Abstract
Large-scale genetic screens in zebrafish have identified thousands of mutations in hundreds of essential genes. The genetic mapping of these mutations is necessary to link DNA sequences to the gene functions defined by mutant phenotypes. Here, we report two advances that will accelerate the mapping of zebrafish mutations: (1) The construction of a first generation single nucleotide polymorphism (SNP) map of the zebrafish genome comprising 2035 SNPs and 178 small insertions/deletions, and (2) the development of a method for mapping mutations in which hundreds of SNPs can be scored in parallel with an oligonucleotide microarray. We have demonstrated the utility of the microarray technique in crosses with haploid and diploid embryos by mapping two known mutations to their previously identified locations. We have also used this approach to localize four previously unmapped mutations. We expect that mapping with SNPs and oligonucleotide microarrays will accelerate the molecular analysis of zebrafish mutations.
[Supplemental material is available online at www.genome.org. The sequence data described in this paper have been submitted to dbSNP under accession nos. 5103507–5105537. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: J. Postlethwait, C.-B. Chien, C. Kimmel, L. Maves, and M. Westerfield.]
Genetic screens in zebrafish (Danio rerio) have isolated several thousand mutations that define the functions of hundreds of essential genes (Driever et al. 1996; Haffter et al. 1996). Identification of the genes disrupted by these mutations can provide molecular entry points into a wide array of biochemical pathways acting in vertebrate development, physiology, and behavior. Although insertional mutagenesis with retroviral vectors has been used in some genetic screens (Golling et al. 2002), the great majority of zebrafish mutations have been induced by the point mutagen ethyl nitrosourea (ENU) (Driever et al. 1996; Haffter et al. 1996). Genes mutated by ENU are identified by the positional cloning and candidate gene approaches, which are critically dependent on knowledge of the mutation's map position (for review, see Talbot and Hopkins 2000). Current methods involving microsatellite markers have localized some of the mutations identified in the first large-scale screens. Most, however, have not been mapped, and many new mutations are being identified in ongoing genetic screens. Therefore, developing strategies and resources to accelerate the genetic mapping of zebrafish mutations is an important goal.
In human and several model systems, the advent of single nucleotide polymorphism (SNP) maps and high-throughput techniques to rapidly score SNPs has accelerated mapping of mutations (Wang et al. 1998; Winzeler et al. 1998; Cho et al. 1999; Lindblad-Toh et al. 2000; Berger et al. 2001; Hoskins et al. 2001; Wicks et al. 2001; Guo et al. 2002; Swan et al. 2002). Although extensive maps of genes and microsatellites have been constructed for the zebrafish (Postlethwait et al. 1998; Geisler et al. 1999; Shimoda et al. 1999; Barbazuk et al. 2000; Woods et al. 2000; Hukriede et al. 2001), no zebrafish SNP map has heretofore been available. We report the construction of a first generation SNP map of the zebrafish genome. In addition, we describe a method for mapping mutations in which hundreds of SNPs can be scored in parallel by hybridization to an oligonucleotide microarray. By facilitating genetic mapping, this SNP map and corresponding oligonucleotide microarray will accelerate the molecular analysis of zebrafish mutations.
RESULTS AND DISCUSSION
To locate SNPs at defined positions throughout the genome, we sequenced about 1000 PCR fragments, most of which were derived from meiotically mapped ESTs (Kelly et al. 2000; Woods et al. 2000), from the divergent inbred strains C32 and SJD. Because these ESTs were mapped on the basis of genetic polymorphisms (Woods et al. 2000), selection of mapped fragments enriched for SNP-containing sequences. PolyPhred analysis identified 1313 SNPs in 191,005 bp (average frequency 1 SNP/145bp) of sequence derived from mapped, polymorphic ESTs (Woods et al. 2000). Inspection of 41,640 bp of sequence from ESTs that were not known previously to contain polymorphisms revealed 190 additional SNPs (average frequency 1 SNP/219 bp). In surveys of other genes of interest, we identified 384 SNPs between C32 and SJD, raising the total of SNPs characterized between the two strains to 1887 (Fig. 1; Web Supplement A). We also noted 178 insertions/deletions (indels) of 1–6 bp (Web Supplement B).
Figure 1.
Zebrafish SNP map. Vertical lines represent the 25 linkage groups, dots represent individual SNPs. Red dots correspond to SNPs represented on the oligonucleotide microarray. The positions of sequenced markers were derived from the meiotic map of Woods et al. (2000). Names, sequences, positions, and strain data for SNPs are shown in Web Supplement A.
To measure the rate of polymorphism in other strains (Johnson and Zon 1999), we sequenced a subset of the fragments described above in the AB, TL, Tü, and WIK strains. This analysis revealed 148 SNPs that were not identified in the survey of C32 and SJD. In any pair-wise comparison of AB, TL, Tü, and WIK, 30% to 43% of the SNP loci sequenced were polymorphic (Table 1). The polymorphism rate increased in comparisons with C32 and SJD (Table 1), reflecting the bias caused by our selection of fragments known to be polymorphic in these strains.
Table 1.
Percentage of Analyzed SNPs That are Polymorphic Between Common Zebrafish Strains
| Strain | C32 | SJD | WIK | Tü | TL |
|---|---|---|---|---|---|
| AB | 41.8% (207/495) | 64.2% (325/506) | 34.2% (149/436) | 30.2% (121/401) | 36.5% (155/425) |
| TL | 44.3% (224/506) | 59.0% (301/510) | 43.0% (187/435) | 29.9% (119/398) | |
| Tü | 46.5% (214/460) | 60.0% (290/483) | 36.2% (149/412) | ||
| WIK | 53.9% (269/499) | 53.8% (275/511) |
SNP-containing fragments from single adult fish of the indicated strains were amplified by PCR and sequenced. Note that the analyzed fragments were prescreened for polymorphisms between C32 and SJD.
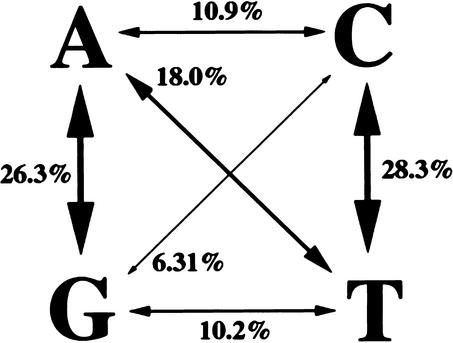
In total, our sequence comparisons identified 2035 SNPs in 712 genes and ESTs (Web Supplement A). Because the great majority of the SNP-containing fragments had been meiotically mapped in previous studies (Woods et al. 2000), we could determine the map positions for 1930 of the SNPs (Fig. 1; Web Supplement A) and 164 of the indels (Web Supplement B) that we identified. The SNPs occupy 430 unique positions on the 3000-cM female meiotic map (Fig. 1). The average distance between groups of SNPs is 6.98 cM (3000 cM/430 map positions), with the biggest gap spanning 57.9 cM at the top of linkage group 7. Transversions comprise 45.4% of the single base pair substitutions and transitions, 54.6% (Fig. 2). Among transversions, A↔T events are over-represented and G↔C events under-represented. These figures are quite similar to those reported for Caenorhabditis elegans (Wicks et al. 2001), but differ from other figures published for Drosophila and mammalian SNPs (Hacia et al. 1999; Petrov and Hartl 1999; Lindblad-Toh et al. 2000; Taillon-Miller and Kwok 2000; Berger et al. 2001).
Figure 2.
Distribution of transitions and transversions among SNPs.
To explore the utility of SNPs in mapping mutations, we developed a strategy using an oligonucleotide microarray to simultaneously score hundreds of SNPs at defined genomic locations. In this method, SNPs are scored by discrimination between perfect matches and single base pair mismatches in hybridization assays with oligonucleotide probes. The microarray comprised oligonucleotide probes (10–26 bp) predicted to have the same Tm (50°C) for all four possible alleles for each of 599 SNPs derived from the C32-SJD sequence survey (Fig. 3A; Web Supplement A). The 599 SNPs define 324 genes/ESTs at 234 unique map positions on the female meiotic map (Fig. 1; Web Supplement A), such that the average distance between groups of SNPs represented on the array is 12.8 cM (3000 cM/234). With respect to the sex-averaged map (Shimoda et al. 1999), which is more relevant for genetic mapping in standard diploid crosses, the average distance between groups of SNPs on the array is 9.8 cM (2300 cM/234).
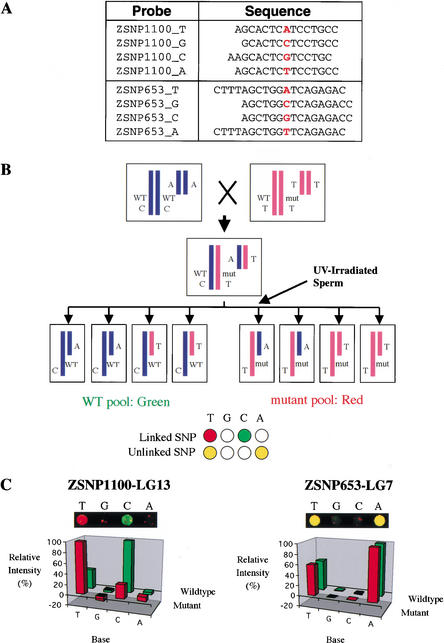
Figure 3.
(A) Oligonucleotide probes specific for two SNPs. (B) Mapping scheme with haploid crosses. Haploid embryos are generated from F1 females heterozygous for the mutation and for many SNPs (two of which are shown). SNPs are scored in pooled wild-type and mutant samples by hybridizing differentially labeled fragments to the same microarray. Alleles of linked SNPs are differentially labeled, whereas unlinked SNPs have both labels on each allele. Putative locations indicated by the microarray analysis are then tested by scoring markers in the region in individuals (not shown). (C) Microarray data mapping the floating head mutation to LG13. A two-color image from the hybridized microarray and a graph of relative hybridization intensity for the four probes of two SNPs are shown. ZSNP1100 on LG 13 exhibited differential labeling characteristic of linked markers. Polymorphic markers at other positions, including ZSNP653 on LG7, showed labeling characteristic of unlinked markers. In another trial of this experiment, we obtained similar results: Differential labeling indicative of linkage was detected for ZSNP1100, and there were no false positive or false negative markers (data not shown).
We used this array to map zebrafish mutations with a variation of the bulked segregant analysis approach that is commonly used in zebrafish mapping projects. In the traditional approach, gel-based markers are scored in pools of wild-type and mutant genomic DNA, and differential amplification of alleles in the two pools identifies linked markers (Postlethwait et al. 1994; Talbot and Schier 1999). In the array approach, fragments encompassing each SNP are amplified by PCR from pools of wild-type and mutant genomic DNA (Fig. 3B). The two pools of DNA fragments are differentially labeled with Cy3 and Cy5, made single-stranded, and simultaneously hybridized to the array (see Methods). The results of the hybridization are then quantified with a microarray scanner, and base calls for each SNP are generated from the relative fluorescence intensity of the four probes for that SNP (Fig. 3C). When haploids are used for mapping, SNP loci linked to the mutation locus exhibit differential labeling of alleles (Fig. 3B; C in wild type, T in mutant), whereas Cy3 and Cy5-labeled DNA hybridize equally well to both alleles of unlinked SNPs (Fig. 3B; T and A alleles). Because not all SNPs are informative in every cross, monomorphic loci are also detected and are visible as a single spot with both labels at one base position on the array.
As a test of the array mapping approach, we mapped floating head (flh), a mutation localized previously to linkage group (LG) 13 (Talbot et al. 1995). We analyzed a haploid mapping cross between the outbred flh line and the DAR strain (Johnson and Zon 1999), which shows a high degree of polymorphism compared with commonly used lab strains (Fig. 3C). Of the 599 SNPs on the array, one SNP (ZSNP1100) near the middle of LG13 displayed differential labeling indicative of linkage. Restriction fragment length polymorphism (RFLP) analysis on the individual embryos contributing to the pooled DNA samples demonstrated that this SNP was located 19 cM from flh. In addition, the array generated unambiguous base calls for 324 other SNPs, which exhibited labeling characteristic of polymorphic unlinked (80) or monomorphic (244) loci. No false positives or false negatives were evident.
The same general principles apply to the use of these microarrays with diploid embryos, although the analysis is slightly different due to the presence of embryos heterozygous for the mutation in the wild-type pool (Fig. 4A). Because of these heterozygotes, SNP alleles linked in cis to the mutation are found in both pools, whereas SNP alleles tightly linked in cis to the wild-type form of the gene are present only in the wild-type pool. Thus, only probes for alleles linked in cis to the wild-type allele of the gene of interest display differential labeling.
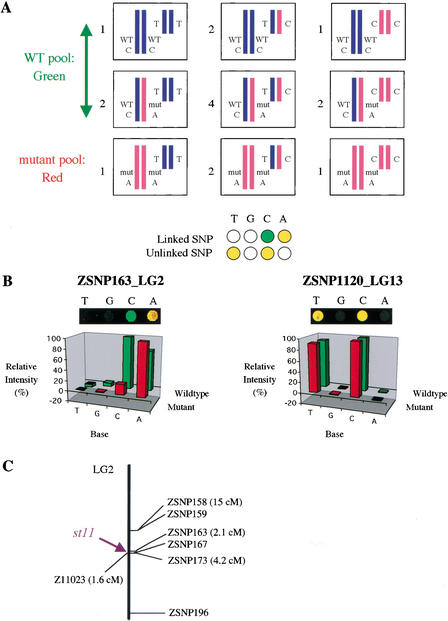
Figure 4.
(A) Mapping scheme for diploid crosses. Boxes represent F2 progeny from an intercross of F1 fish (not shown) heterozygous for the mutation and for many SNPs, two of which are shown. Numbers adjacent to the genotypes refer to the ratios at which they are present among F2 progeny. SNPs from wild-type and mutant pools are differentially labeled as in Fig. 3. For recessive mutations, the SNP allele linked in cis to the mutation is present in both pools due to the presence of heterozygotes in the wild-type pool. Therefore, this allele is labeled with both colors, whereas its wild-type counterpart is labeled only with one color (green in this example). (B) Microarray data mapping the st11 mutation to LG2. A two-color image from the hybridized microarray and a graph of relative hybridization intensity for the four probes of two SNPs are shown. ZSNP163 on LG2 exhibited differential labeling characteristic of linked markers. Polymorphic markers at most other positions, including ZSNP1120 on LG13, showed labeling characteristic of unlinked markers. (C) Map of LG2 showing the positions of st11 and SNPs that showed differential labeling characteristic of linked markers. Analysis of ZSNP158, ZSNP163, ZSNP173, and SSLP marker Z11023 in individual embryos confirmed that st11 resides in this region of LG2 (distances indicated). In another trial of this experiment (data not shown), we detected differential labeling indicative of linkage for markers ZSNP163, ZSNP165, ZSNP171, and ZSNP173 on LG2; no base calls were generated for ZSNP158, ZSNP159, and ZSNP167, or for ZSNP196, which was detected as a false positive in the other trial.
Figure 4 depicts the mapping results for st11, a previously unmapped mutation disrupting notochord differentiation that we identified in a screen for mutants with embryonic lethal phenotypes (I.G. Woods and W.S. Talbot, unpubl.). To map st11, which was isolated in a TL background, we analyzed diploid F2 embryos from a cross constructed with the WIK mapping strain (Johnson and Zon 1999; Nechiporuk et al. 1999). In an analysis of 599 SNPs, six SNPs located on LG2 showed differential labeling between wild-type and mutant pools (Fig. 4B; data not shown). Five of these SNPs cluster in the middle of the linkage group (Fig. 4C). Eleven other SNPs also exhibited differential labeling indicative of possible linkage, but the map positions of these were scattered throughout the genome and three of the eleven were located in the same gene or EST as a SNP with labeling characteristic of unlinked SNPs. All other SNPs that generated unambiguous base calls (450) exhibited labeling characteristic of polymorphic unlinked (56) or monomorphic (394) loci. Hence, analysis of the array data suggested that the st11 gene is located on LG2, and that the putatively linked markers on other linkage groups were false positives. This interpretation was verified by scoring RFLPs caused by three of the LG2 SNPs in individual st11 mutants and wild-type siblings; these markers were located 2–15 cM from the st11 mutation (Fig. 4C). In addition, analysis of SSLP markers on LG2 indicated that st11 was located within the cluster of five linked SNPs (Fig. 4C). The sixth SNP locus on LG2 (ZSNP196) lies ∼70 cM from the mutation and is separated from the mutation by a putatively unlinked SNP, suggesting that it is most likely an additional false positive. Parsons et al. (2002) reported recently that sleepy, a mutation disrupting notochord differentiation, maps to the same region of LG2. Our map assignment and the phenotypic similarity suggest that st11 is a new allele of sleepy.
We have used the microarray strategy to map four other mutations to the correct location in diploid crosses, including iguana, which was mapped previously to LG6 (H. Stickney and W. Talbot, unpubl.), and three other mutations for which no prior map information was available. In these experiments, genotypes were assigned for 60.0% (1361/2269) of the SNPs analyzed. The polymorphism frequency ranged from 17.4%–28.7%, corresponding to an average of 77 polymorphic markers per mapping experiment (range 62–88). One to four linked markers were detected in each experiment, and the false positive rate was 2.5% (34/1361).
We have constructed the first SNP map of the zebrafish genome and developed a new strategy to localize zebrafish mutations by scoring SNPs from pooled genomic DNA samples with oligonucleotide microarrays. Because SNPs can be scored with high-throughput methods (Kwok 2001), SNP maps have accelerated genetic mapping in a variety of model organisms (Winzeler et al. 1998; Cho et al. 1999; Lindblad-Toh et al. 2000; Berger et al. 2001; Hoskins et al. 2001; Wicks et al. 2001; Swan et al. 2002). For example, a mouse SNP map with 1942 mapped markers and a multiplex genotyping assay allow mouse mutations to be mapped in a genome scan with only six genotyping reactions per animal (Lindblad-Toh et al. 2000). Similarly, our zebrafish SNP map comprising 2102 markers enables rapid mapping of zebrafish mutations. The microarray approach that we have developed simultaneously scores hundreds of SNPs in two samples, wild-type and mutant DNA pools, on a single microarray. Differential labeling on alleles of linked markers suggests possible map locations, which can then be tested by scoring markers in the region (e.g., mapped SNPs or SSLPs) on individuals. Analysis of pooled samples with the microarray can detect linkage of markers 10–20 cM from the mutation (Figs. 3 and 4; data not shown). This indicates that, under ideal conditions, analysis of only 60–120 polymorphic SNPs spaced at intervals of 20–40 cM over the 2300 cM sex-averaged map (Shimoda et al. 1999) would be sufficient to identify a marker linked to the average mutation. In practice, it is necessary to score more markers, because the SNPs on the current version of the microarray are not evenly spaced and only a fraction of the SNPs are informative in the outbred backgrounds commonly used for mapping crosses (see Table 1). We expect that future versions of the array will feature improvements in coverage and polymorphism rate of the represented markers. Nonetheless, the current array represents 599 SNPs, and our analysis of a variety of genetic backgrounds indicates that the array will be useful in most crosses with commonly used mapping strains. In combination with the advancing genomic sequence and the ability to rapidly test candidate genes, mapping with SNPs and oligonucleotide arrays will accelerate the molecular analysis of zebrafish mutations.
METHODS
Fish Strains
The derivation of fish strains C32, SJD, AB, TL, Tü, and WIK has been described (Haffter et al. 1996; Johnson and Zon 1999). Genomic DNA samples from C32 and SJD fish were kindly provided by J. Postlethwait (Univ. of Oregon). Genomic DNA from the other strains was prepared from fish stocks maintained in our facility at Stanford. C32 and SJD are partially inbred lines, and previous work showed that these strains are not completely homozygous for all genetic markers (Nechiporuk et al. 1999). Accordingly, we found that a C32 adult was heterozygous for 92 of 1887 SNPs analyzed, and that a SJD adult was heterozygous for 27 of these 1887 SNPs (Web Supplement A).
SNP Sequencing and Detection
To identify SNPs, fragments from C32, SJD, AB, TL, Tü, or WIK genomic DNA were amplified by PCR and directly sequenced on both strands with an ABI377 or ABI3700. Most of the primers were designed from 3′ ESTs for previous mapping experiments (Woods et al. 2000), but some were designed specifically to analyze SNPs of interest. Sequence traces were analyzed with Phred (Ewing et al. 1998) and assembled with Phrap (www.phrap.org). Polyphred (Nickerson et al. 1997) was used to identify puta- tive SNPs. Polyphred ranked 1–3 SNPs were inspected with Consed (Gordon et al. 1998) and called directly from the traces. Some of the traces were also examined using DNAstar software to identify SNPs.
Oligonucleotide Microarrays
The oligonucleotide microarrays were constructed by Protogene, Inc., using ink-jet in situ synthesis and differential surface tension technology (Butler et al. 2001).
Hybridization Assays
We prepared genomic DNA from 16–50 mutant embryos and a similar number of wild-type siblings from a mapping cross as described (Talbot and Schier 1999). PCR primers were designed with Primer 3 (Rozen and Skaletsky 1998) to have a length of 21–27 nucleotides, a Tm of 57–63°C, and a product of 100–175 bp. One primer from each pair was phosphorylated at the 5′ end with T4 polynucleotide kinase. Multiplex (4X) amplification reactions were performed in a 25 μL volume containing ∼13 ng genomic DNA, 0.3 μM primers, 0.6 U Taq DNA polymerase, 100 μM of each dNTP, 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 0.0001% gelatin, and 0.01 mg/mL BSA. Thermocycling was performed under standard conditions consisting of an initial denaturation at 94°C for 2 min, followed by 45 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, and elongation at 72°C for 30 sec, and then a final incubation at 72°C for 5 min.
Following amplification, the PCR products were purified with QIAGEN miniprep columns. Cyanine 3-dUTP or Cyanine 5-dUTP fluorescent labels from NEN Life Science Products, Inc were attached to the 3′ end of the purifed PCR products with Takara terminal deoxynucleotidyl transferase (0.4 mM Cyanine label per 40 pmoles DNA, 2.5 mM CoCl2, 0.2 M potassium cacodylate, 25 mM Tris-HCl, and 0.25 mg/mL BSA; overnight reaction at 37°C). One strand was digested with Lambda exonuclease (5 units Lambda exonuclease per 2 μg DNA, 67 mM Glycine-KOH, 2.5 mM MgCl2, 50 μg/mL BSA; 1-h incubation at 37°C). The single-stranded product was then reduced to less than 3 μL by use of a YM-30 Microcon filter device and brought to a final volume of 35 μL by addition of hybridization solution.
The array was pre-soaked in hybridization solution containing 50 mM 2-[N-morpholino]ethanesulfonic acid (MES), 250 mM NaCl, and 0.1% Tween-20 at room temperature for 20 min, and allowed to air dry. The labeled sample was denatured at 95°C for 10 min, immediately cooled in an ethanol-dry ice bath, and allowed to thaw on ice. The 35-μL sample was then applied to the array and incubated at 42°C for 1 h. After hybridization, the array was washed once at 42°C for 5 min with a solution of 2× SSC and 0.1% Tween-20, followed by a 5-min wash at 42°C with 1× SSC and 0.1% Tween-20. The array was scanned at 532 and 635 nm with a GenePix 4000a microarray scanner.
GenePix software was used to generate a digitized intensity table for each of the features on the array. An Excel macro in combination with visual inspection of the intensity data was then used to score the SNPs. The macro makes base calls by first establishing background levels for each SNP by averaging the intensity levels on the two alleles with the least hybridization. This background level is then subtracted from the hybridization intensity for the two remaining alleles. If the adjusted intensity of the allele with the most hybridization is more than twice that of the adjusted intensity of the remaining allele, the macro calls that allele; otherwise the macro calls both bases.
SSLP Markers
SSLP analysis was performed as described (Talbot and Schier 1999) to confirm locations derived from microarray data for previously unmapped mutations.
Acknowledgments
We thank scientists at Protogene, particularly S. Lott, T. Yang and J. Todd, for microarray construction and advice on hybridization conditions; J. Postlethwait for providing C32 and SJD DNA samples; C.-B. Chien, C. Kimmel, L. Maves and M. Westerfield for supplying mapping crosses; P. Chang and K. Lenkov for technical assistance; and A. Schier for comments on the manuscript. H.L.S. and I.G.W. were supported by predoctoral fellowships from HHMI. This work was supported by NIH grant RR12349 (W.S.T.). W.S.T. is a Pew Scholar in the Biomedical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL talbot@cmgm.stanford.edu; FAX (650) 725-7739.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.777302.
REFERENCES
- Barbazuk WB, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell JA, McPherson JD, Johnson SL. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Suzuki T, Senti KA, Stubbs J, Schaffner G, Dickson BJ. Genetic mapping with SNP markers in Drosophila. Nat Genet. 2001;29:475–481. doi: 10.1038/ng773. [DOI] [PubMed] [Google Scholar]
- Butler JH, Cronin M, Anderson KM, Biddison GM, Chatelain F, Cummer M, Davi DJ, Fisher L, Frauendorf AW, Frueh FW, et al. In situ synthesis of oligonucleotide arrays by using surface tension. J Am Chem Soc. 2001;123:8887–8894. doi: 10.1021/ja003758r. [DOI] [PubMed] [Google Scholar]
- Cho RJ, Mindrinos M, Richards DR, Sapolsky RJ, Anderson M, Drenkard E, Dewdney J, Reuber TL, Stammers M, Federspiel N, et al. Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat Genet. 1999;23:203–207. doi: 10.1038/13833. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Geisler R, Rauch GJ, Baier H, van Bebber F, Brobeta L, Dekens MP, Finger K, Fricke C, Gates MA, Geiger H, et al. A radiation hybrid map of the zebrafish genome. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Guo Z, Gatterman MS, Hood L, Hansen JA, Petersdorf EW. Oligonucleotide arrays for high-throughput SNPs detection in the MHC class I genes: HLA-B as a model system. Genome Res. 2002;12:447–457. doi: 10.1101/gr.206402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robbins CM, et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Phan AC, Naeemuddin M, Mapa FA, Ruddy DA, Ryan JJ, Young LM, Wells T, Kopczynski C, Ellis MC. Single nucleotide polymorphism markers for genetic mapping in Drosophila melanogaster. Genome Res. 2001;11:1100–1113. doi: 10.1101/gr.178001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukriede N, Fisher D, Epstein J, Joly L, Tellis P, Zhou Y, Barbazuk B, Cox K, Fenton-Noriega L, Hersey C, et al. The LN54 radiation hybrid map of zebrafish expressed sequences. Genome Res. 2001;11:2127–2132. doi: 10.1101/gr.210601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Zon LI. Genetic backgrounds and some standard stocks and strains used in zebrafish developmental biology and genetics. Methods Cell Biol. 1999;60:357–359. doi: 10.1016/s0091-679x(08)61910-x. [DOI] [PubMed] [Google Scholar]
- Kelly PD, Chu F, Woods IG, Ngo-Hazelett P, Cardozo T, Huang H, Kimm F, Liao L, Yan YL, Zhou Y, et al. Genetic linkage mapping of zebrafish genes and ESTs. Genome Res. 2000;10:558–567. doi: 10.1101/gr.10.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok PY. Methods for genotyping single nucleotide polymorphisms. Annu Rev Genomics Hum Genet. 2001;2:235–258. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Winchester E, Daly MJ, Wang DG, Hirschhorn JN, Laviolette JP, Ardlie K, Reich DE, Robinson E, Sklar P, et al. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat Genet. 2000;24:381–386. doi: 10.1038/74215. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Finney JE, Keating MT, Johnson SL. Assessment of polymorphism in zebrafish mapping strains. Genome Res. 1999;9:1231–1238. doi: 10.1101/gr.9.12.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL. PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Pollard SM, Saude L, Feldman B, Coutinho P, Hirst EM, Stemple DL. Zebrafish mutants identify an essential role for laminins in notochord formation. Development. 2002;129:3137–3146. doi: 10.1242/dev.129.13.3137. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL. Patterns of nucleotide substitution in Drosophila and mammalian genomes. Proc Natl Acad Sci. 1999;96:1475–1479. doi: 10.1073/pnas.96.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Johnson SL, Midson CN, Talbot WS, Gates M, Ballinger EW, Africa D, Andrews R, Carl T, Eisen JS, et al. A genetic linkage map for the zebrafish. Science. 1994;264:699–703. doi: 10.1126/science.8171321. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H.J. 1998. Primer3. Code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html.
- Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, de Sauvage F, Jacob H, Fishman MC. Zebrafish genetic map with 2000 microsatellite markers. Genomics. 1999;58:219–232. doi: 10.1006/geno.1999.5824. [DOI] [PubMed] [Google Scholar]
- Swan KA, Curtis DE, McKusick KB, Voinov AV, Mapa FA, Cancilla MR. High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 2002;12:1100–1105. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon-Miller P, Kwok PY. A high-density single-nucleotide polymorphism map of Xq25-q28. Genomics. 2000;65:195–202. doi: 10.1006/geno.2000.6165. [DOI] [PubMed] [Google Scholar]
- Talbot WS, Hopkins N. Zebrafish mutations and functional analysis of the vertebrate genome. Genes & Dev. 2000;14:755–762. [PubMed] [Google Scholar]
- Talbot WS, Schier AF. Positional cloning of mutated zebrafish genes. Methods Cell Biol. 1999;60:259–286. doi: 10.1016/s0091-679x(08)61905-6. [DOI] [PubMed] [Google Scholar]
- Talbot WS, Trevarrow B, Halpern ME, Melby AE, Farr G, Postlethwait JH, Jowett T, Kimmel CB, Kimelman D. A homeobox gene essential for zebrafish notochord development. Nature. 1995;378:150–157. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, McCullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ, et al. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- Woods IG, Kelly PD, Chu F, Ngo-Hazelett P, Yan YL, Huang H, Postlethwait JH, Talbot WS. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]