. Thus, identification of the regular clusters of variants within adjacent higher-order repeats allows us to assess the relative rate of the underlying process with an unprecedented clearness. Without evidence for haplotypic repeat lineages, we conclude that the patterns are most compatible with a fast sequence-conversion mechanism contributing directly to ongoing homogenization without the need for additional amplification mechanisms. In principle, such localized clustering could also be explained by repeated rounds of unequal crossing over within very short distances around the particular variant analyzed. Such an isolated process would require additional amplification and loss to significantly contribute to an overall homogenization, and then would be expected to cause uneven variant distributions within the population (Smith 1976), not fitting the comparably fixed ratio of present variants found in this study. In short, it seems that predictions from the unequal crossing-over model are clearly supported, if variation between repeats with a significant evolutionary divergence, say, <97% sequence identity or members of distinct array types with different higher-order repeat structure are analyzed. In contrast, if looking at the current spread within a single homogeneous array (>97% identity and structural homogeneity), evidence for unequal crossing over is poor.
RESULTS
Representative Subregions of DXZ1
Instability of repetitive DNA has been observed in plasmids, cosmids, and YACs (Neil et al. 1990; Schindelhauer et al. 1996). We therefore analyzed representation of α-satellite DNA in segment 1 of the human PAC library derived from an MboI partially digested, male genome (Ioannou et al. 1994). Because the higher-order repeats usually contain 2–6 MboI sites, similar to the average genome, a relevant cloning bias was not expected. Stringent hybridization using the 2-kb higher-order repeat probe X5 (pBamX5 kindly provided by P. Warburton; Willard et al. 1983) revealed a total of 347 clones, which were divided into groups A, B, C, and D with decreasing signal intensity (strong after 3, 30, 300 min of exposure, and weak at 300 min, respectively). Hundreds of very faint signals were excluded. The groups contained 40, 74, 93, and 140 clones, respectively. For 12 clones of each group, chromosomal origin was analyzed with restriction nucleases BamHI, EcoRI, and XbaI, diagnostic for higher-order repeats of Chromosomes X, 17, and 11, respectively (see Table 1). Assuming a fraction of 2% of homogeneous α-satellite DNA per diploid genome of 6,000 Mb, that is, 2.6 Mb per chromosome, the 85,000 clones of the library segment would contain 1,700 α-satellite clones from 46 chromosomes, and 184 clones from Chromosomes X, 17, and 11. The extrapolated 244 clones (see Table 1) indicate normal coverage and suggest hybridization of most, if not all parts of the arrays (Mahtani and Willard 1990), using probe X5. The average pulsed-field size of α-satellite inserts was 109 kb (data not shown), comparing well with 110 kb for the total library (http://www.chori.org/bacpac/humalmaleall.htm). To assess the risk of deletion or rearrangement during bacterial growth, we reanalyzed two separate colonies of 12 large PACs (120–200 kb) of Chromosomes X and 17 (6 of each). All 12 clones were stable in size; one showed a rearranged BamHI fragment and was excluded (data not shown). Five stable PACs of Chromosome X (120–160 kb) were used in this study. PAC end sequence pairs show the same orientation of α-satellite DNA and presence of junction MboI or BamHI sites at positions typical for X alphoid DNA, indicating a high cloning accuracy in all cases. The actual array size and localization of the PACs within the array are unknown. Assuming an array size of 1–4 Mb, each PAC would represent between 3% and 16% of DXZ1.
Table 1.
Representation of α-Satellite Arrays in a PAC Library
| Hybridization groups | Signal intensity | Number of signals | Analyzed clones | Origin of analyzed clones | Calculated clone number in library segment 1 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr. X (BamHI) | Chr. 17 (EcoRI) | Chr. 11 (XbaI) | Other | Chr. X | Chr. 17 | Chr. 11 | ||||
| A | Very strong | 40 | 12 | 9 | — | — | 3 | 30 | — | — |
| B | Strong | 74 | 12 | 3 | 6 | 3 | — | 18 | 38 | 18 |
| C | Medium | 93 | 12 | — | 7 | 5 | — | — | 54 | 39 |
| D | Weak | 140 | 12 | — | 1 | 3 | 8 | — | 12 | 35 |
| Sum | 48 | 104 | 92 | |||||||
| Total | 347 | 244 | ||||||||
PAC Ranking
Estimates from ethidium bromide staining and Southern analysis showed that almost all of the PAC inserts were cut into 2-kb BamHI higher-order repeats (data not shown). All different-sized nonvector fragments (Table 2) also hybridized to X5, and most likely represent cloning boundaries and divergent repeats (rare RFLPs, or structural changes caused by unequal crossing over or insertion). We subcloned samples of 10 higher-order repeats (2-kb BamHI) of each of the 5 PACs and sequenced both ends covering roughly 10 kb of each PAC. We found four pairs of identical sequences, one in PAC A7, one in PAC A8, and two in PAC A6, compatible with duplicate cloning. At least 46 individually subcloned higher-order repeats were isolated. Of those, one of PAC A8 matched one of PAC A10, possibly indicating overlap or extensive homology in the sequenced portion. Pairwise comparison confirmed high overall homogeneity exceeding 97%, and revealed slight differences between PAC samples (Table 2). To trace an ongoing homogenization process, it was unclear whether actual homogeneity would be a good measure, and which kind of homogeneity would be relevant. Local disruption of an otherwise homogeneous array could lead to exclusion from turnover or could be compensated by sufficient medium identity. Therefore, we determined the medium homology and the maximum deviation of higher-order repeats within PAC samples, and estimated the incidence of unusual BamHI fragments from ethidium bromide-stained PAC digestions (Table 2). Clearly, all parameters gave a similar order. PACs A7, A8, and A10 are highly homogeneous, and PACs A6 and B11 are slightly more diverged. Because ongoing homogenization should efficiently eliminate the majority of de novo mutations (which cannot be distinguished from rare variants), we determined the frequency of variants occurring only once within the 45 individual BamHI higher-order repeats analyzed in this study. Strikingly, the PACs with higher homogeneity have fewer such rare variants, indicating a rough correlation between present homogeneity and recent homogenization (Table 2). Suiting a strong non-Mendelian molecular drive component of α-satellite evolution, the majority of differences between individual higher-order repeats is not caused by de novo mutations, which are responsible for <5% of differences within the most homogeneous sections (Table 2).
Table 2.
PAC Ranking
| PAC | Insert size (pulsed-field) | Fraction of 2-kb BamHI fragments of insert size (%) | Total number of 2-kb BamHI fragments | Size of unusual BamHI fragments in kilobases and number (n) as estimated from total BamHI digests and partial mapping or deduced from PAC end sequencing (*) | Medium homology of sequence pairs (% nucleotides) | Maximum deviation of sequence pairs (% nucleotides) | Frequency of single variants (% nucleotides) |
|---|---|---|---|---|---|---|---|
| A6 | 160 kb | 82 | 63 | 0.9 (1*), 1.7 (2), 2.2 (1), 2.9 (5), 4.9 (1) | 98.83 | 2.1 | 0.33 |
| B11 | 125 kb | 91 | 56 | 0.55 (1*), 2.9 (2), 4.9 (1) | 98.58 | 2.1 | 0.20 |
| A7 | 145 kb | 99 | 70 | 0.9 (1*) | 99.12 | 1.5 | 0.14 |
| A10 | 130 kb | 99 | 64 | 0.9 (1*) | 99.11 | 1.3 | 0.07 |
| A8 | 120 kb | 98 | 59 | 0.9 (1*), 1.7 (1) | 99.25 | 1.3 | 0.08 |
Nucleotide Variants and Fixation in the Population
To identify putatively fixed nucleotide variants, we assumed their broad distribution throughout the array. Using the arbitrary criterion: occurrence in at least three higher-order repeats of at least two PAC samples, we identified a set of 18 common (and abundant) variants (9 transitions, 8 transversions, 1 deletion of 3 bp) within the sequenced portions (see variants in Fig. 1 and in the sequence alignment in Supplementary Fig. 1; available online at http://www.genome.org). To examine the actual transition state on the population level, we used a 0.5-kb DXZ1-specific PCR (Warburton and Willard 1992) containing 10 of the 18 common variants, 5 of which affect the restriction sites HinfI, DraIII, Cac8I, and AciI. Analysis of 10 male Europeans revealed relatively constant, variant typical ratios (individual variants have constant ratios between 5/1 and 1/1) of cut to uncut fragments in all genomic PCRs, consistent with a fixed transition state (data not shown). As a control on the sequence level, 9 PCR fragments of one, and 4 of three other individuals were subcloned. All 13 clones were different in sequence. The 9 subclones of one individual showed a medium identity of 97.9% and a maximum deviation of 3.3%. This slightly higher variability compared with the same 365-bp region of 9 subclones from the 5 PACs (98.8% and 2.7%, respectively) possibly indicates broader representation by PCR. Restriction sites in the PCR fragments were found affected by the expected nucleotide variants, indicating uniformity throughout the population. Of the 10 PAC variants within the 0.5 kb, 8 were also found in the PCR fragments, undermining the usefulness of PAC samples of one individual to identify common variants with a fixed transition state. Three PCR variants were present in one PAC sample only, and one was uniform within PACs, indicating that most, but not all, common variants were identified. To better estimate the abundance of a fixed variant within whole individual arrays, we used BamHI, which cuts homogeneous arrays into 2-kb higher-order repeats, and the unique DraIII restriction variant, which generates subfragments of 0.5 and 1.5 kb if present. On a genomic Southern blot using probe X5, all 6 random Europeans analyzed showed a ratio of approximately 3/1 (cut to uncut) as estimated from different exposures. Within the limits of quantifiability, the result appeared similar to the ∼2/1 ratio observed in the same 6 individuals (and in 4 others) analyzed by array-specific PCR (data not shown).
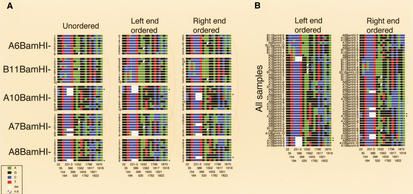
Figure 1.
Variant box. For haplotype analysis, the combinations of a set of 18 “common” variants were diagramed for each of the individual 2-kb BamHI subclones from the 5 PAC samples. Little squares were drawn for each nucleotide using the ABI color code (see inset). Variant positions (bottom) belong to the numbering of the satellite sequence in this work (see Supplementary Fig. 1; available online at http://www.genome.org). The 3-bp deletion variant at position 231–233 spans three squares. The variants initially were isolated because they were abundant within a single X-chromosome array and typically revealed a fixed transition state within the population. Within the most homogeneous PACs A7, A8, and A10, such common variants account for the majority of the total variation. The slightly lower homogeneity of PACs A6 and B11 is mainly attributable to a higher fraction of regional variants, possibly owing to a reduced participation in the overall homogenization process (spatially or temporally). The overall sequence divergence of PACs A6 and B11 could well be considerably higher, because a significant fraction of dispersed BamHI fragments with sizes other than 2 kb were not analyzed (Table 2). Of the 3 most homogeneous PACs, only A8 contains such a disruption of the regular BamHI higher-order repeat structure (data not shown). Ordering the combinations of the 18 common variants derived from PAC samples (A) or from all PACs (B) along their left or right ends, neither led to regular patterns reminiscent of a limited number of major haplotypes, nor revealed any obvious relatedness of haplotypes with respect to PAC origin. Of the 46 combinations from individual subclones, 40 are unique and none occurs three times. The six pairs of combinations occurring twice were found within the highly homogeneous PAC A7 (2×), the less homogeneous PAC B11 (containing a duplication of 35–50 kb), between the two homogeneous PACs A10 and A8, or between the homogeneous PAC A7 and diverged PAC A6, as indicated by arches. Asterisks indicate individual clones with identical end sequences. Several combinations of PAC B11 seem to be more related to each other than to the combinations of other PACs, which to some extent could be explained by a somewhat reduced information from the set of 18 variants in this PAC. Interestingly, according to partial restriction mapping, PACs B11, A6, and A7 cannot extensively overlap with each other, and all three cannot extensively overlap with PACs A8 and A10 (which could overlap almost entirely). Nevertheless, ordering the combinations from all PACs results in many possible relationships within and between the PACs. This analysis indicates a lack of major haplotypes within the array, not supporting homogenization via amplification and replacement. In addition, similar haplotype analyses within narrow regions derived from the two or three DraIII and non-DraIII clusters of PAC A7 (see Supplementary Figs. 3, 5; available online at http://www.genome.org) indicate that small regions of amplified haplotypes are also very unlikely.
Variants Occur Sequence-Independently
To judge whether variant positions would depend on the primary sequence, we modified one primer of the X-array-specific PCR and amplified closely related X-chromosome α-satellite DNA of three unrelated male lowland gorillas. Of the 5 human restriction variants analyzed within the 0.5-kb PCR, only DraIII revealed a possible transition state. An initial sequencing analysis of subcloned α-satellite fragments of gorilla revealed that 11 fragments (of which 3 sequences originating from two gorillas were identical) shared approximately 92% –95% with a human consensus sequence. However, two of the fragments (also sharing 92% –95% with the human consensus) had only 91% –92% in common with the other gorilla sequences, indicating representation of two array types of gorilla X chromosomes (Durfy and Willard 1990), which complicates restriction analysis. Both fragments of the second type showed a mutation 2 nucleotides apart from the human variant affecting DraIII, whereas all fragments of the first type contained the intact DraIII site. The small sample of 7 nonidentical sequences of the first type revealed only 1 common variant (twice in 7 gorilla sequences). In addition, 11 single variants (which likely are common too) were identified. Only two of these could be identified in all our human sequences, one being a single, and one a minor variant only present in PAC A7. Thus, the positions of human and gorilla variants (albeit possibly varying in their regional density within the higher-order repeats) do not appear to be related to any obvious sequence motif or to binding boxes for centromere protein B (Earnshaw et al. 1987; Masumoto et al. 1989). This random occurrence of variants within the conserved α-satellite sequences of closely related species rules out any major sequence-dependent mutation mechanism recurrently generating variants de novo.
Lack of Haplotypic Higher-Order Repeats
To analyze whether higher-order repeats within the homogeneous array segments would belong to distinct major haplotypes, we compared the combinations of the 18 common and abundant variants within individual higher-order repeats. Of 45 independent sequences, 40 different haplotypes were identified (Fig. 1A). Apart from incidental co-occurrence of more frequent variants, combinations appear largely random. Attempts to order the 40 combinations by arranging them along their left- or right-end variants neither resulted in special haplotypic groups nor revealed a strict relatedness of higher-order repeats derived from a given PAC (Fig. 1B). Examples for close relatives can be found between any two PACs, indicating related combinations in medium to long distances (>0.26 Mb). In addition, a particular combination can hardly be found more frequently than any other within one or different PACs covering at least 0.51 Mb of the array (see below). Together with the lack of haplotypic variant combinations within short distances of less than a few tens of kilobases within DraIII and non-DraIII clusters, and within 12 kb of an HindIII cluster (see below), variant spread clearly occurs interindependently. On the array level, the local abundance of certain variants might vary widely, as seen in the slightly less homogeneous PACs A6 and B11, which harbor a number of local variants absent in other PAC samples. However, small sample sizes do not allow an accurate determination of variant frequencies within PACs. Small sample sizes also suggest that true de novo mutations are much rarer than the observed frequency of single variants (0.07%–0.33%).
PAC-Based Partial Restriction Mapping of Common Variants
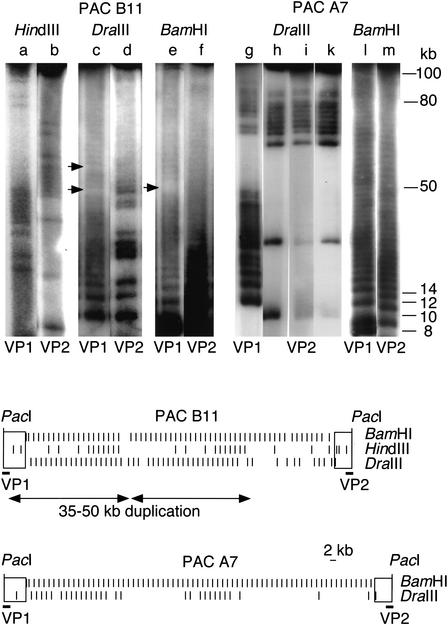
As frequently cutting nucleases produce a smear rather than a resolved ladder on a pulsed-field gel, identification of the single-cutting restriction variants HindIII and DraIII gave us the unique opportunity to study exact patterns of variant spread throughout whole PACs. Mapping revealed a highly nonrandom variant distribution. The HindIII variant within PAC B11 presents two identical clusters of 7 neighboring higher-order repeats in a distance of ∼50 kb (Fig. 2, top panel, lanes a,b). In a similar distance, PAC A7 shows two regular DraIII clusters of 7 and 10 higher-order repeats (Fig. 2, top panel, lanes g,h). Interestingly, the borders of the clusters tend to present gaps of single higher-order repeats, indicating a regionally confined, but saltatory spread, rather than a strict lateral movement. To check whether clustering would be an artifact of bacterial DNA metabolism during the period from library production until arrival in our laboratory, we replated two single colonies of PAC A7 every day for 6 d (400 generations). After this period, identical patterns were found (Fig. 2, top panel, lanes i,k). This proves the human origin of the clusters and further demonstrates the unprecedented stability of PAC-cloned α-satellite DNA in the recA–Escherichia coli strain DH10B (Ioannou et al. 1994). In addition to the variants, we mapped the constant BamHI site to analyze array structure (Fig. 2, top panel, lanes e,f,l,m). Whereas, within the limits of resolution, PAC A7 presents a 2-kb ladder throughout the entire insert, PAC B11 shows a disruption of the otherwise homogeneous ladder. Integrated restriction maps are presented (Fig. 2). Interestingly, a BamHI spacing (4.9-kb fragment) is localized between the two identical HindIII patterns of PAC B11, possibly marking out-of-frame recombination.
Figure 2.
(Top) Large-scale partial restriction mapping. Intact PAC DNA was linearized using the unique PacI site in vector DNA, and electroeluted (Biotrap, Schleicher Schüll). Partial digestions, using 0.1–3 U and 1–2 min of incubation time, were run on a CHEF DRII pulsed-field gel apparatus (Biorad) under conditions separating fragments up to 80–100 kb (switch 2.8 sec, 6 V/cm, 25 h). Gels were Southern-blotted and hybridized with end probes VP1 and VP2. X-Ray images were photographed and processed on a Nicon Cool Scan III. Each lane (a–m) represents one selected condition. PAC B11 (lanes a–f) shows clustered occurrence of the HindIII variant common within this array segment. Two clusters of 7 higher-order repeats containing the HindIII variant are arranged in tandem, at a distance of ∼50 kb (lanes a,b). The slightly more diverged array of PAC B11 shows disruption of the 2-kb BamHI higher-order repeat structure (lane e) and of the DraIII variant (lane c), possibly marking sites of unequal recombination (little arrows). The highly homogeneous PAC A7 (lanes g–m) is entirely composed of 2-kb BamHI higher-order repeats (lanes l,m). The DraIII variant, which is presently in a fixed transition state, occurs in clusters (lanes g–k), indicating a rather localized component of spread. Two additional, independent colonies were consecutively replated for 6 d (400 generations), which did not alter the DraIII restriction patterns (lanes i,k), further demonstrating the high stability of α-satellite DNA cloned in PACs. Integrated restriction maps of PACs B11 and A7 are shown at the bottom. Left (VP1) and right (VP2) end probes (bars) are indicated below PAC vectors (open rectangles). Variants presently in transition (HindIII, DraIII) occur in a highly nonrandom fashion, forming clusters of varying size. PAC B11 presents a duplicated pattern of 35–50 kb (duplicate arrows).
Analysis of the other PACs revealed that A10 and A8 showed only few HindIII variants in total PAC DNA digestions (none in the sequenced samples), as well as uniformity of the DraIII and BamHI ladders throughout the entire inserts, except a single BamHI spacing smaller than 2 kb in A8 (data not shown), making them uninformative. The mapping data obtained allow us to exclude extensive overlap of PACs A6, B11, A7, and A10/A8, but not between A10 and A8, indicating that the PACs represent at least 0.51 Mb of DXZ1, and, consequently, that the sequences sample a minimum region in excess of 0.26 Mb (assuming cloning stability). An attempt to map rare-cutter restriction sites as potential anchorage points for future PAC and BAC contig building revealed that none of the inserts contained NotI, AscI, MluI, BssHII, PvuI, or PacI, and only B11 contained an SalI site. Because SalI maps close to, or within the 4.9-kb BamHI fragment marking the large-scale duplication, we subcloned the ∼60-kb SalI fragment containing the PAC vector and end-sequenced over the SalI cloning site. A single nucleotide exchange within an otherwise typical X alphoid sequence (only one side analyzed) seems to have caused this SalI site.
Ongoing Conversion Overrides Unequal Crossing Over
Likely, the duplicated HindIII clusters within PAC B11 represent a large-scale unequal crossing-over event. Large-scale duplicative transposition (often onto other chromosomes), as known from pericentromeric regions (Horvath et al. 2000; Eichler 2001), might also be an explanation; however, owing to the lack of a genomic map position, we cannot decide whether the slightly more diverged PAC B11 might represent a pericentric border at all. Interestingly, HindIII-containing DXZ1 sequences have been shown to be polymorphic in some individuals (Durfy and Willard 1987). Including the partial BamHI and DraIII patterns of this PAC, a putative segment with a size between 35 and 50 kb could have been duplicated (Fig. 2). To analyze whether higher-order repeats within the duplicated HindIII clusters would contain identical sequences, we subcloned 18 from the pool of 12 existing 2-kb HindIII higher-order repeats of PAC B11. End sequences of the 18 subclones and of the 3 previously sequenced BamHI clones containing an HindIII site revealed at least 9 distinct higher-order repeats within the duplicated hexa-clusters, indicating divergence since the duplication event. Interestingly, all 9 cases of different HindIII higher-order repeat types are attributable to the turnover of common variants. Single variants were also found, not allowing a formal exclusion of de novo mutations since the duplication event, but in no case were single variants the sole reason for the differences between higher-order repeats. Obviously, conversion of variants continued, and random de novo mutagenesis did not have sufficient time to significantly add to the divergence (or more likely, mutations were efficiently converted). Clearly, the large number of at least 9 differing higher-order repeats does not support simple amplification as the clustering mechanism. In addition, the overall structure of the duplicated segment was stable and evidenced no expansion or deletion since the duplication event, whereas common variants within at least 3 of the 2-kb HindIII repeats were converted. However, the slightly lower overall homogeneity of PAC B11 makes it difficult to extend conclusions from the observed patterns to present homogenization. To analyze whether the clusters of the evenly fixed DraIII variant within the highly homogeneous PAC A7 could be explained by amplification of major haplotypic repeat types, we subcloned and end-sequenced 19 of the pool of 21 existing 2-kb DraIII higher-order repeats of this PAC. In agreement with expected duplicate cloning, 4 pairs of identical end sequences were found. Ignoring single variants, the 15 nonidentical DraIII higher-order repeats showed 12 distinct combinations of common variants (see Supplementary Figs. 2 and 3; available online at http://www.genome.org). Thus, the variants from clustered higher-order repeats of this homogeneous PAC also occur rather interindependently, indicating a lack of haplotypes even within the narrow regions of clusters, reminiscent of sequence conversion. Assuming that clustering would be a general feature of variant spread, clusters of different variants would be expected to overlap within given array subregions. This could lead to local co-occurrence of certain variants, independently of their distance within the higher-order repeats; however, on the single array and population level, individual variants would spread independently, as has been found within and between the 5 PAC samples. For example, see Figure 1: Whereas variants 22 and 231–233 suggest dependence within and between PACs A8 and A10, the same variants suggest independence between PACs A7 and A8/A10, as is the case for variants 1821 and 1822 between PACs B11 and A10.
Intrachromosome Homogenization and Interchromosome Fixation
Sequence conversion can take place within and between chromosomes. To construct an interchromosome mechanism leading to regular clusters, some force would be required to repeatedly target a particular variant to the same locus, regardless of differences in the overall structure and size of the interacting arrays. Dramatically reduced conversion efficiency is known from the double-strand-break repair pathway (Szostak et al. 1983; for review on recombination, see Bollag et al. 1989) in human cells if divergence exceeds a few percent (Taghian and Nickoloff 1997; Elliott et al. 1998; Johnson and Jasin 2000). To exclude patches of abnormal homology within PAC A7, we subcloned 15 additional BamHI higher-order repeats lacking DraIII, resulting in 21 out of the pool of 40 existing non-DraIII higher-order repeats, of which 18 showed individual end-sequence pairs. Ignoring single variants, the 18 nonidentical higher-order repeats showed 16 distinct combinations of common variants, indicating sequence conversion during formation of non-DraIII clusters, too (see Supplementary Figs. 4 and 5; available online at http://www.genome.org). Pairwise comparison of the 365-bp overlap of the sequenced portions of DraIII and non-DraIII subclones revealed a medium identity of 98.9%. Within the groups of DraIII and non-DraIII sequences, the medium identity was 98.8% (n = 18) and 99.2% (n = 18), respectively, indicating normal homogeneity throughout DraIII and non-DraIII clusters. Moreover, the maximum divergence between any two of the 365-bp sequences of PAC A7 was 2.7%, which was found within the DraIII group, whereas between DraIII and non-DraIII sequences, the maximum divergence did not exceed 2.2%. In addition to the high sequence homogeneity, the uniform 2-kb ladder of the BamHI higher-order repeats (Fig. 2, top panel, lanes l,m) indicates perfect structural homogeneity throughout DraIII and non-DraIII higher-order repeats in PAC A7. Interestingly, the frequency of rare variants within the compared 365-bp region appeared slightly higher within DraIII clusters than within non-DraIII clusters (0.14% and 0.03% respectively), possibly indicating more recent homogenization of non-DraIII repeat sections within this array segment. For comparison, the 365-bp region showed a frequency of rare variants of 0.03%–0.45% in the other PAC samples (n = 8–10). Taken together, the data strongly contradict recurrent, targeted interarray spread, indicating that clustering is the result of a localized, intrachromosomal process (including sister chromatids).
DISCUSSION
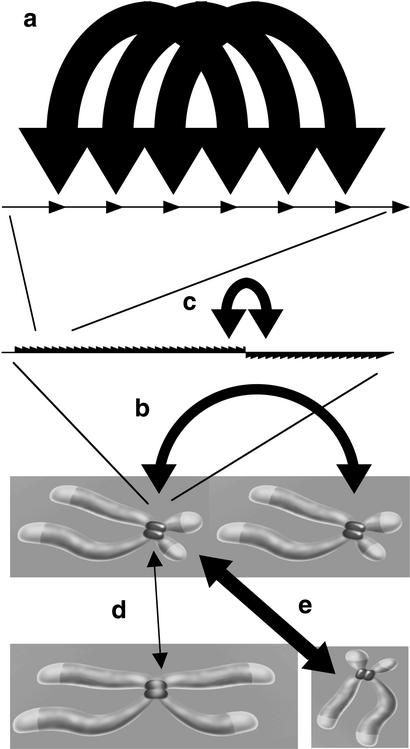
In this paper we characterize the distribution of nucleotide variants within large stretches of highly homogeneous α-satellite DNA. In contrast to many studies of satellite DNA, which included variants belonging to several categories of transition states between and within subsets of α-satellite sequences (Waye and Willard 1986; Warburton and Willard 1990, 1995), this study concentrates on variants, which presently are in transition within a single homogeneous array. This led to the discovery of regularly clustering variants within otherwise highly homogeneous array segments without evidence for haplotypic repeats. This highly nonrandom pattern needs to be explained. Clearly, intrachromosome clustering cannot explain fixation, which, on the other hand, is very efficient between arrays of the same type, as compared with the lower extent between a number of closely related array types with distinct higher-order repeat structure (Warburton and Willard 1995). However, any proposed fixation mechanism continuously contributing to the concerted evolution (nontargeted interchromosome exchange) would either disintegrate regular clusters or lead to haplotypes (unequal crossing over between homologs, rolling circle amplification, and transposition). Because variants in this study are presently in a fixed transition state and therefore have been exposed to all the ongoing molecular drive mechanisms, we conclude that the clustering mechanism must be fast, easily overriding the mechanisms causing fixation (Fig. 3a,b). This is different from the fast exchange mechanism of Warburton and Willard (1995) based on the fact that intrachromosome spread of some variants can precede the complete process of fixation or occurrence in a distinct, related array type (Fig. 3b,c). Furthermore, we assume that the clusters reflect the fastest ongoing mechanism (putative occasional mass amplifications excluded), and not a complicated mixture of similarly fast mechanisms of which some would homogenize and others degrade, but all would somehow contribute to the same nonrandom patterns. Thus, the definition of “fast” is a direct consequence of observing clusters that withstand fixation, based on a mechanistic view and not on evolutionary reasoning. Analysis of two levels of distance between higher-order repeats from 5 unmapped PACs representing medium and large distances (2–160 kb and up to >0.26 Mb), and the higher-order repeats belonging to clusters that represent short distances of <30 kb (including <12-kb HindIII higher-order repeat hexamers), revealed largely interindependent variant occurrence. Particularly within the most homogeneous regions, no evidence for the present spread of major haplotypic higher-order repeats has been found. These results are fully compatible with and typical for simple sequence conversions. It has to be remarked that this fast conversion mechanism does not necessarily have an important influence on the concerted evolution in general, which also includes interactions between multiple distinct, closely related α-satellite families on single chromosomes (Fig. 3c), loosely related array types on different chromosomes (Fig. 3d), virtually identical array types on different chromosomes (Fig. 3e), and that of diverged monomers within higher-order repeats. Even other repetitive sequence elements within the genome can have profound influences on the long-term α-satellite evolution, for example, if high copy number outweighs poor homology (allowing exchange) or if transposition into a homogeneous array impedes or redirects other mechanisms. Nevertheless, it would be bewildering if the fast, intrachromosomal conversion mechanism described here would not at least indirectly serve the concerted evolution, not only by erasing de novo mutations (homogenization), but also by increasing the probability of a variant entering the next round of a fixation-driving mechanism.
Figure 3.
Schematic diagram of several possible trajectories of molecular drive. (a) Localized clustering of variants within neighboring higher-order repeats within highly homogeneous array segments must be the fastest ongoing molecular drive mechanism. The (efficient) interchromosome process, causing fixation in the population (b), is too slow to disintegrate the clusters. Intrachromosomal exchange between distinct array types is generally inefficient, allowing complete fixation of two or more homogeneous array types at a centromere. Sometimes, if closely related array types share sufficient sequence homology, intrachromosomal interarray exchange can be comparably fast evolutionarily (c), preceding complete fixation (Warburton and Willard 1995). Exchange between highly homogeneous (>97%), structurally indistinguishable array types on different chromosomes can be very efficient, as known from pairs of acrocentrics (Greig et al. 1993), or Chromosomes 5 and 19 (e). Any rare exchange between distinct arrays (d) is irrelevant for our mechanistic point of view, which concentrates on the ongoing homogenization within a single homogeneous array, but might well be relevant for the concerted evolution of all the loosely related α-satellite DNA families in general.
In theory, the same patchwork-like snapshot of the clusters could also be the result of unequal crossing over admixing two evolutionarily slightly distinct array types in the population, one containing the first morph of the variant, and one the second. Moreover, unequal crossing over between sister chromatids within a single homogeneous array, at least if very short tracts were repeatedly exchanged, could also result in clustered variants. However, both possibilities are not well supported from our data. Classically, in order to drive homogenization and not only admixture of (old) DNA segments, unequal crossing over requires amplification and loss of repeats to overcome random mutagenesis (Smith 1976). This would be accompanied by an uneven distribution of variants within the array and within the population, as described for some variants of Chromosome 17 (Warburton and Willard 1995). However, despite acknowledged array size polymorphisms in the population (Mahtani and Willard 1990), the set of variants in this study, which have been isolated because they were in transition within one single array, also show fixation. Moreover, 5 out of 5 variants of this set showed a relatively fixed variant ratio, indicating a rather uniform distribution in the population. Fine mapping of such a variant showed that the actual spread is highly nonrandom, indicating that fixed variants are still subject to clustering. If the clustering mechanism were not on average several times faster than any interchromosome exchange mechanism (Fig. 3a,b), variant distribution would become random.
The data indicate that a fast sequence-conversion process significantly contributes to the ongoing concerted evolution within a homogeneous α-satellite DNA array. Although proposed in the past (Dover 1982; Durfy and Willard 1989), no strong data have yet been presented supporting a significant role of sequence conversion within homogeneous satellite DNA. Moreover, genetics textbooks, in order to explain the concerted evolution of tandem repeat arrays, often depict the unequal crossing-over model of Smith (1976), saying that a process solely based on random mutagenesis, unequal crossing over, and expansion is sufficient. After 25 years, the model is almost perceived as valid, and sequence conversion is usually only called in to explain the concerted evolution of interspersed repeats, such as alu elements. Some variants within array types of Chromosome 17 indicated exchanges taking place relatively faster within the array than throughout the population or between two distinct, but closely related array types of the same chromosome (Warburton and Willard 1995). However, without fine mapping of variants within homogeneous array segments, it was impossible to relate the spread directly to a spatially confined (exclusively intrachromosomal) conversion mechanism. Instead, a number of analyses supported the hypothesis of Smith. If sequence conversion would take place without significantly changing the predicted sequence outcome, the crossing-over fixation model would stand without the need to emphasize the role of sequence conversion. However, there is no reason that the present variants within segments of highest homogeneity analyzed in this study should not adhere to the main predictions of the unequal crossing-over model, such as the spread of haplotypes caused by homogenization by amplification, which would be expected to be accompanied by uneven transition states in the population. We found (1) exchanges of common variants within a duplicated HindIII cluster without evidence for amplification because the overall structure appeared stable; (2) a lack of amplified haplotypes within the few groups of adjacent 2-kb higher-order repeats derived from regular variant clusters; (3) fixed and even transition states (variant ratios) for 5 out of 5 restriction variants isolated from a single array; (4) a number of differing variant combinations close to the possible maximum, indicating a general lack of major haplotypes; and (5) grossly random occurrence of similar variant combinations. Interestingly, if looking at the variation between evolutionarily distinct array types (rarely interacting), the majority of present differences might be attributable to former variants, which have already approached a uniform state within their array. The variation between two such (for whatever reason) rarely interacting arrays would confirm the haplotype prediction of the unequal crossing-over model, even if the actual spread involved sequence conversion. This might in some cases provide an explanation for the apparent discrepancy between the present patterns within homogeneous array segments and the acknowledged variation between distinct array types or at diverged borders.
Considering the paucity of de novo mutations and the general lack of haplotypic higher-order repeats, which hampers ordering and assembly of unmapped sequence reads, the data and large scale mapping technique might be useful to assist in PAC- and BAC-based contig building to map and sequence through centromeres. Contiguous sequencing of homogeneous arrays would provide a more complete snapshot of ongoing turnover, and in addition would allow resequencing of centromeres from human artificial chromosomes, in order to directly measure absolute turnover rates. This might give new insight into molecular drive mechanisms (Dover et al. 1982; Dover 2000a,b) and their impact on genome variability and evolution.
METHODS
Subcloning and Sequencing
Restriction digestion was carried out inside low-melting-point agarose. Plug DNA was run on an agarose gel; 2-kb BamHI higher-order repeats of PACs were electroeluted from a gel slice and cloned into plasmid pBS-II-SK (Stratagene). HindIII fragments of PAC B11 were subcloned into the HindIII site. To subclone DraIII higher-order repeats, the DraIII restriction site in the pBS-II-SK backbone was eliminated using S1 nuclease and blunt-end ligation. To introduce a DraIII cloning site, the synthetic oligonucleotide (5′-GATCGCCCGGCACTGGGT GATTCG-3′ annealed to 5′-AGCTCGAATCACCCAGTGCC GGGC-3′) was cloned into HindIII and BamHI of pBS-II-SK, maintaining white–blue selectability. Because all 34 BamHI higher-order repeats containing a DraIII site showed the same overhang, we used only this overhang in the synthetic site. Insert and plasmid size was checked using BamHI and EcoRI to exclude double insert clones. Primers X-3A and X-4A were used for a 0.5-kb PCR specific for the DXZ1 array. To increase representation, a large input of genomic DNA (0.5–1 μg per 50-μL reaction) and only 15–20 cycles were used (Warburton and Willard 1992). To amplify the corresponding gorilla X-chromosome array, the human primer X-4A was replaced by gorilla primer X-4Ag 5′-TGTGAAGATAAAGCCTTTTCC-3′, derived from GGSATG (acc. no. X56887). To sufficiently amplify gorilla DNA for subcloning, 25–30 cycles were required. PCR products were subcloned into pGEM-TA (Promega), and clones were size checked by PCR using primers T7 and SP6.
Ends of subclones were cycle-sequenced using primers T7, T3, or SP6 on plasmid DNA (QIAGEN spin columns) and run on ABI sequencers. Individual electropherograms were hand-checked, and low-quality parts were cut off. Of the total of 180 sequences, 18 were resequenced with end primers or internal primers X-4A (n = 6) or X-3A (n = 7), owing to low quality and small G peaks of the G/C variant at position 1617. Finally, sequences were rechecked by aligning electropherograms. Approximately 6.4 kb of the sequences were derived from PCR subclones that had been sequenced on both strands. In this case, alignment of validated single strands did not reveal any undetected mistake, indicating that single-strand end sequencing was accurate. Excluding sequences of potential doublet clones and sections sequenced twice (total 23.677 kb), a total of 95.817 kb has been analyzed by pairwise comparison (see Supplementary Material; available online at http://www.genome.org). For illustration, four color sequence alignments (unordered satellite tartans) comprising the sequenced sections of the 5 PACs were assembled (see Supplementary Figs. 1, 2, 4; available online at http://www.genome.org). To check whether common variants belong to certain haplotypes, color-based variant boxes were created. The variant boxes, only depicting the selection of common variants, were either used to compare randomly subcloned repeats from different PACs (Fig. 2), or to compare repeats subcloned from two or three narrow groups of higher-order repeats derived from DraIII or non-DraIII clusters (Supplementary Figs. 3, 5; available online at http://www.genome.org). Direct whole PAC end-sequencing was carried out on circular PAC DNA purified from an agarose plug (QIAGEN), on electroeluted NotI-linearized insert DNA containing SP6 and T7 priming sites at its ends, and from derived subclones in a telomerized PAC vector. All procedures gave consistent results. For end-sequencing the 60-kb SalI subclone of PAC B11, primer P86, 5′-TGCGATCTGCCGTTTCGA-3′, has been designed.
Large-Scale Partial Restriction Mapping
We previously have developed a method to isolate intact PAC DNA (Schindelhauer and Cooke 1997) that facilitates large-scale partial restriction mapping. We linearized 2–5 μg of supercoiled PAC using PacI (NEBiolabs), electroeluted (Biotrap, Schleicher Schüll) from a pulsed-field gel slice (without UV), and subdivided using wide bore tips. Partial digestions were carried out using 0, 0.1, 0.4, 0.8, 1.6, and 3 U for 1 min (2 min for HindIII). Samples of 20 μL were loaded on a pulsed-field gel. For end-fragment detection, PAC vector probes from both PacI ends were PCR-amplified using primers VP1f: 5′-CGATGTCAATTCAGAACATCATTG-3′, and VP1r: 5′-TAG CCCGTCTAACACCTATTGC-3′, and primers VP2f: 5′-TCG CTAAAGCCTGTGGTTTCC-3′, and VP2r: 5′-ATGTATGCGT AGATGCTTGTAC-3′ (from pCYPAC2N acc. no. U09128) and random primed labeling with P-32 dCTP (Amersham) and Klenow polymerase (Roche). All hybridizations were carried out at 65°C in the presence of dextran sulfate followed by stringent washing at 60°C in 0.1× SSC, 0.1% SDS.
WEB SITE REFERENCES
http://www.chori.org/bacpac/humalmaleall.htm; RPCI-1 library, Pieter de Jong, Children's Hospital Oakland Research Institute.
Acknowledgments
The underlying work has been carried out over a period of several years in laboratories of the Children's Hospital, LMU Munich (Thomas Meitinger), at the HGU, MRC, Edinburgh (Howard Cooke), and at the Institute of Human Genetics, TU, Munich. We thank all lab members for fruitful discussions. We thank Christian Roos at the German Primate Center, Göttingen, for gorilla samples; Peter Warburton for probe X5; and Wilfried Berning, Institute of Radiology, Münster, for support with figure processing. For helpful discussions, we thank G. Dover, Leicester, and W. Stephan, Munich. Special thanks to Elizabeth Schroeder-Reiter, Botanical Institute, Munich, for review of the manuscript, and Thomas Meitinger for long-lasting backup and support. The work was financed mainly from DFG grants on human artificial chromosomes to D.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dirk@pedgen.med.uni-muenchen.de; FAX 49 89 5160 4780.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.451502.
REFERENCES
- Alexandrov IA, Mitkevich SP, Yurov YB. The phylogeny of human chromosome specific α satellites. Chromosoma. 1988;96:443–453. doi: 10.1007/BF00303039. [DOI] [PubMed] [Google Scholar]
- Alexandrov IA, Medvedev LI, Mashkova TD, Kisselev LL, Romanova LY, Yurov YB. Definition of a new α satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res. 1993;21:2209–2215. doi: 10.1093/nar/21.9.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry AE, Howman EV, Cancilla MR, Saffery R, Choo KHA. Sequence analysis of an 80 kb human neocentromere. Hum Mol Genet. 1999;8:217–227. doi: 10.1093/hmg/8.2.217. [DOI] [PubMed] [Google Scholar]
- Bollag RJ, Waldman AS, Liskay RM. Homologous recombination in mammalian cells. Annu Rev Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Choo KH. Centromere DNA dynamics: Latent centromeres and neocentromere formation. Am J Hum Genet. 1997a;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— . The centromere. Oxford, UK: Oxford University press; 1997b. pp. 98–108. [Google Scholar]
- ————— Why is the centromere so cold? Genome Res. 1998;8:81–82. doi: 10.1101/gr.8.2.81. [DOI] [PubMed] [Google Scholar]
- Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of α satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991;19:1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. New goals for the U.S. Human Genome Project: 1998–2003. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: A cohesive mode of species evolution. Nature. 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- ————— How genomic and developmental dynamics affect evolutionary processes. BioEssays. 2000a;22:1153–1159. doi: 10.1002/1521-1878(200012)22:12<1153::AID-BIES13>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- ————— . Dear Mr Darwin; Letters on the evolution of life and human behaviour. London, England: Weidenfeld and Nicolson; 2000b. [Google Scholar]
- Dover G, Brown S, Coen E, Dallas J, Strachan T, Trick M. The dynamics of genome evolution and species differentiation. In: Dover GA, Flavell RB, editors. Genome evolution. London: Academic Press; 1982. pp. 343–372. [Google Scholar]
- Durfy SJ, Willard HF. Molecular analysis of a polymorphic domain of α satellite from the human X chromosome. Am J Hum Genet. 1987;41:391–401. [PMC free article] [PubMed] [Google Scholar]
- ————— Patterns of intra- and interarray sequence variation in α satellite from the human X chromosome: Evidence for short-range homogenization of tandemly repeated DNA sequences. Genomics. 1989;5:810–821. doi: 10.1016/0888-7543(89)90123-7. [DOI] [PubMed] [Google Scholar]
- ————— Concerted evolution of primate α satellite DNA. Evidence for an ancestral sequence shared by gorilla and human X chromosome α satellite. J Mol Biol. 1990;216:555–566. doi: 10.1016/0022-2836(90)90383-W. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-α-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE. Repetitive conundrums of centromere structure and function. Hum Mol Genet. 1999;8:151–155. doi: 10.1093/hmg/8.2.151. [DOI] [PubMed] [Google Scholar]
- ————— Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- Elder JF, Jr, Turner BJ. Concerted evolution of repetitive DNA sequences in eukaryotes. Quart Rev Biol. 1995;70:297–320. doi: 10.1086/419073. [DOI] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig GM, Warburton PE, Willard HF. Organization and evolution of an α satellite DNA subset shared by human chromosomes 13 and 21. J Mol Evol. 1993;37:464–475. doi: 10.1007/BF00160427. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Horvath JE, Viggiano L, Loftus BJ, Adams MD, Archidiacono N, Rocchi M, Eichler EE. Molecular structure and evolution of an α satellite/non-α satellite junction at 16p11. Hum Mol Genet. 2000;9:113–123. doi: 10.1093/hmg/9.1.113. [DOI] [PubMed] [Google Scholar]
- Horz W, Zachau HG. Characterization of distinct segments in mouse satellite DNA by restriction nucleases. Eur J Biochem. 1977;73:383–392. doi: 10.1111/j.1432-1033.1977.tb11329.x. [DOI] [PubMed] [Google Scholar]
- Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen AL, Bostock CJ, Bak AL. Chromosome-specific subfamilies within human alphoid repetitive DNA. J Mol Biol. 1986;187:185–196. doi: 10.1016/0022-2836(86)90227-5. [DOI] [PubMed] [Google Scholar]
- Jorgensen AL, Laursen HB, Jones C, Bak AL. Evolutionarily different alphoid repeat DNA on homologous chromosomes in human and chimpanzee. Proc Natl Acad Sci. 1992;89:3310–3314. doi: 10.1073/pnas.89.8.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Mahtani MM, Willard HF. Pulsed-field gel analysis of α-satellite DNA at the human X chromosome centromere: High-frequency polymorphisms and array size estimate. Genomics. 1990;7:607–613. doi: 10.1016/0888-7543(90)90206-a. [DOI] [PubMed] [Google Scholar]
- Manuelidis L, Wu JC. Homology between human and simian repeated DNA. Nature. 1978;276:92–94. doi: 10.1038/276092a0. [DOI] [PubMed] [Google Scholar]
- Mashkova T, Oparina N, Alexandrov I, Zinovieva O, Marusina A, Yurov Y, Lacroix MH, Kisselev L. Unequal cross-over is involved in human α satellite DNA rearrangements on a border of the satellite domain. FEBS Lett. 1998;441:451–457. doi: 10.1016/s0014-5793(98)01600-7. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil DL, Villasante A, Fisher RB, Vetrie D, Cox B, Tyler-Smith C. Structural instability of human tandemly repeated DNA sequences cloned in yeast artificial chromosome vectors. Nucleic Acids Res. 1990;18:1421–1428. doi: 10.1093/nar/18.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelhauer D, Cooke HJ. Efficient combination of large DNA in vitro: In gel site specific recombination (IGSSR) of PAC fragments containing α satellite DNA and the human HPRT gene locus. Nucleic Acids Res. 1997;25:2241–2243. doi: 10.1093/nar/25.11.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelhauer D, Hellebrand H, Grimm L, Bader I, Meitinger T, Wehnert M, Ross M, Meindl A. Long-range map of a 3.5-Mb region in Xp11.23-22 with a sequence-ready map from a 1.1-Mb gene-rich interval. Genome Res. 1996;6:1056–1069. doi: 10.1101/gr.6.11.1056. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Southern EM. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975;94:51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: Cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Taghian DG, Nickoloff JA. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Sylvester JE, Gonzalez IL, Costanzi CC, Gillespie D. Definition of a second dimeric subfamily of human α satellite DNA. Nucleic Acids Res. 1989;17:2769–2782. doi: 10.1093/nar/17.7.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C, Floridia G. Many paths to the top of the mountain: Diverse evolutionary solutions to centromere structure. Cell. 2000;102:5–8. doi: 10.1016/s0092-8674(00)00004-0. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Gimelli G, Giglio S, Floridia G, Pandya A, Terzoli G, Warburton PE, Earnshaw WC, Zuffardi O. Transmission of a fully functional human neocentromere through three generations. Am J Hum Genet. 1999;64:1440–1444. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Willard HF. Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of α-satellite DNA. J Mol Biol. 1990;216:3–16. doi: 10.1016/s0022-2836(05)80056-7. [DOI] [PubMed] [Google Scholar]
- ————— PCR amplification of tandemly repeated DNA: Analysis of intra- and interchromosomal sequence variation and homologous unequal crossing-over in human α satellite DNA. Nucleic Acids Res. 1992;20:6033–6042. doi: 10.1093/nar/20.22.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Interhomologue sequence variation of α satellite DNA from human chromosome 17: Evidence for concerted evolution along haplotypic lineages. J Mol Evol. 1995;41:1006–1015. doi: 10.1007/BF00173182. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Chromosome-specific α satellite DNA: Nucleotide sequence analysis of the 2.0 kilobase pair repeat from the human X chromosome. Nucleic Acids Res. 1985;13:2731–2743. doi: 10.1093/nar/13.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Structure, organization, and sequence of α satellite DNA from human chromosome 17: Evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol. 1986;6:3156–3165. doi: 10.1128/mcb.6.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick R, Willard HF. Long-range organization of tandem arrays of α satellite DNA at the centromeres of human chromosomes: High-frequency array-length polymorphism and meiotic stability. Proc Natl Acad Sci. 1989;86:9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF, Smith KD, Sutherland J. Isolation and characterization of a major tandem repeat family from the human X chromosome. Nucleic Acids Res. 1983;11:2017–2033. doi: 10.1093/nar/11.7.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TP, Hansen SK, Oishi KK, Ryder OA, Hamkalo BA. Characterization of a cloned repetitive DNA sequence concentrated on the human X chromosome. Proc Natl Acad Sci. 1982;79:6593–6597. doi: 10.1073/pnas.79.21.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]