Abstract
Many chromosome regions in the human genome exist in four similar copies, suggesting that the entire genome was duplicated twice in early vertebrate evolution, a concept called the 2R hypothesis. Forty-two gene families on the four Hox-bearing chromosomes were recently analyzed by others, and 32 of these were reported to have evolutionary histories incompatible with duplications concomitant with the Hox clusters, thereby contradicting the 2R hypothesis. However, we show here that nine of the families have probably been translocated to the Hox-bearing chromosomes more recently, and that three of these belong to other chromosome quartets where they actually support the 2R hypothesis. We consider 13 families too complex to shed light on the chromosome duplication hypothesis. Among the remaining 20 families, 14 display phylogenies that support or are at least consistent with the Hox-cluster duplications. Only six families seem to have other phylogenies, but these trees are highly uncertain due to shortage of sequence information. We conclude that all relevant and analyzable families support or are consistent with block/chromosome duplications and that none clearly contradicts the 2R hypothesis.
The hypothesis that chromosome duplications, or even genome doublings, have contributed to the expansion of the vertebrate genome has been debated intensely during the past few years (Pennisi 2001). A recent article in Genome Research by Hughes et al. (2001) aimed to test the chromosome/genome duplication hypothesis by studying gene families with members on two or more of the human Hox-bearing chromosomes 2, 7, 12, and 17 to investigate whether the duplications may have occurred concomitantly. Hughes et al. studied 42 gene families and reported that 32 of these provided evidence against simultaneous duplication with the Hox clusters, as based on phylogenetic trees and deduced time points for gene duplications. They concluded in their article title that “Ancient genome duplications did not structure the human Hox-bearing chromosomes.” A commentary in the same issue stated that the authors “scrutinize the hypothesis with a series of the most rigorous tests to date,” and that these were “even more sophisticated” than previous tests (Makalowski 2001). However, close inspection of these 42 gene families reveals that most have complications that invalidate the authors' conclusion and that many of the families actually support the chromosome duplication hypothesis.
A group of similar-looking chromosome segments, located on different chromosomes, has been given the term paralogon (Coulier et al. 2000). Such sets of paralogous regions are assumed to have arisen by duplications of an intact chromosome segment, so-called block duplications. If many block duplications occurred simultaneously, they are more likely to have resulted from complete chromosome duplications or even whole genome doubling, that is, tetraploidization. The hypothesis that two rounds of tetraploidization have occurred in early vertebrate evolution is called the 2R hypothesis (Hughes 1999).
Hughes et al. (2001) based their analyses of the 2R hypothesis on the assumptions (Hughes 1999) that gene families support chromosome/genome duplication only if: (1) the vertebrate members of the gene family can be shown to have duplicated within the vertebrate lineage, and (2) the gene family phylogeny shows double-forked tree topology, that is, so-called 2 + 2 or (AB)(CD) topology. However, the first assumption is oversimplified, and in contrast to what Hughes et al. argue, the 2R hypothesis is indeed compatible with additional duplications either before or after the proposed chromosome duplications (Holland 2002). The second assumption is incomplete, as it requires that the members of each gene family must have similar evolutionary rates. Furthermore, the 2 + 2 topology also requires that sufficient time has elapsed between the chromosome/genome duplications to allow the duplication events to be resolved, but available data suggest that the two proposed tetraploidizations were close in time at the origin of vertebrates (Furlong and Holland 2002).
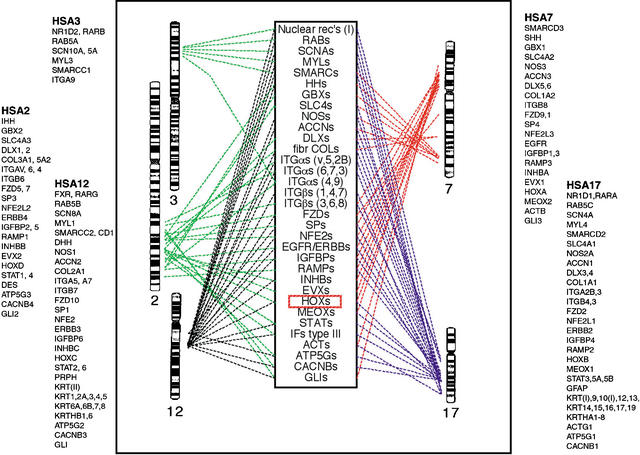
Another objection of great importance is that it cannot generally be assumed that gene families present today on the human Hox-bearing chromosomes have remained linked since the duplications of the Hox clusters, because many chromosomal rearrangements are known to have taken place (Chowdhary et al. 1998; Murphy et al. 2001; Gregory et al. 2002). This is particularly clear for two of the four human Hox-bearing chromosomes that differ from those of the mammalian ancestor. Hsa2 is the result of a fusion of two different chromosomes in the primate lineage, and Hsa12 was rearranged during primate evolution (Murphy et al. 2001). Interestingly, part of Hsa2p belongs to a different paralogon than that consisting of the four extended Hox clusters. In addition, parts of 12p belong to two non-Hox paralogons, as do several genes on 17p, probably due to rearrangements that took place before the origin of mammals. Similarly, Hsa7 has genes that seem to belong to a paralogon different from the Hox paralogon. The Hox-chromosome duplications are postulated to have taken place some 500 Myr ago, and many rearrangements may have occurred since then, as shown by comparative chromosome maps in chicken (Groenen et al. 2000), zebrafish (Postlethwait et al. 2000; Woods et al. 2000), and pufferfish (Aparicio et al. 2002). Thus, the mere presence of a gene family on two of the four Hox chromosomes does not mean that this family can be used to test whether the entire human Hox chromosomes arose by chromosome duplication. As we show in Figure 1, the regions of the four human Hox chromosomes that carry genes with ancient linkage to the Hox clusters may actually be quite limited, particularly for Hsa 12 and 17, but also for Hsa2, where only the q arm seems to be involved. These aspects were not considered in the above-mentioned article by Hughes et al.
Figure 1.
Gene families with members on the human Hox-bearing chromosomes. Note that many of these gene families cluster to restricted regions of the chromosomes. A subset of genes are located on chromosome 3, probably due to a translocation. ACCN, amiloride-sensitive cation channels; ACT, actins; ATP5G, ATPase; CACNBs, calcium channel β subunits; fibr COL, fibrillar associated collagens; DLX, distal-less homeo box; EGFR/ERBB, epidermal growth factor receptor/erythroblastoma; EVX, even-skipped homeo box; FZD, frizzled; GBX, gastrulation brain homeo box; GLI, glioma-associated oncogene homolog belonging to Krüppel family; HH, hedgehog; HOX, homeo box (antennapedia-like); IFs type III, intermediate filaments type III; IGFBP, insulin-like growth factor-binding protein; INHB, inhibins; ITGA, integrin α chains; ITGB, integrin β chains; MEOX, mesenchyme homeo box; MYL, myosin light chains; NFE2, nuclear factor erythroid; NOS, nitric oxide synthase; Nuclear rec's, nuclear hormone receptors; RAB, member of Ras-oncogene family; RAMP, receptor activity-modifying protein; SCNA, sodium channel α subunits; SLC4, solute carrier family 4 (anion exchangers); SMARC, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin; SP, transcription factor Sp; STAT, signal transducers and activators of transcription.
In addition to the formal complications mentioned above, the phylogenetic analyses performed by Hughes et al. were based on sequence matrices with mammalian overrepresentation and very few sequences from other classes of vertebrates. For some gene families, mammals were the only vertebrate representatives. Importantly, very little information was used from those gnathosomes that are most distantly related to mammals, namely actinopterygian fishes and cartilaginous fishes. This is particularly regrettable since these classes diverged shortly after the Hox-cluster duplications. Furthermore, molecular phylogeny as a tool to test relatedness is complicated by the fact that several of the sequences used are from species that have undergone additional tetraploidizations. A basal tetraploidization took place in teleost fishes (Taylor et al. 2001) and was followed by more recent independent tetraploidizations in salmonids and goldfish. Xenopus laevis has undergone an independent tetraploidization. After duplications, the resulting gene duplicates seem to have a higher evolutionary rate (Iwabe et al. 1996; Nembaware et al. 2002), and in many instances the daughter genes seem to have evolved at different rates (Ohta 1991; Larhammar and Risinger 1994; Cerdá-Reverter and Larhammar 2000; Málaga-Trillo and Meyer 2001; Van de Peer et al. 2001; Conlon 2002), perhaps as a result of subfunctionalization (Force et al. 1999), although the generality of these observations is questioned by some reports (Hughes and Hughes 1993; Robinson-Rechavi and Laudet 2001; Wallis 2001). Indeed, high bootstrap values have been observed for false phylogenies for paralogous genes and were therefore suggested not to be a good indicator of the validity of the analysis (Abi-Rached et al. 2002). Considering these issues, tree topology information should be used with great caution when testing hypotheses such as the 2R hypothesis.
Here we reanalyze each of the families studied by Hughes et al. and conclude that as many as half of the gene families actually support or are at least consistent with duplications concomitant with the Hox clusters, whereas many others are irrelevant (as they do not belong to the Hox paralogon) or unclear regarding this hypothesis. It should be noted that some of the reinterpretations described here were possible thanks to sequence information that became available after Hughes et al. performed their analyses. The figures and tables in the paper by Hughes et al. are referred to by the abbreviations H-Fig and H-Table. We have used the gene abbreviations used in OMIM and show those used by Hughes et al. in parentheses whenever different.
RESULTS
Acetylcholine Receptor—ACHR
This gene family was found to have members on only two of the four Hox chromosomes. Although included in H-Table 1, it is suprisingly not dealt with in the paper. Two genes are on Hsa2 on the same arm as the HoxC cluster, but both of the genes on Hsa17 are on the p arm, whereas the extended HoxB cluster is on the q arm (Fig. 1). It is possible that this could be due to a pericentric inversion, but in the absence of data from other vertebrates supporting linkage to HoxB, it is unclear whether the ACHR gene family has anything to do with the Hox clusters. We conclude that this gene family is not relevant for testing the hypothesis of duplication concomitant with the Hox cluster.
Acetyl-coA Carboxylase
This gene family too was found to have members on only two of the four Hox chromosomes, Hsa12 and Hsa17, and the family did not evolve in a clock-like manner. Thus, we agree with Hughes et al. that it is uninformative.
Actins—ACT
Functionally, actins are classified as cytoskeletal, sarcomeric, and smooth muscle. Chromosomes 7 and 17 carry the cytoskeletal actin genes ACTB and ACTG1 (ACTG), and these may have arisen as a result of chromosome duplication. The divergence time estimated in H-Fig. 3, 226 Myr, does not take into account that ACTB has been found in chicken, goose, frog, and pufferfish, and what appears to be ACTG1 has been described in chicken and Xenopus laevis (P53505). Although the true subtype identities of the two latter sequences are still uncertain, it appears that the duplication took place well before the origin of amphibians some 350 Myr ago.
The actin gene ACTG2 (ACTH, P12718) encodes a smooth muscle actin and is on the wrong arm of Hsa2, the p arm, and therefore does not seem to be part of the extended Hox cluster. Interestingly, ACTG2 together with ACTA2 (ACTSA) on Hsa10 are located in a separate paralogon, namely Hsa4, 5, 8(2), 10(13) (F. Hallböök, L.-G. Lundin, and D. Larhammar, in prep.).
Two additional actin genes, ACTA1 and ACTC, were included in the phylogenetic analyses by Hughes et al. These are located on 1q and 15q in regions that share several other gene families, suggesting that they too arose by chromosome duplication. These chromosome segments seem to belong to the paralogon consisting of Hsa1, 11, 12 (14, 15), 19, where Hsa14 and 15 carry members of some gene families that appear to have been translocated from Hsa12 (Popovici et al. 2001; F. Hallböök, L.-G. Lundin, and D. Larhammar, in prep.).
Thus, all three pairs of related actin genes are consistent with chromosome duplications, and the ACTB and ACTG1 genes seem to agree with the Hox duplications.
Acyl-coA Dehydrogenase—ACAD
At least seven ACAD genes are found in human. Four genes are located on Hox chromosomes (two on Hsa17), but two of these genes are located outside of the Hox regions: ACADS is in 12q24.31 (HoxD and most of the linked genes are in 12q13), and ACADVL is in 17p (HoxB is on the q arm). ACADL in 2q34 and ACOX (COA-OXP) in 17q25 seem to be near Hox clusters, but these two genes are the most distantly related in the whole ACAD tree analyzed by Hughes et al., which includes sequences from Caenorhabditis elegans and several prokaryotes. Thus, ACADL and ACOX probably arose long before the Hox duplications. In consideration of the large number of members in this gene family, the possibility that two members have become independently associated with the Hox regions cannot be ruled out. Until chromosome mapping data from other vertebrates are available, the evolutionary history of this gene family remains unclear, and we conclude in contrast to Hughes et al. that it is not informative.
ADP-Ribosylation Factors—ARF
These genes comprise a large family with at least 13 members in mammals. Phylogenetic analysis shows that the duplications of the genes found on Hsa 7, 12, and 17 took place before the divergence of protostomes and deuterostomes (H-Fig. 1). Two genes located on Hsa2 probably arose before the protostome-deuterostome divergence (Jacobs et al. 1999). The gene duplications that seem to have occurred in the deuterostome lineage do not fit with the Hox chromosomes. Thus, if additional duplicates arose in the chromosome duplications, these seem to have been lost, thereby making it difficult to evaluate this large and ancient gene family with respect to Hox duplications.
Anion Exchanger—SLC4A (AE)
Three members of the anion exchanger family, called SLC4A for solute carrier 4A, are located in chromosomal regions that are fully consistent with the chromosome duplication hypothesis, but the tree topology calculated by Hughes et al. disagreed with that of the Hox clusters, although it was consistent with three other gene families (H-Fig. 5). However, very few taxa are available for each of the three genes, and six of the nine sequences are from mammals and two are from chicken, making the basal branching order uncertain. Therefore, we disagree with the conclusion by Hughes et al. that this tree provides evidence against duplication concomitant with the Hox clusters.
Aquaporins—AQP
The aquaporin family has at least ten members in the human genome, but only two Hox-bearing chromosomes are involved. Two family members were mentioned by Hughes et al. (H-Table 1 and H-Fig. 3), namely AQP1 on Hsa7 and AQP2 on Hsa12, although the latter carries four AQP genes. A recent phylogenetic analysis (Zardoya and Villalba 2001) showed that evolutionary rates differ greatly between family members and that only mammalian sequences are known for AQP2. This makes time estimates highly uncertain. Indeed, the tree presented by Zardoya and Villalba (2001) gives a divergence date for AQP1 and AQP2 that seems consistent with vertebrate origins, rather than the 1600 Myr reported by Hughes et al. Thus, in contrast to the conclusion drawn by Hughes et al., this family can hardly be used to investigate the relationships of Hsa7 and Hsa12 until information becomes available from additional species.
Arrestin—ARR
Four family members are known in human, two of which are on Hox chromsomes, namely β-arrestin 2, abbreviated ARRB2 (ARR2) on Hsa17p13 and SAG (S-ARR) on 2q37.1, but the former location is on the opposite arm of Hsa17 compared to the Hox region, suggesting that these genes were not part of the same ancestral chromosomal region. The previously published phylogenetic analysis (Craft and Whitmore 1995) as well as that of Hughes et al. are consistent with gene duplications at the dawn of vertebrate evolution, and invertebrate sequences branch outside the vertebrate subtypes, but it is still unclear whether the localization of the vertebrate genes are consistent with any known paralogon.
Brain Amiloride-Sensitive Sodium Channel—ACCN (BNAC)
The neuronal sodium channel genes on Hsa12 and Hsa17 are consistent with duplication concomitant with the Hox clusters. Naturally, no analysis could be performed with only two family members regarding phylogenetic consistency with the Hox clusters. However, a third member, ACCN3 on Hsa7q36.1, adds further support for duplications concomitant with the Hox-cluster regions.
Cyclin-Dependent Kinases—CDK
Ten human family members were included in the analysis in H-Fig. 1. Five were said in H-Fig. 2 to pertain to the Hox chromosome duplications, but the chromosomal localization of CDK7 is on the wrong arm of Hsa2, outside the extended Hox cluster. Furthermore, in the human genome sequence, CDK7 is found on Hsa5p13.3. The remaining four CDK genes do seem to be associated with the Hox regions; CDK2, CDK3, CDK4, and CDK5. Among these, CDK4 and CDK5 are very distantly related to each other and seem to have originated before the radiation of eukaryotes. The remaining two genes, CDK2 on Hsa12 and CDK3 on Hsa17 are more closely related and may be the result of a chromosome duplication. However, it should be noted that the phylogenetic analysis reveals quite uneven evolutionary rates between the family members [e.g., CDK5 and PCTAIRE-1 (STPK1) compared to PCTAIRE-3 (STPK3)] as well as over time for individual genes (CDK7 and CDK1, the latter called CDKH by Hughes et al.). Taken together, these observations make the duplication-time estimates for CDK3 and CDK4 (H-Fig. 3) questionable and unsuitable for testing the chromosome duplication hypothesis. It is unclear to us why those authors chose to show the CDK3-CDK4 duplication time point in H-Fig. 3 and not that of CDK2 (Hsa12) and CDK3 (Hsa17), which does fall within the Hox duplication time range and would support the 2R hypothesis. In conclusion, we find the CDK family too complex to provide a test of the 2R hypothesis.
Enolase—ENO (ENOL)
Three enolase genes are known in the human genome, two of which were included in this analysis, ENO2 (γ) on 12p13 and ENO3 (β) on 17p13.1. However, both of these genes are on the wrong chromosome arm. They seem to belong to a different paralogon, namely the one involving Hsa1, 3, 12, and 17 (Popovici et al. 2001; F. Hallböök, L.-G. Lundin, and D. Larhammar, in prep.). The duplication time was estimated by Hughes et al. as 382 Myr ago. However, both α-enolase and β-enolase have been discovered in different classes of fishes, thus showing that the gene duplications leading to the three isozymes occurred before the origin of osteichthyes and perhaps even gnathostomes (Tracy and Hedges 2000).
ERBB Receptor Protein-TK—ERBB
The four family members of the ERBB family in human are located in chromosomal regions that are fully consistent with the chromosome duplication hypothesis. According to H-Fig. 1, the origin of two other related genes on Hsa7, EPB4 and MET (list of included sequences kindly provided by A. Hughes), predated the divergence of protostomes and deuterostomes. The ephrin receptors EPHB1–4 form a separate family with members on chromosomes 1 and 3 and thus do not seem to have anything to do with the duplications of the Hox cluster. Likwise, the MET-related gene MST1R (RON) is located on Hsa3, further indicating unrelatedness to the Hox duplications. It is unclear to us why Hughes et al. chose to show this early divergence in H-Fig. 1 rather than the ERBB quadruplication shown in H-Fig. 4c. The latter H-figure showed that the internal relationships of the four ERBB genes disagree with the Hox relationships, suggesting a different order of duplications. However, the ERBB analysis included very few taxa (seven of nine sequences were from mammals) and would thus be unlikely to detect any rate differences between family members or taxa or over time.
Even-Skipped—EVX
Only two EVX genes are known in the human genome, EVX1 on Hsa7 and EVX2 on Hsa2, and they were found not to evolve in a clock-like manner and thus were regarded by Hughes et al. as uninformative. However, their close proximity to the Hox clusters makes them virtually as likely as the members of each Hox cluster to be part of a chromosome region that has been duplicated, as discussed by Pollard and Holland (2000), thus supporting the block duplication hypothesis. Hughes et al. do not seem to question that the Hox clusters themselves were duplicated as blocks. EVX1 is 45 kb away from HoxA13, and EVX2 is only 13 kb upstream from HoxD13; each Hox cluster spans approximately 100 kb. Hughes et al. did not report which species were included in the analysis that lead to the rejection of a molecular clock for the EVX genes.
Frizzled—FZD
The two frizzled genes FZD1 (FR1) on Hsa7q21 and FZD7 (FR7) on Hsa2q33 were found not to evolve in a clock-like manner and thus were regarded as uninformative (note that the chromosomal localizations were reversed in the paper by Hughes et al.). However, together with FZD2 on Hsa 17q21.31 (Zhao et al. 1995) they form a triplet of genes linked to Hox clusters that seem to have duplicated at the same time as the Hox clusters (Koike et al. 1999), thus supporting the block duplication hypothesis. Again, it was not clear in the article by Hughes et al. which species were included in their analysis of FZD1 and FZD7 that led to their conclusion.
GLI Zinc-Finger Protein—GLI
The family of Krüppel-like zinc-finger-containing transcription factors GLI (for glioma-associated oncogene homolog) was found by Hughes et al. to have duplicated in the same time period as the Hox clusters (H-Fig. 3), but gave a different internal phylogeny (H-Fig. 5). However, this analysis was based on only human, mouse, and Xenopus laevis sequences and therefore should be evaluated with great caution and cannot be used to reject duplications simultaneously with the Hox clusters.
Glucagon—GCG
The glucagon gene on 2q24.2 is related to the GIP (glucose-dependent insulinotropic peptide) gene on 17q21.3. The genes were found by Hughes et al. to have duplicated 949 Myr ago, too early to be consistent with the duplication of the Hox clusters. However, GIP has been sequenced only in mammals, and the branch leading to the human and mouse sequences diverged just basal to the glucagon tree that includes mammalian and actinopterygian sequences. It should also be noted that glucagon seems to have a slower replacement rate in mammals than in other vertebrates (Irwin 2001), thus giving an impression of early origin. We conclude that the duplication most likely took place at the dawn of vertebrate evolution, as recently reported by others (Irwin 2002). The use of short peptide sequences or peptide precursor sequences for phylogenetic analyses was previously found to be highly problematic (Dores et al. 1996) because the different parts of the prepropeptide sequences differ dramatically in their evolutionary rates.
Glucose Transporter—SLC2A (GLUT)
This solute carrier family for glucose was one of the four that Hughes et al. found could have been duplicated concomitantly with the Hox clusters as it agreed with the timepoint of Hox cluster duplications (H-Fig. 3). However, SLC2A3 (GLUT3) is on 12p13.3 and SLC2A4 (GLUT4) is on 17p13, and thus both genes are on the wrong chromosome arm relative to the Hox-bearing arms. These SLC2A genes are more likely to belong to the paralogon Hsa1, 3, 12, and 17 (Popovici et al. 2001; F. Hallböök, L.-G. Lundin, and D. Larhammar, in prep.).
G Protein-Coupled Receptor—GPR
This is one of the largest gene families in the human genome. Previous sequence analyses have shown that many of the gene duplications took place before the divergence of protostomes and deuterostomes. H-Table 1 listed seven family members, but the phylogenetic analysis included as many as 40 sequences plus a few invertebrate sequences. However, four of the seven receptors selected for comparison of chromosomes 2, 7, and 17 have distinct ligands, strongly suggesting that they arose before the origin of vertebrates, as most types of ligands seem to have done. Both IL8 receptor genes are located on Hsa2 and probably arose through a recent local duplication, and comparison with other mammals reveals rapid evolution. CCR7 (CKR7) and GRP37 are orphan receptors, and it is therefore difficult to determine when these might have arisen from a common ancestral gene. The TACR1 (NK-1R) sequence was recently mapped to Hsa2p12 and is thus on the wrong arm. Included among the 40 sequences were also two NPY-family receptors (on non-Hox chromosomes). These arose before the proposed chromosome duplications, although they still bind the same ligands (Wraith et al. 2000). Other NPY-family receptors (Wraith et al. 2000) as well as dopamine receptors D1 and D5 and adrenergic receptors support chromosome duplications, albeit a different paralogon than the one discussed here. Thus, the analysis performed by Hughes et al. cannot be taken as evidence for or against block duplications.
G Nucleotide-Binding Protein—GNB
The GNB family has at least four members in human. However, only two of these are on Hox chromosomes, one of which is on the wrong arm (GNB3 in Hsa12p13.31). Thus, there is no reason to assume that this gene family has anything to do with the evolution of the Hox clusters. They do seem to be part of the paralogon Hsa1p (GNB1), 3q (GNB4), 12p (GNB3; the fourth region is 17p but GNB2 is on 7q22.1; Popovici et al. 2001; F. Hallböök, L.-G. Lundin, and D. Larhammar, in prep.) and thereby support the chromosome duplication or tetraploidization hypothesis.
Hedgehog—HH
This gene family was the third for which Hughes et al. found data supporting duplication concomitant with Hox clusters. The genes IHH and SHH on Hsa2 and 7, respectively, seemed to have duplicated in the same time period as the Hox clusters. A third member, DHH, was not listed in H-Table 1 although it was included in their phylogenetic analyses. The DHH gene is located in 12q13.1, which adds further support for duplication concomitant with the Hox clusters.
Hepatocyte Nuclear Factor—TCF (HNF)
Hughes et al. found that the TCF (HNF) genes did not evolve in a clock-like manner and were therefore uninformative. In addition, the TCF genes 1 and 2 (HNF A and B) are in Hsa12q24.2 and 17q12, and thus TCF1 is some distance away from HoxD on Hsa12, similar to ACADS described above; the connection of TCF genes 1 and 2 with Hox evolution is unclear.
Immunoglobulin-Related—IG
Four immunoglobulin (IG)-related genes were mentioned. The genes for CD4 and CD7 were found to have duplicated in the same time period as the Hox clusters (H-Fig. 3). However, the CD4 gene is on 12p13.31, which is the wrong arm of Hsa12. The IG-related genes form a huge gene family, and it is difficult to evaluate these four members without more information than that mentioned in the Hughes et al. article.
Inhibin—INHB
The four inhibin genes listed by Hughes et al. are located in the same chromosome regions as the Hox clusters, with both INHA and INHBB on Hsa2. The INHB gene family is not mentioned in the article except in H-Tables 1 and 2. The genes INHBA, INHBB, and INHBC do seem to be the result of chromosome duplications, whereas the INHA gene is much more distantly related and is located more than 100 Mb from INHBB on Hsa2.
Insulin-like Growth Factor-Binding Protein—IGFBP (IGBP)
This family is represented in all four Hox chromosome regions, but was found by Hughes et al. to have a phylogeny inconsistent with that of the Hox clusters (H-Fig. 5). However, only human and mouse sequences were included in the analysis, making it difficult to detect any differences in evolutionary rates. Two IGFBP genes are present on Hsa2 and Hsa7, and the phylogenetic analysis suggests that a local duplication preceded the chromosome duplications, after which one copy seems to have been lost in each pair on Hsa12 and Hsa17.
Integrin α—ITGA (INTA)
The six integrin α genes ITGA (INTA) were found to have duplicated too early (H-Fig. 2) and to have phylogenies inconsistent with the Hox clusters (H-Fig. 5). However, these early duplications most likely reflect local duplication events that generated three integrin α genes on the ancestral vertebrate chromosome or even in the common ancestor of deuterostomes and protostomes, after which the vertebrate chromosome duplications copied this cluster. Hsa2 still has three ITGA genes, whereas Hsa12 and Hsa17 seem to have retained two and lost the third. Hsa7 has no ITGA gene, but ITGA9 (INTA9) on Hsa3p22.3 may have been translocated from Hsa7 (along with MYL and SCN gene families, see below). It remains to be shown whether ITGA8 (INTA8) on Hsa10p13 may also have been translocated from Hsa7.
Integrin β—ITGB (INTB)
The integrin β family has at least eight members. Four genes were listed in H-Table 1, one in each of the four Hox regions. In addition, ITGB4 (INTB4) is located on Hsa17. As for the ITGA family, the duplications seemed to be too early compared to the Hox clusters (H-Fig. 2), but this actually concerns only ITGB4 relative to the other four, due to branching of invertebrate sequences in between these. However, low bootstrap values make this conclusion uncertain, and among vertebrates, only mammalian and a few chicken and Xenopus laevis sequences are avilable, making it difficult to detect any fluctuations or differences in evolutionary rates among the eight ITGB family members. The four genes ITGB3, 5, 6, and 8 are more closely related to each other than to other members of the family. Three of these are located on Hox chromosomes and therefore support chromosome duplications. The fourth member, ITGB5, is on Hsa3q. The remaining four family members are more difficult to interpret. Two are on Hox chromosomes, ITGB7 on Hsa12 and ITGB4 on Hsa17, but the latter is more divergent from all human ITGB sequences according to Hughes et al., and the last two members (ITGB1 and ITGB2) are on non-Hox chromosomes.
Intermediate Filament—IF
The huge IF family has multiple keratin members on each of chromosomes 12 and 17 as well as a peripherin gene on Hsa12 and a desmin gene on Hsa2. Many of the keratin duplicates appear to be of quite recent origin in the phylogenetic analysis. However, some duplications seem to have preceded the vertebrate radiation, and Hughes et al. concluded that some duplications took place before the divergence of the cephalochordate and gnathostome lineages (H-Fig. 1). Three sequences are known from amphioxus, one of which is from Branchiostoma lanceolatum and two are from Branchiostoma floridae, and these three differ greatly from each other, but no sequences are yet available from cyclostomes, making it difficult to evaluate this highly complex gene family. Some of the duplications seem to be compatible with a chromosome duplication scenario, but data from additional species are required before more definitive conclusions can be drawn.
Myosin Light Chain—MYL
Three MYL family members were listed in H-Table 1: MYL1 and MYL3, located on Hsa2, and MYLE on Hsa17. These genes were found not to evolve in a clock-like manner. However, the MYL1 and MYL3 genes seem to be confused in some databases. A review article (Oota and Saitou 1999) described five human myelin light chain genes with MOHUA2 in 2q34, MOHUSA and MOHU6M in 12q13.3 in the Hox-cluster region, MOHU4E (= MYLE) in 17q21.32, and MOHU3V in 3p21.31. Sequence comparisons from mammals and chicken as well as with invertebrate myosin light chains suggested that the five human subtypes arose after the protostome-deuterostome divergence (Oota and Saitou 1999). This gene family appears to be consistent with duplications concomitant with the Hox clusters, with one extra gene on Hsa12. MYL3 (P06741) is located on Hsa3p21.31 whereas one MYL gene is missing from Hsa7, suggesting a translocation similar to that of the INTA and SCN families.
NAB Transcriptional Regulator—NAB
This gene family, also called EGR, has at least four members in human, one of which is on Hsa2q and one on 12q. The two latter genes were found by Hughes et al. not to display clock-like evolution. The other two genes are on chromosomes unrelated to the Hox-cluster regions. The phylogeny of the four genes has been difficult to resolve (Martin 2000) with very different tree topologies. Until the phylogeny has been clarified, this gene family cannot be used to argue for or against duplications concomitant with the Hox-cluster regions.
NRAMP—SLC11A (NRAMP)
The natural resistance-associated mapcrophage protein (NRAMP) family is now called SLC11A for solute carrier family 11 (NRAMP2 is also called the duodenal metal transporter). SLC11A1 (NRAMP1) is in 2q35, and SLC11A2 is in 12q13. Again, the genes were found by Hughes et al. not to display clock-like evolution. Sequences from teleost fishes display higher identity to mammalian SLC11A2 than to SLC11A1, suggesting that the gene duplication took place before the divergence of actinopterygians and sarcopterygians and that the teleost ortholog of SLC11A1 has not yet been discovered (or has been lost). Thus, the presently available information is consistent with gene duplication concomitant with the Hox clusters.
Nuclear Hormone Receptor—NHR
This highly complex gene family has been divided into subfamilies (Maglich et al. 2001), several of which are represented in the Hox-bearing regions. Phylogenetic analyses suggest that these subfamilies arose before the protostome-deuterostome divergence, in agreement with H-Fig. 1. Hughes et al. also found that some duplications seem to have taken place later, but before the urochordate divergence. However, as there are several NHR genes on each of the Hox chromosomes, it is unclear exactly which genes were used by Hughes et al. to determine the duplication timepoints in H-Fig. 1 as well as in H-Fig. 3 (the two THRA genes listed by Hughes et al. are splice variants of the same gene). We find that some duplications seem to have taken place concomitantly with the Hox regions, namely RARA on 17q12 and RARG on 12q13 as well as NR4A1 (called NOFIP by in H-Table 1) on 12q13 and NR4A2 (= NURR1, called NOT2 in H-Table 1) on 2q22–23. It is possible that RARB and THRB on 3p24 may also have arisen through block duplication but subsequently have been translocated from Hsa7, similarly to ITGA, MYL, and SCN.
Tachykinin (Neurokinin)—TAC (NKN)
The two neurokinin genes TAC1 and TAC3 on Hsa7 and Hsa12, respectively, were found to have duplicated only 106 Myr ago. However, this conclusion was based on a tree containing only different splice variants of mammalian TAC1 compared with a goldfish sequence (tree provided by A. Hughes). Mature peptides from the TAC1 precursor have been sequenced from chicken, alligator, and Burmese python (Conlon et al. 1997), and neurokinin B from the TAC3 prepropeptide has been sequenced from a Rana frog (O'Harte et al. 1991), thus showing that the gene duplication took place before the radiation of tetrapods and thereby disqualifying the basis for the conclusion drawn by Hughes et al.
Nitric Oxide Synthase—NOS
The three NOS genes were found by Hughes et al. to be phylogenetically inconsistent with the Hox cluster tree (H-Fig. 5). However, it is unclear which species were included in this analysis, and as discussed below, the Hox tree can take different shapes depending on how the analysis is performed. The chromosomal locations of the three NOS genes agree with duplications concomitant with the Hox clusters. One phylogenetic analysis (Wang et al. 2001) indicates that one gene duplication might have taken place before the divergence of protostomes and deuterostomes. However, this analysis lacks many crucial animal groups with eNOS sequences only from mammals, nNOS sequences only from mammals and Xenopus laevis, and iNOS from mammals, chicken, and two teleosts. Furthermore, this would imply that one locus has been lost from all protostomes (which is possible). Thus, this data set is too limited to refute duplications in the vertebrate lineage that receive support from chromosomal localization.
Olfactory Receptor—OR
This gene family is one of the largest in the human genome and is also very large in other species, and thus does not lend itself easily to evolutionary comparisons between groups of animals. Recent reviews suggested duplications of an ancestral olfactory receptor gene cluster as well as subsequent local duplications (Glusman et al. 2001; Zozulya et al. 2001). We agree with Hughes et al. that it is too early to draw conclusions about vertebrate genome evolution from presently available data (they did not comment on this gene family).
Pancreatic Polypeptide/Neuropeptide Y
This family of neuroendocrine peptides was found to lack a molecular clock and therefore was regarded as uninformative. The members of this family differ greatly in their evolutionary rates, but thanks to the many species for which sequences have been reported, it has been possible to conclude that neuropeptide Y (NPY) and peptide YY (PYY; not studied by Hughes et al.) most probably arose by duplication from a common ancestral peptide gene in early vertebrate evolution concomitant with the Hox chromosomes. Pancreatic polypeptide arose by tandem duplication of PYY, probably in an early tetrapod. Additional members exist that may be due to separate duplication events, namely PY in certain teleost fishes and a second PYY-like peptide in lampreys. The evolution of this family has been reviewed (Larhammar 1996; Cerdá-Reverter and Larhammar 2000).
Peroxidase
The two peroxidase genes investigated by Hughes et al. were found to lack a molecular clock. As the TPO gene is in 2p25, that is, the wrong arm of Hsa2, it seems unlikely that the evolutionary history has anything to do with the Hox cluster on this chromosome.
Proteasome β Subunit—PSMB
The two genes PSMB-ϑ and PSMBD were found to have duplicated before the divergence of fungi and animals (H-Fig. 1). The PSMBD gene is in 17p13, which is the wrong arm of this chromosome, and cannot be considered part of the Hox-cluster region. Furthermore, this gene family contains many more members, making its evolutionary history difficult to deduce with the presently available information.
RAD52
The RAD52 gene is on 12p13-p12.2, that is, the wrong arm compared to the Hox cluster. The RAD52 pseudogene is only known in the human genome, and its high sequence identity to RAD52 suggests that it arose in the primate lineage. Thus, this gene family is irrelevent for testing the chromosome duplication hypothesis.
Ras-Related—RASR
This is a huge gene family with at least 60 members (Stenmark and Olkkonen 2001), which makes evaluation exceedingly difficult. Hughes et al. found that some duplications took place before the divergence of fungi and animals (H-Fig. 1). However, their phylogenetic tree also showed some duplications that have taken place in deuterostomes after the divergence from protostomes. For instance, human RALA and RALB genes may support the block duplication hypothesis, but were not discussed. More information is needed before the evolution of this complex gene family can be correlated with the evolution of the various groups of organisms.
Sodium Channel—SCN
Three SCN genes were listed in H-Table 1, two on Hsa2 and one on Hsa17. A single sequence from the urochordate Halocynthia roretzi suggested that a gene duplication took place before the split of this group from the lineage leading to vertebrates (H-Fig. 1). The duplication leading to SCNA2 on Hsa2 and SCNA4 on Hsa17 was found to be within the same time range as the Hox duplications (H-Fig. 3). A more extensive analysis of ten human SCN genes (Plummer and Meisler 1999) suggested that the genes did indeed arise by chromosome duplications followed by local duplications on two of the chromosomes, although the phylogenetic analysis considered only the human genes and two Drosophila melanogaster genes. Two of the human SCN genes were most likely translocated from Hsa7 to Hsa3 along with members of a few other gene families (see above). Sequences from additional taxa are required before definitive conclusions can be drawn, but the presently available data agree with duplications concomitant with the Hox clusters.
Synaptobrevin—SYB
The two genes SYB1 and SYB2 studied by Hughes et al. were found to have duplicated some 888 Myr ago, well before the Hox duplications (H-Fig. 3). However, these genes are on the wrong arm of Hsa12 and Hsa17, respectively. Furthermore, this gene family consists of at least ten more members, making its evolution difficult to analyze based on two members.
Wnt-related—WNT
This is another very large gene family with several gene duplications apparently of ancient origin, whereas others seem to be more recent. The genes WNT10B on 12q13 and WNT10A on 2q35 could have duplicated concomitantly with the Hox clusters. However, information from more taxa is required before the evolution of this large family can be compared with the evolution of animal groups.
DISCUSSION
The four human Hox clusters are generally accepted to have resulted from duplications of a single ancestral cluster, as shown by their high conservation of sequences and organization across vertebrates. The Hox-cluster duplications are assumed to have taken place in the lineage leading to vertebrates after divergence from cephalochordates, based on the observation that amphioxus has a single Hox cluster (Garcia-Fernàndez and Holland 1994), as do most other invertebrates. The question therefore is how large the duplicated regions might have been, that is, how many flanking genes were duplicated along with the Hox clusters. To address this question, gene families with members on the Hox-bearing chromosomes in the human genome have been analyzed by several investigators to determine whether their phylogeny is consistent with that of the Hox-cluster genes themselves. Hughes et al. (2001) analyzed 42 gene families and concluded in the Abstract that 32 of these provided evidence against duplication simultaneously with the Hox clusters. In the Discussion, those authors wrote that 29 gene families were inconsistent with simultaneous duplication (p. 777). After repeated reading of the article we are able to identify 26 gene families that those authors interpreted as providing evidence against simultaneous duplication (Table 1).
Table 1.
Summary of Our Conclusions and of the Analyses Reported in the Paper by Hughes et al. (2001)a
| Gene family genes | Abbrev in OMIM | Hughes' abbrev (when different) | Phylogenetic analyses H-Fig. 1 | Duplication time estimates H-Fig. 3 | Phylogenetic consistency H-Fig. 4+5 | Total Hughes et al | Our conclusion |
|---|---|---|---|---|---|---|---|
| 16 gene fam | 15 gene fam | 7 gene fam | 42 gene fam | 42 gene fam | |||
| Acetylcholine rec (nicot.) | ACHR | N.A. | * | ||||
| Acetyl-coA carboxylase | ± | ± | |||||
| Actin | ACT | − | − | − | |||
| ACTB-ACTG1 | + | ||||||
| ACTG2 (ACTH)-ACTA2 (ACTSA) | * (+) | ||||||
| ACTA1-ACTC | * (+) | ||||||
| Acyl-coA dehydrogenase | ACAD | − | − | ± | |||
| ADP-ribosylation factor | ARF | − | − | ± | |||
| Anlon exchanger (SLC4A) | SLC4A | AE | − | − | + (◊) | ||
| Aquaporin | AQP | − | − | ± | |||
| Arrestin | ARR | − | − | * | |||
| Brain amilor, sens. Na ch | ACCN | BNAC | + | + | + | ||
| Cyclin-dependent kinase | CDK | − | − | − | ± | ||
| Enolase | ENO | ENOL | − | − | * (+) | ||
| ERBB receptor protein-TK | ERBB | − | − | − | + (◊) | ||
| Even-skipped | EVX | ± | + | ||||
| Frizzled | FZD | ± | + | ||||
| GLI zinc-finger protein | GLI | + | − | − | + (◊) | ||
| Glucagon | GCG | − | − | + | |||
| Glucose transporter (SLC2A) | SLC2A | GLUT | + | + | * (+) | ||
| G protein-coupled receptor | GPR | − | − | ± | |||
| G nucleotide binding prot. | GNB | − | − | * (+) | |||
| Hedgehog | HH | + | + | + | |||
| Hepatocyte nuclear factor | TCF | HNF | ± | ± | |||
| Immunoglobulin-related | IG | + | + | ± | |||
| Inhibin | INHB | N.A. | + | ||||
| Insulin-like GF-BP | IFGBP | IGBP | − | − | + (◊) | ||
| Integrin α | ITGA | INTA | − | − | − | ||
| ITGAv, 5 and 2B | + | ||||||
| ITGA3, 6 and 7 | + | ||||||
| ITGA4 and 9 | + | ||||||
| Integrin β | ITGB | INTB | − | − | − | ||
| ITGB3, 6 and 8 | + | ||||||
| ITGB1, 4 and 7 | + | ||||||
| Intermediate filament | IF | − | − | ± | |||
| Myosin light chain | MYL | ± | + | ||||
| NAB transcriptional regul. | NAB | ± | ± | ||||
| NRAMP (SLC11A) | SLC11A | NRAMP | ± | + | |||
| Nuclear hormone receptor | NHR | − | − | − | |||
| RARA-RARG | + | ||||||
| NR4A1-NR4A2 | + | ||||||
| Tachykinin (neurokinin) | TAC | NKN | − | − | + | ||
| Nitric oxide synthase | NOS | − | − | + (◊) | |||
| Olfactory receptor | OR | N.A. | ± | ||||
| Pancr. polypeptide/NPY | NPY | ± | + | ||||
| Peroxidase | ± | * | |||||
| Proteasome β subunit | PSMB | − | − | * | |||
| RAD52 | RAD52 | − | − | * | |||
| Ras-related | RASR | − | − | ± | |||
| Sodium channel | SCN | − | + | − | + (◊) | ||
| Synaptobrevin | SYB | − | − | * | |||
| Wnt-related | WNT | − | − | ± | |||
| Total supporting | 0+ | 6+ | 0+ | 4+ | 20+ (6 ◊) | ||
| Total inconsistent | 16− | 9− | 7− | 26− | 0− | ||
| Total not informative | 9± | 13± | |||||
| 3 N.A. | 9* (3 of which +) Including actins: 11* (5 of which +) |
In their figures 1, 3, and 4+5.
+, The analysis supports chromosome duplication vs. individual gene duplication; −, the analysis rejects chromosome duplication; ±, the analysis is not informative. N.A., the family was not analyzed. The last column shows our interpretation for each gene family considering the totality of evidence, including chromosomal localization. (◊) The gene family's phylogeny is not consistent with that of the Hox gene family; however, in all of these cases very few taxa were included. *, The gene family members are located outside of the extended Hox cluster. “(+)” indicates support for duplication concomitantly with large chromosome regions other than those bearing the Hox clusters (i.e., they belong to a different paralogon).
The first important requirement for an analysis of block duplications is that the linkage of the gene families with the Hox clusters is ancestral. The human Hox-bearing chromosomes have clearly undergone rearrangements and thus harbor many genes that have arrived by translocation. As shown in Figure 1, most gene families with members on three or four of the Hox chromosomes are located in very close proximity to the Hox clusters on Hsa12 and 17 and on the q arm of Hsa2. Only on Hsa7 are the gene families distributed on both arms. Nine of the gene families studied by Hughes et al. do not seem to have ancestral linkage with the Hox clusters (Table 1), namely ACHR, ARR, ENO, SLC2A (GLUT), GNB, peroxidase, PSMB, RAD52, and SYB. All of these have only two members on the Hox-bearing chromosomes, suggesting that these arrived by translocations. In fact, the families ENO and SLC2A seem to belong to a different paralogon, namely the one involving Hsa1, 3, 12, and 17 (F. Hallböök, L.-G. Lundin, and D. Larhammar, in prep.). The RAD52 gene duplication took place as late as in the primate lineage and is not informative.
Many other gene families included in the analysis have large numbers of members and therefore have very complicated phylogenetic histories. Some of these families have several duplications preceding the origin of vertebrates, for instance AQP, CDK, GPR, IG, RASR, and WNT, whereas others have duplications that took place after the vertebrate radiation, primarily IF. The enormous OR family probably had multiple duplications both before and after the Hox-cluster duplications. In total, we consider 13 gene families too complex to address the hypothesis of extended Hox-cluster duplications with the presently available information on sequences, taxa, and chromosomal localization, as shown with the symbol +/− in Table 1 (Hughes et al. found a total of nine families uninformative). Nevertheless, some of the 13 complicated families have duplications that do seem to coincide with the Hox-cluster duplications such as AQP1-AQP2, CDK2-CDK3, and WNT10A-WNT10B. However, we have refrained from counting these as supportive evidence in Table 1.
The remaining 20 gene families seem to have their members in close proximity to the Hox clusters. Two families, ERBB and IGFBP, are represented on all four Hox chromosomes. Ten families have members on three of the Hox chromosomes, namely SLC4A (AE), FZD (only two members were listed by Hughes et al.), GLI, HH (two listed by Hughes et al.), INHB, ITGA, ITGB, MYL, NOS, and SCN. Three of these families have a fourth member on Hsa3p, and these genes may have been block-translocated together from Hsa2 (ITGA, MYL, and SCN). The remaining eight families have members on two of the Hox chromosomes, namely ACT, ACCN (BNAC), EVX, GCG, SLC11A (NRAMP), NHR, TAC (NKN), and NPY. Of the 20 families, only ACCN and HH were found by Hughes et al. to be compatible with the Hox-cluster duplications. The others were concluded by Hughes et al. to have duplication times inconsistent with the Hox-cluster duplications, or if having been duplicated during the same time period as the Hox clusters, they had conflicting tree topologies as analyzed by molecular phylogeny.
However, the phylogenetic analyses performed by Hughes et al. are very difficult to evaluate because sequence information is missing from several vertebrate classes, particularly actinopterygian fishes and cartilaginous fishes. Many trees were in fact based on sequences from mammals with only scattered representatives from chicken or Xenopus laevis. This makes it impossible to detect any deviations or fluctuations in evolutionary rates. Due to the lack of taxon representation, we conclude that not a single one of these trees can be considered to be clearly incompatible with duplications concomitant with the Hox clusters. In fact, several gene families have a phylogenetic distribution that conflicts with the duplication timepoints calculated by Hughes et al. (H-Fig. 3), most notably the families NKN, ENO, and ACTB-ACTG1. More complete taxon representation shows that the GCG and ARR families also have duplication timepoints that coincide with early vertebrate evolution (although ARR does not seem to belong to the Hox paralogon). Our analyses suggest that 14 of the 20 analyzable and relevant families are consistent with the Hox clusters and the remaining six are uncertain. None clearly contradicts duplication concomitant with Hox. Some gene families actually give twofold (ITGB, NHR) or even threefold (ITGA) support for block/chromosome duplication, as they consist of subfamilies that duplicated along with the Hox clusters. Note that of the four families interpreted by Hughes et al. to support duplication concomitant with the Hox clusters, two do not hold up to scrutiny, namely SLC2A (GLUT) and IG.
Even the Hox clusters themselves have been found to display different phylogenetic relationships depending on how the analysis is performed (Bailey et al. 1997). The homeoboxes are unreliable due to their short and highly conserved sequences, and the remaining parts of the Hox proteins are known in only a few species. As pointed out above, the Hox-cluster duplications (or tetraploidizations) may have been very close in time, making it questionable as to whether phylogenetic analyses of gene families can give consistent results. Furthermore, the time period before “diploidization” after such a tetraploidization is likely to involve crossing-over and perhaps gene conversion that scrambles the sequences (Angers et al. 2002).
It might be argued that the short chromosomal regions near the Hox clusters constitute a selected data set that cannot be considered sufficient for discussion of duplications of entire chromosomes, let alone tetraploidizations and the 2R hypothesis. However, the extended Hox clusters that seem to constitute the duplicated unit may nevertheless represent a significant proportion of a chromosome, particularly when considering that several other gene families exist in the Hox-cluster regions that were not analyzed by Hughes et al. but which seem consistent with duplications concomitant with the Hox clusters (Fig. 1; Popovici et al. 2001; F. Hallböök, L.-G. Lundin, and D. Larhammar, in prep.). Comparative chromosome mapping suggests that chromosome rearrangements have occurred after the origin of the four Hox clusters and before Hox-bearing chromosomes arrived at their present organization in human (Chowdhary et al. 1998; Groenen et al. 2000; Postlethwait et al. 2000; Woods et al. 2000; Murphy et al. 2001). One may therefore add that many additional gene families could have been part of the duplicated cluster, but their traces have been eliminated by gene losses and translocations.
It is generally agreed that the four Hox clusters in the human genome arose by duplication of a single ancestral cluster in early vertebrate (or pre-vertebrate) evolution, that is, by a block duplication. Based on the data discussed here and elsewhere (Pollard and Holland 2000; Murphy et al. 2001; Popovici et al. 2001; F. Hallböök, L.-G. Lundin, and D. Larhammar, in prep.), we conclude that the duplicated Hox-cluster regions contained numerous other genes, making it likely that a very large block or an entire chromosome was duplicated. Overall, the evidence for duplications of an extended Hox cluster, as shown by the chromosomal localization of many gene families, seems much stronger than the argument against this from incomplete and uncertain phylogenetic trees. Together with the observation that many other paralogons exist (Popovici et al. 2001; F. Hallböök, L.-G. Lundin, and D. Larhammar, in prep.), a parsimonious explanation would be that the entire genome underwent two tetraploidizations, that is, the 2R hypothesis. This appears particularly likely because we know that extensive gene loss (Gu and Huang 2002) may take place after such events.
METHODS
Chromosome localization data were retrieved from the Online Medelian Inheritance in Man database (www3.ncbi.nlm.nih.gov/omim/) and the human genome database at the University of California Santa Cruz (http://genome.ucsc.edu/).
Phylogenetic data were obtained from already published reports. The phylogenetic trees underlying the conclusions presented in the paper by Hughes et al. (2001) were kindly provided by Austin L. Hughes.
WEB SITE REFERENCES
http://www3.ncbi.nlm.nih.gov/omim/; Online Medelian Inheritance in Man.
http://genome.ucsc.edu/; The human genome database at the University of California Santa Cruz.
Acknowledgments
We thank Dr. Austin Hughes for providing the phylogenetic trees on which the article by him and his coauthors was based. D.L. and F.H. are supported by grants from the Swedish Research Council and the Wallenberg Research Foundation, Consortium North.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Dan.Larhammar@neuro.uu.se; FAX 46-18-511540.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.445702.
REFERENCES
- Abi-Rached L, Gilles A, Shiina T, Pontarotti P, Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nature Genet. 2002;31:100–105. doi: 10.1038/ng855. [DOI] [PubMed] [Google Scholar]
- Angers B, Gharbi K, Estoup A. Evidence of gene conversion events between paralogous sequences produced by tetraploidization in Salmoninae fish. J Mol Evol. 2002;54:501–510. doi: 10.1007/s00239-001-0041-x. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit AF, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Bailey WJ, Kim J, Wagner GP, Ruddle FH. Phylogenetic reconstruction of vertebrate Hox cluster duplications. Mol Biol Evol. 1997;14:843–853. doi: 10.1093/oxfordjournals.molbev.a025825. [DOI] [PubMed] [Google Scholar]
- Cerdá-Reverter JM, Larhammar D. Neuropeptide Y family of peptides: Structure, anatomical expression, function, and molecular evolution. Biochem Cell Biol. 2000;78:371–392. [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T, Frönicke L, Sherthan H. Emerging patterns of comparative genome organization in some mammalian species as revealed by zoo-FISH. Genome Res. 1998;8:577–589. doi: 10.1101/gr.8.6.577. [DOI] [PubMed] [Google Scholar]
- Conlon JM. The origin and evolution of peptide YY (PYY) and pancreatic polypeptide (PP) Peptides. 2002;23:269–278. doi: 10.1016/s0196-9781(01)00608-8. [DOI] [PubMed] [Google Scholar]
- Conlon JM, Adrian TE, Secor SM. Tachykinins (substance P, neurokinin A and neuropeptide γ) and neurotensin from the intestine of the Burmese python, Python molurus. Peptides. 1997;18:1505–1510. doi: 10.1016/s0196-9781(97)00232-5. [DOI] [PubMed] [Google Scholar]
- Coulier F, Popovici C, Villet R, Birnbaum D. MetaHOX gene clusters. J Exp Zool. 2000;288:345–351. doi: 10.1002/1097-010X(20001215)288:4<345::AID-JEZ7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Craft CM, Whitmore DH. The arrestin superfamily: Cone arrestins are a fourth family. FEBS Lett. 1995;362:247–255. doi: 10.1016/0014-5793(95)00213-s. [DOI] [PubMed] [Google Scholar]
- Dores RM, Rubin DA, Quinn TW. Is it possible to construct phylogenetic trees using polypeptide hormone sequences? Gen Comp Endocrinol. 1996;103:1–12. doi: 10.1006/gcen.1996.0088. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong RF, Holland PWH. Were vertebrates octoploid? Phil Trans R Soc Lond B. 2002;357:531–544. doi: 10.1098/rstb.2001.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernàndez J, Holland PWH. Archetypal organization of the amphioxus Hox gene cluster. Nature. 1994;370:563–566. doi: 10.1038/370563a0. [DOI] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- Gregory SG, Sekhon M, Schein J, Zhao S, Osoegawa K, Scott CE, Evans RS, Burridge PW, Cox TV, Fox CA, et al. A physical map of the mouse genome. Nature. 2002;418:743–750. doi: 10.1038/nature00957. [DOI] [PubMed] [Google Scholar]
- Groenen MAM, Cheng HH, Bumstead N, Benkel BF, Briles WE, Burke T, Burt DW, Crittenden LB, Dodgson J, Hillel J, et al. A consensus linkage map of the chicken genome. Genome Res. 2000;10:137–147. doi: 10.1101/gr.10.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Huang W. Testing the parsimony test of genome duplications: A counterexample. Genome Res. 2002;12:1–2. doi: 10.1101/gr.214402. [DOI] [PubMed] [Google Scholar]
- Holland PWH. More genes in vertebrates? In: Meyer A, editor. Genome evolution. Lancaster, UK: Kluwer; 2002. (in press). [Google Scholar]
- Hughes AL. Phylogenies of developmentally important proteins do not support the hypothesis of two rounds of genome duplication early in vertebrate history. J Mol Evol. 1999;48:565–576. doi: 10.1007/pl00006499. [DOI] [PubMed] [Google Scholar]
- Hughes AL, da Silva J, Friedman R. Ancient genome duplications did not structure the human Hox-bearing chromosomes. Genome Res. 2001;11:771–780. doi: 10.1101/gr.160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MK, Hughes AL. Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol Biol Evol. 1993;10:1360–1369. doi: 10.1093/oxfordjournals.molbev.a040080. [DOI] [PubMed] [Google Scholar]
- Irwin DM. Molecular evolution of proglucagon. Regul Peptides. 2001;98:1–12. doi: 10.1016/s0167-0115(00)00232-9. [DOI] [PubMed] [Google Scholar]
- Irwin DM. Ancient duplications of the human proglucagon gene. Genomics. 2002;79:741–746. doi: 10.1006/geno.2002.6762. [DOI] [PubMed] [Google Scholar]
- Iwabe N, Kuma K-i, Miyata T. Evolution of gene families and relationship with organismal evolution: Rapid divergence of tissue-specific genes in the early evolution of chordates. Mol Biol Evol. 1996;13:483–493. doi: 10.1093/oxfordjournals.molbev.a025609. [DOI] [PubMed] [Google Scholar]
- Jacobs S, Schilf C, Fliegert F, Koling S, Weber Y, Schürmann A, Joost H-G. ADP-ribosylation factor (ARF)-like 4, 6, and 7 represent a subgroup of the ARF family characterized by rapid nucleotide exchange and a nuclear localization signal. FEBS Lett. 1999;456:384–388. doi: 10.1016/s0014-5793(99)00759-0. [DOI] [PubMed] [Google Scholar]
- Koike J, Takagi A, Miwa T, Hirai M, Terada M, Katoh M. Molecular cloning of Frizzle-10, a novel member of the Frizzled gene family. Biochem Biophys Res Comm. 1999;262:39–43. doi: 10.1006/bbrc.1999.1161. [DOI] [PubMed] [Google Scholar]
- Larhammar D. Evolution of neuropeptide Y, peptide YY, and pancreatic polypeptide. Regul Pept. 1996;62:1–11. doi: 10.1016/0167-0115(95)00169-7. [DOI] [PubMed] [Google Scholar]
- Larhammar D, Risinger C. Molecular genetic aspects of tetraploidy in the common carp, Cyprinus carpio. Mol Phylogenet Evol. 1994;3:59–68. doi: 10.1006/mpev.1994.1007. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Sluder A, Guan Z, Shi Y, McKee DD, Carrick K, Kamdar K, Willson TM, Moore JT. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2001;2:29.21–29.27. doi: 10.1186/gb-2001-2-8-research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski W. Are we polyploids? A brief history of one hypothesis. Genome Res. 2001;11:667–670. doi: 10.1101/gr.188801. [DOI] [PubMed] [Google Scholar]
- Málaga-Trillo E, Meyer A. Genome duplications and accelerated evolution of Hox genes and cluster architecture in teleost fishes. Amer Zool. 2001;41:676–686. [Google Scholar]
- Martin AP. Choosing among alternative trees of multigene families. Mol Phylogenet Evol. 2000;16:430–439. doi: 10.1006/mpev.2000.0818. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Stanyon R, O'Brien SJ. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. 2001;2:5.1–5.8. doi: 10.1186/gb-2001-2-6-reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nembaware V, Crum K, Kelso J, Seoighe C. Impact of the presence of paralogs on sequence divergence in a set of mouse-human orthologs. Genome Res. 2002;12:1370–1376. doi: 10.1101/gr.270902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Harte F, Bockman CS, Abel PW, Conlon MJ. Isolation, structural characterization and pharmacological activity of dog neuromedin U. Peptides. 1991;12:11–15. doi: 10.1016/0196-9781(91)90159-m. [DOI] [PubMed] [Google Scholar]
- Ohta T. Multigene families and the evolution of complexity. J Mol Evol. 1991;33:34–41. doi: 10.1007/BF02100193. [DOI] [PubMed] [Google Scholar]
- Oota S, Saitou N. Phylogenetic relationship of muscle tissues deduced from superimposition of gene trees. Mol Biol Evol. 1999;16:856–867. doi: 10.1093/oxfordjournals.molbev.a026170. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Genome duplications: The stuff of evolution? Science. 2001;294:2458–2460. doi: 10.1126/science.294.5551.2458. [DOI] [PubMed] [Google Scholar]
- Plummer NW, Meisler MH. Evolution and diversity of mammalian sodium channel genes. Genomics. 1999;57:323–331. doi: 10.1006/geno.1998.5735. [DOI] [PubMed] [Google Scholar]
- Pollard SL, Holland PWH. Evidence for fourteen homeobox gene clusters in human genome ancestry. Curr Biol. 2000;10:1059–1062. doi: 10.1016/s0960-9822(00)00676-x. [DOI] [PubMed] [Google Scholar]
- Popovici C, Leveugle M, Birnbaum D, Coulier F. Homeobox gene clusters and the human paralogy map. FEBS Lett. 2001;491:237–242. doi: 10.1016/s0014-5793(01)02187-1. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan Y-L, Kelly PD, Chu F, Hill-Force A, Walbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;2000:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Laudet V. Evolutionary rates of duplicate genes in fish and mammals. Mol Biol Evol. 2001;18:681–683. doi: 10.1093/oxfordjournals.molbev.a003849. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Braasch I, Meyer A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Phil Trans R Soc Lond B. 2001;356:1661–1679. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy MR, Hedges SB. Evolutionary history of the enolase gene family. Gene. 2000;259:129–138. doi: 10.1016/s0378-1119(00)00439-x. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Taylor JS, Braasch I, Meyer A. The ghost of selection past: Rates of evolution and functional divergence of anciently duplicated genes. J Mol Evol. 2001;53:436–446. doi: 10.1007/s002390010233. [DOI] [PubMed] [Google Scholar]
- Wallis M. Episodic evolution of protein hormones in mammals. J Mol Evol. 2001;53:10–18. doi: 10.1007/s002390010187. [DOI] [PubMed] [Google Scholar]
- Wang T, Ward M, Grabowski P, Secombes CJ. Molecular cloning, gene organization and expression of rainbow trout (Oncorhynchus mykiss) inducible nitric oxide synthase (iNOS) gene. Biochem J. 2001;358:747–755. doi: 10.1042/0264-6021:3580747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods IG, Kelly PD, Chu F, Ngo-Hazelett P, Yan Y-L, Huang H, Postlethwait JH, Talbot WS. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wraith A, Törnsten A, Chardon P, Harbitz I, Chowdhary BP, Andersson L, Lundin L-G, Larhammar D. Evolution of the neuropeptide Y receptor family: Gene and chromosome duplications deduced from the cloning of the five receptor subtype genes in pig. Genome Res. 2000;10:302–310. doi: 10.1101/gr.10.3.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardoya R, Villalba S. A phylogenetic framework for the aquaporin family in eukaryotes. J Mol Evol. 2001;52:391–404. doi: 10.1007/s002390010169. [DOI] [PubMed] [Google Scholar]
- Zhao ZY, Lee CC, Baldini A, Caskey CT. A human homologue of the Drosophila polarity gene frizzled has been identified and mapped to 17q21.1. Genomics. 1995;27:370–373. doi: 10.1006/geno.1995.1060. [DOI] [PubMed] [Google Scholar]
- Zozulya S, Echeverri F, Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2:18.11–18.12. doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]