Abstract
A machine that employs a novel reagent delivery technique for biomolecular synthesis has been developed. This machine separates the addressing of individual synthesis sites from the actual process of reagent delivery by using masks placed over the sites. Because of this separation, this machine is both cost-effective and scalable, and thus the time required to synthesize 384 or 1536 unique biomolecules is very nearly the same. Importantly, the mask design allows scaling of the number of synthesis sites without the addition of new valving. Physical and biological comparisons between DNA made on a commercially available synthesizer and this unit show that it produces DNA of similar quality.
There is a growing need for automated, high-throughput chemical synthesizers in biotechnology. Uses of these machines include combinatorial library synthesis and high-throughput synthesis of biopolymers. This need is dramatically illustrated by the large and growing demand for single-stranded DNA or oligonucleotides (often simply referred to as “oligos”) (Williams et al. 1988; Niece et al. 1991; Ivanetich et al. 1993; Pon et al. 1994; Hager et al. 1999; Goforth 2002). Oligos are used as primers in the polymerase chain reaction (PCR) technique (Meldrum 2000a), for the de novo chemical synthesis of genes (Kenneth et al. 1988; Dietrich et al. 1998), and for spotted DNA microarrays (Meldrum 2000b). Their use has risen significantly over the last decade (Jaklevic et al. 1999) and is expected to increase even more, driven largely by the genome sequencing of a number of organisms. As a particular example, even if only two PCR primers (forward and reverse) were needed to amplify all ∼35,000 genes (Lander et al. 2001; Venter et al. 2001) in the human genome, ∼70,000 PCR primers would be needed in total. The increasing use of chemical synthesis of genes and the expanding use of RNAi (Tuschl 2001) will only increase this demand. The design of commercially available DNA synthesizers is inconsistent with this demand; these machines have largely reached the limit of their economy of scale. A significant increase in throughput leads to only marginal benefits in total synthesis time and/or cost, but is invariably associated with a significant increase in the complexity of the machine, notably valves, and an associated increase in probability of malfunction. As a result, our development of the machine described in this paper, which has a much larger economy of scale, was focused on its use as a DNA synthesizer. Increasing the total number of oligos synthesized with this machine (e.g., from 96 to 384 to 1,536) has only a minor effect on total synthesis time without any notable increase in the complexity of the machine.
The standard chemistry used in DNA synthesis, phosphoramidite (PA) chemistry, consists of repeating a synthesis protocol of four basic steps: deblock, couple, cap, and oxidize (Gait 1984). Each cycle through these steps adds one base [adenine (A), guanine (G), cytosine (C), or thymine (T)] to an oligo. When many oligos, each immobilized on a solid support [e.g., controlled-pore glass (CPG)] at a unique synthesis site, are being made in parallel, there must be a way to deliver reagents to some oligos and not others. That is, each site must be addressable, without the possibility of cross-contamination between sites, because otherwise each oligo will be identical. State-of-the-art synthesizers fall into two main categories, distinguished by their method of reagent delivery. Machines from the first category synthesize into stationary columns (each column is a unique synthesis site), each with its own system of valves and tubing [e.g., the design employed by an Applied Biosystems 394 synthesizer (Models 392 and 394 DNA/RNA Synthesizers User's Manual, 1992)]. Although this design is highly reliable, scaling up this design gives limited benefits. For example, although scaling up a one-column machine to a two-column machine cuts the time to synthesize two oligos in half, the overall cost to make an oligo is unaffected. Further, this requires the addition of an entirely new set of valves and tubing for the new column, significantly increasing (i.e., roughly doubling) the complexity of the machine.
DNA synthesizers that fall into the second category synthesize in standard multiwell plate formats (each well is a unique synthesis site) and currently offer the highest throughput. In this design, the plate is typically mounted on a combination of linear motion tables which position each well of the plate in succession below the outputs of a set of stationary reagent delivery valves, one for each synthesis reagent [Lashkari et al. 1995 (AMOS); Rayner et al. 1998 (MerMade)]. To increase the throughput of this design, the total number of wells in the plate can be increased. However, this leads to an increase in the total number of movements (and thus total synthesis time) needed to position each well below the delivery heads, without a decrease in cost. The total synthesis time can be decreased by the addition of more delivery valves for each reagent, but this adds unwanted complexity to the machine.
The current design of the new synthesizer is also plate-based. However, a key to its operation is the separation of the addressing of individual synthesis sites from the delivery of reagents to those sites. Instead of moving each well of a plate below each reagent output, a given reagent is simply poured, or flooded, over all synthesis sites at the same time. To address a reagent to a set of specific synthesis sites and no others, a mask is placed over the sites before the reagent is flooded. Reagent poured over the mask goes through the holes in the mask and ends up only at the synthesis sites sitting below. The techniques of flooding and masking are embodied by the two separate components of the synthesizer, the mask-making machine and the reagent-delivery machine.
RESULTS AND DISCUSSION
Overview
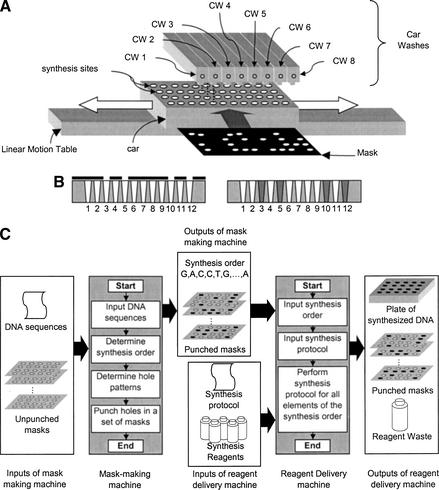
The basic operation of the reagent-delivery machine is illustrated in Figure 1A. Individual synthesis sites (i.e., one for each unique DNA sequence) are immobilized on a plate referred to as the car. The car is mounted on a single linear motion table that moves the car back-and-forth below a set of reagent delivery heads referred to as the car washes(CW); one CW for each reagent. CW heads do not address individual synthesis sites, but flood reagent over all sites. To address a reagent to individual sites, a mask is placed over the sites. A mask has holes in it (Fig. 1B) that allow a flooded reagent to flow only to those sites which sit below the holes (reagent which does not go through a hole is discarded as waste). Masks have holes punched in them by the mask-making machine. Once all the masks for a synthesis run are made by the mask-punching machine, they are transferred to the reagent-delivery machine, and once the reagent-delivery machine is done with them the masks will typically be discarded.
Figure 1.
Essence of the synthesizer. (A) Basic idea behind masking and flooding. (B) Cross-section through one row of the car (left) with a mask over the car and (right) after flooding and mask removal (only sites 3, 5, 10, and 12 received reagent). (C) Inputs and outputs of the mask-making and reagent-delivery machines.
Figure 1C shows the inputs and outputs of the mask-making machine and the reagent-delivery machine when used for DNA synthesis (here the deblock steps are masked; the alternative would be to mask coupling steps). The mask-making machine accepts as input a text file description of up to 384 unique oligonucleotide sequences and a stack of unpunched masks. [Currently each oligo sequence is a combination of the bases A, G, C, and T but the addition of other bases (e.g., dU, modified bases, etc.) requires only straightforward changes to the synthesizer]. This machine determines the synthesis order, that is, which base will be delivered during the first coupling step, during the second, and so on (e.g., G, A, C, C, T, G, … , A) and then punches a hole pattern in each mask—one mask for each step. In the first mask, there are holes only above those sites which will need the first base of the synthesis order (e.g., G), the second mask has holes only above those sites which will need the second base of the synthesis order (e.g., A), and so on. The reagent-delivery machine accepts as input the synthesis order and the stack of punched masks as well as the synthesis reagents and the synthesis protocol. Once all of the steps of the synthesis protocol are carried out for every base in the synthesis order, a synthesized plate of DNA, a stack of used masks, and a container of reagent waste have been generated. Unless some set of oligos resulting from a permutation of the masks (an unexpected situation) is to be made, the masks are now discarded along with the reagent waste.
The CBI synthesizer has been used for 59 DNA synthesis runs, of which 47 were successful. These runs were performed on an either 96-well or 384-well scale, and the percentage of successful sequences and the quality of those sequences varied significantly for each run. Under current operation, the highest percentage of successful sequences obtained was ∼92% as determined by mass spectrometry. This is not unexpected, since the instrument is a first-of-its-kind prototype designed to perform proof of principle experiments.
Table 1 compares the reagent usage and cost to synthesize 384 unique 20-mers (oligos which are 20 bases long) on the new CBI synthesizer and on a MerMade that is run in-house on a nearly daily basis for making oligos for use as PCR primers and for gene building. The CBI synthesizer can synthesize a single 384-well plate of oligos at one time, whereas the MerMade can synthesize, at most, two 96-well plates at once. Currently, the MerMade has an advantage in the overall cost (and thus the cost-per-base): 8 cents versus 13.5 cents. It should be noted that CBI was recently able to negotiate a purchase price of 25 cents per base for an order of 5000, 100-nmolar 35-mers (private quotation), and although this cost includes overheads not included in the comparison costs shown here, it does give a feel for the current state of the market. However, the CBI synthesizer is still only a prototype, and it is expected that design changes (discussed below) will drive down both the reagent usage and thus the cost-per-base. Another distinction between the CBI synthesizer and currently available synthesizers (whose synthesis time, as noted above, increases essentially linearly with the number of oligos being synthesized) is the total time required to synthesize a given number of oligos, as illustrated in Figure 2 by a comparison between the CBI synthesizer and the MerMade. The MerMade can run a single plate of 20-mers in ∼5 h and two plates in ∼7 h. For three plates, it must perform a two-plate run followed by a single plate run, and so on. Even increasing the number of wells in the plates does not resolve this increase in synthesis time, because each well must be addressed individually, which increases the total number of table movements. The CBI synthesizer can increase the number of synthesis sites without any dramatic increase in the synthesis time because of the way individual sites are addressed (i.e., the masking technique). The dependence of synthesis time on the number of oligos produced is based on a linear extrapolation of calculations, discussed further below.
Table 1.
Comparison of the CBI Synthesizer and the MerMade
| CBI | MerMade | |
|---|---|---|
| Reagent volumes (milliters) | ||
| Deblock | 1,500 | 1,228 |
| Each phosphoramidite | 455 | 200 |
| Activator | 1,550 | 1,160 |
| Cap A | 785 | 840 |
| Cap B | 785 | 700 |
| Oxidizer | 1,250 | 584 |
| Acetonitrile | 15,000 | 12,800 |
| Total (waste) | 22,690 | 18,112 |
| Other items | ||
| Number of Masks | 60 | NA |
| Argon (ml) | 27,000 | 24,000 |
| Labor (hours) | 4 | 4 |
| CPG (mg) | 768 | 768 |
| Costs | ||
| Reagents | $848.69 | $494.00 |
| Other items | $185.63 | $120.60 |
| Total | $1,034.32 | $614.60 |
| Cost-per-base | $0.135 | $0.080 |
These numbers are for the synthesis of 384 20-mers on a 60-nmol starting synthesis scale. The CBI synthesizer can make 384 20-mers in a single run, whereas the MerMade can make one or two 96-well plates at a time. The masks constitute 5.8% of the total cost to run the CBI machine, and the phosphoramidite concentration has been taken to be 5 g/200 mL of acetonitrile.
Figure 2.
Time comparison between the CBI synthesizer and the MerMade. This figure illustrates the advantage the former gives over any synthesizer whose synthesis time increases linearly with the number of oligos. These numbers are based on synthesis of unique 20-mers. The number of wells in the CBI machine's plate changes, whereas the MerMade is expected to run one or two 96-well plates at a time. Further, it is assumed that the mask-making machine and reagent-delivery machine are run in parallel.
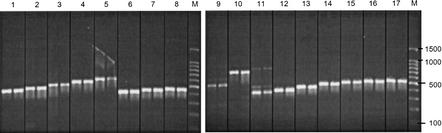
Sixty-eight PCR primers (two copies of 34 unique primers, 17 forward and 17 reverse) were made on the new synthesizer and compared to identical primers made on our in-house MerMade (this run of the CBI synthesizer was the “best” in terms of percentage of successful sequences and the quality of those sequences). Primers from both machines were used in PCR reactions to amplify Mycobacterium tuberculosis (TB) genes. A side-by-side comparison of the agarose gel analysis of these reactions is shown in Figure 3. Although the new synthesizer had an average stepwise yield of only 97.7% (compared to ∼99.5% on the MerMade) with OD readings that varied significantly throughout the plate, this figure clearly shows that the oligos made by the CBI synthesizer can be used as PCR primers with results similar to those made on the MerMade. The variation of OD readings may be due to inconsistencies in the synthesis columns (which for this synthesis run were hand-made from pipette tips, see below) that led to some synthesis reagents draining more rapidly than others, or inconsistencies in reagent delivery to the columns (which may be rectified by the design of the CW heads). Further, we suspect that the stepwise yield of the new machine can be brought to the 98%–99% level by reducing the volume of both the argon chamber and the plumbing to restrict exposure of synthesis reagents to water vapor.
Figure 3.
Comparison of primers made on the new synthesizer (left lanes) and a MerMade (right lanes). Primers were used in a standard 100-μL PCR reaction with 10 ng of TB genomic DNA as a template. A sample of each PCR reaction was analyzed on a 1.5% agarose gel. The lane labeled “M” is a marker lane with markers at the masses indicated on the right (measured in base pairs). The “slanted” bands in lane 5 and the “nonslanted” bands in lane 11 are consistent with nonspecific amplification. Because this amplification occurs with products from both machines, this is attributable to an artifact of the PCR reaction itself and not improper operation of either machine.
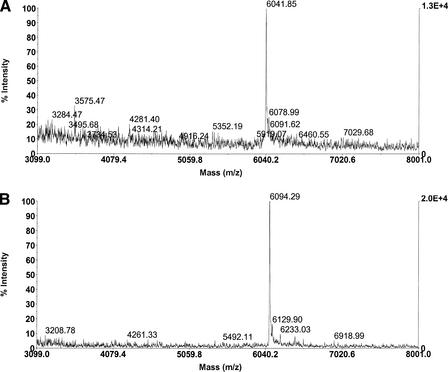
Further analysis of DNA synthesized on the CBI synthesizer shows that masks that do not lead to cross-contamination between wells have been successfully tested. In Figure 4, MALDI spectra (matrix-assisted laser desorption time-of-flight mass spectroscopy) for two unique DNA sequences synthesized in adjacent wells is shown. There is no evidence that the full-length product expected in one well is evident in the other.
Figure 4.
MALDI (mass spectrometer) spectra of two 20-mer oligos made in adjacent wells on the new synthesizer. Theoretical peaks for these spectra are (A) 6,043.0 and (B) 6,098.0 [all peaks are measured in Daltons/(electronic charge)]. The differences between the observed and theoretical peaks are due to slight calibration errors. Both spectra are very “clean” (i.e., they contain insignificant impurities).
Reagent-Delivery Machine Design
The reagent-delivery machine is shown in detail in Figures 5 and 6. Unique oligos are synthesized at specific locations (currently 96 or 384, but any number is possible) on a synthesis chuck referred to as a car (Figs. 5A, 6A). The oligos are linked to solid support [controlled-pore glass (CPG) has been used] which is loaded into the car prior to the start of a synthesis run in one of two forms: (1) prepacked between, or immobilized above, fluid-porous filter membranes, referred to as frits, either in commercially available DNA synthesis columns (volume above the top frit of ∼220 μL) or in custom-made columns consisting of a pipette tip that has had a frit placed into it by hand (volume above the top frit of ∼60 μL), or (2) prepacked between, or immobilized above, frits placed in each well of a custom-made 384-well car (volume above the top frit of ∼75 μL). There are eight CW heads (Figs. 5B, 6B), one each for deblock, acetonitrile (ACN), cap A and cap B combined, oxidizer, and one for each combination of activator (Act) and phosphoramidite (dA + Act, dG + Act, dC + Act and T + Act). Each CW head has a single inlet plus many outlets, typically equal to the number of rows in the car (i.e., 16 for a 384-well car). Reagent flow through the head is controlled by a set of solenoid valves whose configuration depends on the reagent delivered by the CW (Figs. 5C, 6C). All arrangements allow the CW to be flushed with acetonitrile. Two methods of reagent delivery have been tested using the CWs, one in which a CW delivers reagent continuously as the car goes by underneath it, and another in which the CW delivers only when the car is in a set of discrete positions (i.e., 12 for a 96-well plate, 24 for a 384-well plate, etc.), called the stop-and-squirt method. The car is rigidly mounted on a vacuum chuck (Figs. 5D, 6D), which itself it mounted on the “x” linear motion table (Figs. 5E, 6E) so that reagents can be removed from the synthesis sites by pulling vacuum below all of them at once. Reagents flow past the porous frit; the solid support cannot. A prototype mask-changing device automatically places a mask, which it retrieves from a stack of new masks (Figs. 5F, 6F), on top of the car prior to a deblock step, and removes the mask and places it in a stack of used masks (Fig. 5J) following this step (this device currently only handles a single 96-well mask). This device consists of a linear motion table (the “y-table”; Figs. 5G, 6G) that provides movement perpendicular to the x-table, an air cylinder (Figs. 5H, 6H) that provides vertical motion, and a pneumatic gripper (Figs. 5I, 6I) that is used to grab a mask. Synthesis is performed in an inert Argon atmosphere (∼12% relative humidity) enclosed by an airtight Plexiglas chamber. The current design of this chamber has gloves (Fig. 6J) built into one wall to allow for manual manipulation of materials inside the chamber. The current reagent-delivery machine prototype itself has a footprint of 168 cm × 69 cm × 196 cm (length × width × height).
Figure 5.
Reagent delivery machine schematic. Components include (a) the car, (b) the CWs, (c) sets of solenoid valves, (d) the vacuum chuck, (e) the x-table, (f) stack of new masks, (g) the y-table, (h) an air cylinder, (i) a pneumatic gripper, and (j) the stack of new masks. For clarity, the end view focuses on the mask-changing device (i.e., the CW heads are not shown).
Figure 6.
The reagent-delivery machine. Components include (a) the car (b), the CWs, (c) set of solenoid valves, (d) the vacuum chuck, (e) the x-table, (f) stack of new masks, (g) the y-table, (h) the air cylinder, (i) the pneumatic gripper, and (j) the pair of gloves.
Reagent-Delivery Machine Operation
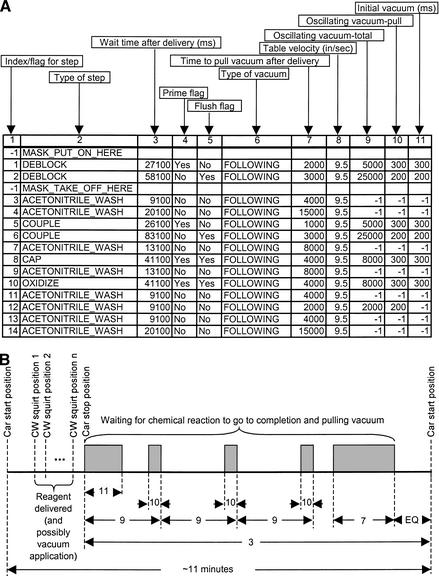
The operation of the reagent-delivery machine is driven by a custom Visual Basic program that repeats all the steps of the synthesis protocol (Fig. 7A) for every base of the synthesis order, automates most of the pre- and postsynthesis procedures, and allows the user to perform simple maintenance operations. Each step of the synthesis protocol is described by 11 different items, illustrated in Figure 7A: (1) an index which doubles as a flag that indicates when a mask is to be placed over or removed from the car, (2) the reagent to be delivered (i.e., deblock, acetonitrile, etc.), (3) how long to wait (in msec) after the reagent has been delivered, (4) a flag that indicates if a CW head is to be primed prior to reagent delivery, (5) a flag that indicates if a CW head is to be flushed following delivery, (6) the table speed in inches per sec, and several items which constitute a detailed description of how the vacuum is to be pulled below the car. Figure 7B schematically shows the components of a single pass through the protocol. The car starts at “car start position”, moves to a succession of CW squirt positions (1, 2, … , n where n = 12 for a 96-well plate, 24 for a 384-well plate, etc.) and at each, the necessary CW head squirts. Vacuum can be applied below the car during reagent delivery, following reagent delivery, during and following reagent delivery, or not at all (as specified by the flag in Fig. 7A, column 6). Pulling vacuum after reagent delivery starts as soon as the car arrives at “car stop position” and can be done in a combination of several different ways; immediately following the arrival of the car at “car stop position” for a time given in Figure 7A, column 11, in a cyclic or oscillating fashion [i.e., on for a while (for a time given by Fig. 7A, column 10) and then off for a while (for a time given by the difference between Fig. 7A, columns 9 and 10)] or after a long wait (Fig. 7A, column 7). After vacuum is pulled, the pressure in the vacuum chuck is equalized with that inside the Ar chamber for a period of time “EQ” which is set inside the computer program and is typically 5000 msec (depending on the columns being used, etc). For the sake of simplicity, prime and flush steps performed prior to the car arriving at the car start position and during the wait time following delivery respectively are not shown in Figure 7B.
Figure 7.
Synthesis protocol (stop-and-squirt). (A) Steps of the protocol with descriptions of each column, and (B) the general time profile of most components of the protocol (prime and flush operations are excluded). The numbers in (B) refer to the column indices of (A); shaded regions indicate when vacuum is being pulled, and EQ refers to vacuum equalization time. The steps of the protocol shown in (A) take ∼11 min to perform.
The protocol shown in Figure 7A was developed by trial-and-error; a protocol was used to synthesize oligos, those oligos were tested (using MALDI and/or HPLC), and a new protocol was developed based on the results of these tests until the “best” one was found. Consideration of a single step of this protocol will serve to illustrate the information contained in the columns described above. During step 2, deblock is flooded over the top of the car, which has already been covered by a mask. Once the car makes it to the “car stop position”, several things occur simultaneously: (1) a timer starts counting down 58,100 msec, (2) the “Yes” flush flag for this step leads to the car beginning its movement to a position that allows acetonitrile to be passed through the deblock CW head and dumped into waste, thereby cleaning, or flushing, the CW head, (3) vacuum is applied under the car and will continue to be applied for 300 msec (Fig. 7A, column 11), and (4) another timer, one to time the application of oscillating vacuum, is started. After this oscillating vacuum timer has waited 24,800 msec (= 25,000 msec − 200 msec; Fig. 7A, columns 9 and 10, respectively), vacuum is applied for 200 msec (Fig. 7A, column 10). This same timer triggers another oscillating vacuum pulse at 49,800 msec (= 2 × 25,000 msec − 200 msec). Before this timer can trigger another, the final vacuum pull of 3000 msec (Fig. 7A, column 7) has already come and gone, having started at 53,100 msec (= 58,100 msec − 5000 msec, Fig. 5A, column 3, and EQ, which was given above as typically 5000 msec), and the vacuum chuck has equalized itself. Once the equalization is complete, the mask is removed and an acetonitrile wash step (index = 3) begins. The sum of all the wait times in the protocol of Figure 7A is 379,400 msec (= 6.32 min), but in practice a single pass through the protocol takes ∼11 min to complete due to the times needed to perform table movements, deal with the mask, etc., which are not included in the protocol.
Mask Design
An essential aspect to the design of the new synthesizer is the way in which masks form a seal over synthesis sites to stop cross-contamination between sites. A number of techniques were investigated, but only two deserve description here. The first involved a 384-well polypropylene (PP) mask (which can currently be made at a cost of $1.00/mask for a quantity of 7500 masks) cast from a custom-made mold (which cost ∼$10,000 to design and manufacture, Project Design Services) that was pressed down tightly into the wells drilled into the 384-well car. However, because (1) the useful lifetime of several synthesizer components was shortened due to the force required to press the mask, and (2) inherent cross-contamination problems with this design existed, this technique was abandoned.
The immobilization of synthesis sites in individual columns (either commercially available or custom-made from pipette tips), the columns themselves immobilized in the wells of a car (Fig. 8A), inherently reduces the possibility of cross-contamination between wells, because a more circuitous path must be traversed by a reagent that has escaped from one well before it can enter another. Instead of a flat plane between wells, a reagent must now either (1) exit the top of a column, travel down its outside edge, across the surface of the car, up the outside edge and then into the next column, or (2) exit the top of a column, travel along the underside of the mask and then enter another column. A further advantage to this method is that a mask can simply be placed on top of the sites without any significant force required to form a seal. Two mask orientations were tested. In the first, the mask is oriented as in Figure 8B—the wells of the mask are “concave-up” and do not penetrate into the tops of the columns. This was successfully tested using the 384-well PP masks mentioned above sitting on top of 96 custom-made columns made of pipette tips (it was using this technique that the PCR primers analyzed in Fig. 3 were synthesized). Little or no cross-contamination was observed using this technique, which was probably partly due to the large spacing between pipette tips (the masks have 384 wells and only 96 pipette tips were used, spaced on a 384-well spacing). Another masking technique, a lid-and-column design, was successfully tested in two implementations: (1) a set of 384 modified pipette tips with 384-well PP masks that were “upside-down” (Fig. 8C), and (2) a set of 96 commercially available columns (slightly modified for this test—a lip running around the top of each column was removed prior to the start of the run using a milling machine) with a custom-made mask (Fig. 8D). The successful tests of the lid-and-column design at two well densities (96 and 384) implies that this design is scalable to higher numbers of wells, in particular, 1536.
Figure 8.
Cross-sections of various mask designs using columns (masks are black in B–D). (A) Individual columns are placed into the wells of a car by hand. A 384-well PP mask (B) sitting on top of 96 columns custom-made from pipette tips, and (C) used in a “lid-and-column” design on top of 384 pipette tips. (D) A 96-well machined Delrin mask used in a “lid-and-column” design on top of 96 columns (with upper and lower frits) whose top lips have been removed with a milling machine.
Mask-Making Machine Design
The mask-making machine (Figs. 9 and 10) has been specifically designed to punch holes in the cast 384-well PP masks described above. The heart of this device is a custom-built stainless steel chuck (Figs. 9A, side view and 10A) with 384 spring-return stainless steel pins (Figs. 9B, side view and 10B) arranged in an array of 16 × 24 on centers which are 0.178" apart (i.e., the same pattern as the 384 wells in the PP mask mold). This chuck has a drawer (Figs. 9C, side view and 10c) that slides below the spring-return pins which accommodates a mask (Figs. 9D, side view and 10D). With the drawer pulled out (pushed in), a mask can be placed on the drawer (the mask sits below the spring-return pins). The chuck is mounted on a linear motion table (Figs. 9E, side view and 10E) that allows it to be moved below a set of 16 air cylinders (Figs. 9F, side view and 10F) that are used to drive specific pins through wells of the mask. There is one air cylinder for each row, A–P, of a 384-well mask arranged in the pattern shown in Figure 9 (top) and (detail). Sixteen individually controllable three-way valves (arranged as two manifolds of eight apiece; Figs. 9G, side view and 10G) are used to control the supply of air to the air cylinders.
Figure 9.
Schematic of the mask-making machine. Components in the side view include (a) the stainless steel chuck, (b) 384 spring-return pins, (c) the drawer (shown pushed all the way under the pins), (d) mask, (e) a linear motion table, (f) air cylinders, (g) three-way valve. (Top) The linear motion table, spring-return pins and air cylinders. (Detail) Arrangement of the air cylinders with respect to the 384 pins.
Figure 10.
The mask-making machine. Components include (a) the stainless steel chuck, (b) 384 spring-return pins, (c) the drawer (shown pushed half way under the pins with a mask sitting on top), (d) mask (only one is shown), (e) a linear motion table, (f) 16 air cylinders, (g) two manifolds of eight three-way valves (one on either side of the linear motion table) and (h) the relays. A 12" ruler is shown for scale.
Mask-Making Machine Operation
Operation of the mask-making machine is controlled by a custom Visual Basic program. This program has procedures to determine the synthesis order for up to 384 unique oligo sequences, allow the user to inspect the hole pattern for each individual mask, and automate the punching process for each mask. To punch holes in a mask, a mask is placed into the drawer by hand. The stainless steel chuck is automatically moved to 44 discrete positions below the air cylinders (44 due to the distance between the air cylinders as shown in Fig. 9, detail). At each position, those air cylinders which sit above a well that needs to be punched are fired, one at a time. Once the mask is complete, the user removes the punched mask from the drawer and replaces it with an unpunched one until the entire set has been punched. This entire process takes ∼3 min per mask on the current implementation of this machine. In future designs, a mask-changing device similar to the one already developed for the reagent-delivery machine will automate placement and removal of the mask from the drawer.
Mask Hole Patterns
It is of benefit to any operator of the synthesizer to minimize the total synthesis time in order to increase the total number of oligos that can be synthesized in a given period of time, and to minimize the total amount of reagent usage so as to minimize cost. Because the total synthesis time consists of the time for presynthesis procedures (which includes hooking up reagent bottles, checking that all CW heads fire correctly, and purging the Ar chamber of air), plus the time needed to repeat every step of the synthesis protocol for every mask, and the time for postsynthesis procedures (which includes flushing all CW heads with acetonitrile after the run and collecting the synthesized oligos), both the time issue and the reagent use issue can be addressed by minimizing the number of times the synthesis protocol needs to be repeated. Each repetition of the protocol calls for a single mask, and thus judiciously choosing the hole pattern for each mask can minimize the number of repetitions.
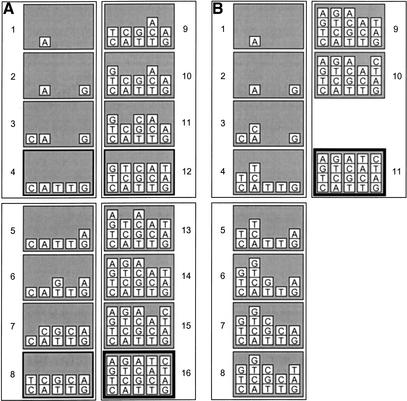
A relatively simplistic algorithm for determining the hole pattern in each mask, referred to as the naive algorithm, leads to a mask set that contains four times the length of the oligos being synthesized regardless of the total number of oligos (given that all are completely random, all have the same length and >∼24 oligos are being synthesized; see below). Thus, 80 masks will be needed for synthesis of either 384 20-mers or 1536 20-mers. Such an algorithm adds bases in a repetitive order of the four bases (e.g., AGCTAGCT…) and ensures that all oligos have their first base added before any of them have their second base added, and so on (i.e., all oligos have their mth base before adding their (m + 1)th). A non-naive algorithm adds bases in a synthesis order that also consists of repetitions through the bases A, G, C, and T. However, here a base is added to any oligo in need of it (the mth base of one is added at the same time as the (m + 1)th of another). Both algorithms skip a base in the order if none of the oligos need it. The distinction between these algorithms is illustrated in Figure 11 by the synthesis of five 4-mers; CTGA, ACTG, TGCA, TCAT, and GATC. The naive algorithm uses the synthesis order AGCTAGCTAGCTAGCT whereas the non-naive algorithm arrives at a synthesis order of AGCTAGC TACT (i.e., three repetitions of AGCT without the G in the last cycle). The naive algorithm uses 16 steps as opposed to 11 in the non-naive. In practice, the non-naive algorithm is employed, but instead of repetitions of AGCT, all possible synthesis orders consisting of repetitive sequences of the 24 (= 4! = 4 × 3 × 2 × 1) possible cyclic permutations of these four bases (AGCTAGCTAGCT… , AGTCAGTCAGTC… , etc.) are tested in silico prior to the start of the run to determine which of them leads to the smallest number of steps in the synthesis order. Such a technique is referred to as a non-naive-with-four-base-permutation-search algorithm (NN4S). The one with the least number of steps is used as the synthesis order. Other techniques to determine the synthesis order include (1) maximizing the total number of oligos that receive a base during a given step, and (2) testing permutations of groups of five (or more) bases (i.e., all permutations of AAGCT, AGGCT, AGCCT, and/or AGCTT). These either do not give any advantage over the NN4S method or have not yet been tested.
Figure 11.
Synthesis steps for five oligonucleotide sequences (CTGA, ACTG, TGCA, TCAT, and GATC) for two different mask-punching algorithms. The method used in (A) requires all oligos to have their first base before any of them receive their second. The method used in (B) allows the oligos to be grown at different rates. Bases were added in the order AGCTAGCT… for both algorithms. The “hole” between steps 10 and 11 in (B) indicates that no oligos needed G at that point.
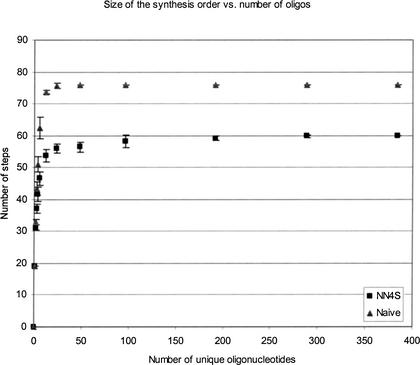
The advantage that the NN4S algorithm has over the naive algorithm is illustrated in Figure 12. There, the number of steps (an average over five individual data sets for each data point) in the synthesis order required to synthesize 0, 1, 2, 3, 4, 6, 12, 24, 48, 96, 192, 288, and 384 unique 20-mers (generated using the random number generator Rnd in Microsoft VB) has been empirically determined using a naive and an NN4S algorithm. (These oligos were assumed to start on standard CPG, and thus the first base is already attached). The behavior of these two curves separates above and below ∼24 oligos. Above ∼24 oligos, the naive algorithm always has 4 × 19 = 76 steps in the synthesis order, whereas the NN4S algorithm assumes the form of a slowly increasing function with only 60 steps in the synthesis order for 384 unique 20-mers. Modeling this function as a line leads to only ∼65 steps in the synthesis order for 1536 unique 20-mers. This is a significant improvement over the 76 required by the naive algorithm. The behavior of both curves below ∼24 oligos has its root in the fact that the probability that a given base in each cycle through the bases of the synthesis order (e.g., through A, G, C, and T, in the synthesis order AGCTAGCT…) can be skipped is no longer almost identically zero. This is most easily illustrated using the naive algorithm. To synthesize a single oligo using the naive algorithm, a 19-step synthesis order identical to the sequence of the oligo (i.e., without the first base because the first base is on the CPG) is used, and every one-well mask has a hole in it. That is, three of the four bases in each cycle of the repetitive order can be skipped (e.g., if a C is called for, then A, G, and T can be skipped). The synthesis of two oligos does not require 2 × 19 = 38 steps, because now either three or two of the four bases for every cycle of the repetitive order can skipped (e.g., if both oligos 1 and 2 need a C, then A, G, and T can be skipped but if oligo 1 needs a C and oligo 2 needs a T, then only A and G can be skipped). Given that some base is to be added to the first oligo (e.g., A), the probability that the second oligo does not need that base (e.g., G, C, or T) and thus requires a different mask, is 75%. Thus, if a synthesis order with 19 steps is required to synthesize one oligo, two should require 1.75 × 19 = 33.25, which is within one standard deviation of the 32.8 ± 0.84 steps found empirically. Similar analyses can be done for more and more oligos until finally the “above-24 oligos” behavior is obtained.
Figure 12.
Number of steps in the synthesis order for 0, 1, 2, 3, 4, 6, 12, 24, 48, 96, 192, 288, and 384 unique 20-mers when the order is calculated using naive and non-naive-with-four-base-permutation-search (NN4S) algorithms. Five independent sets of oligos were calculated at each data point. Error bars indicate standard deviations.
Scalability and Complexity
Reconfiguring the synthesizer to make different numbers of oligos (i.e., to scale the machine) is not complicated and is not restricted to conventional plate dimensions or densities (i.e., 96, 384, and 1536). First and foremost, the reagent-delivery machine does not need any more valves, any more linear motion tables, or any more electromechanical relays. Further, none of the hardware that does need to be changed has moving parts. This includes the car, the mask, and all CWs because, in the current implementation of the machine they all depend on the total number of rows (8, 16 or 32) in the array of oligos (8 × 12 for 96, 16 × 24 for 384, 32 × 48 for 1536). The software needs to be altered relative to the change so that the appropriate synthesis protocol will be used and, further, if the stop-and-squirt reagent-delivery technique is used, the correct number of squirt positions are employed. The mask-making machine would require straightforward changes reflecting the alteration in mask dimensions. These include changing the number of spring-loaded pins (e.g., from 384 to 1536), and the number of air cylinders (e.g., from 16 to 32) and associated three-way valves and relays.
Scalability and Time
As illustrated in Figure 2, the time required to synthesize 96, 384, or 1536 oligos is very nearly the same. The reason for this is twofold. First, because the reagent-delivery machine repeats the steps of the synthesis protocol for every mask and because the total number of masks is nearly independent (see Fig. 12) of the total number of oligos being synthesized, the operation of the reagent-delivery machine is nearly independent of the total number of synthesis sites. A single run of the reagent-delivery machine (including pre- and postsynthesis procedures) for the synthesis of 384 unique 20-mers is ∼13 h with the current protocol [11 h for actual synthesis (11 min per cycle through the synthesis protocol and 60 cycles) plus 2 h for pre- and pos-synthesis operations]. This is similar to commercial synthesizers (see Fig. 2). To make 1536 unique 20-mers will require ∼14 h (11 min per cycle, 65 cycles plus pre- and postsynthesis operations), a dramatic improvement over existing synthesizers whose total time essentially scales linearly with the total number of oligos produced (the MerMade requires ∼63 h). The slow increase in synthesis time as a function of the total number of oligos produced by the new synthesizer will last so long as the mask-making machine can punch a set of masks quicker than the reagent-delivery machine can go through all the required steps of the protocol. That is, as long as the two machines can be run in parallel, a new run on the reagent-delivery machine starting as soon as one finishes, the time to synthesize a given number of oligos will be solely dictated by the reagent-delivery machine. Thus, even though the time required to punch a set of masks does scale essentially linearly with the total number of oligos, this benefit will still yield an advantage for more than 1536 oligos. The current prototype of the mask-making machine takes ∼3 h to punch a set of 60 384-well masks, which leads to ∼13 h to punch 65 1536-well masks. Straightforward improvements in the mask-making machine (including increasing table speed, firing more than one air cylinder at once, and automating the mask-changing process) will decrease this time significantly.
Scalability and Cost and Synthesis Scale
As noted above, oligos (100-nmol scale) can currently be bought commercially at $0.25/base when sufficient quantities are purchased (see above). This is useful as a rough benchmark (since it includes overheads not in our costs) to compare with the current production cost per base for a run of the new synthesizer, performed on the 60-nmol scale, which is ∼$0.17 (this includes all chemical and mask costs (5.8% of the total cost) and the cost of a single technician who spends 4 h per day running the machine). This cost is low enough, despite the wasting of a significant portion of the flooded reagents, to make this machine competitive with existing technologies. Simple modifications to this machine can reduce this significantly. First, reduction of the phosphoramidite concentration (from 5g/125mL (of acetonitrile) to 5g/200 mL—the concentration used on the MerMade used in-house) will reduce the cost per base to ∼$0.135 (which is what is reported in Table 1). (Our concentration is currently so high to insure that reactions go to completion on our prototype). Second, optimizations to the synthesis protocol and modifications to the synthesizer design (most importantly a reduction in the dead volume of the fluidic lines, which is currently ∼10 mL from the outlet of the reagent bottle, through the valve system and through the CW head) are expected to reduce this cost per base to <$0.10. Purchasing synthesis chemicals in bulk will lead to even further reductions.
Another important scalability issue is exposed when one considers using, for example, the same volume of synthesis reagents required to synthesize 384 unique oligos to synthesize 1536. Although this reduces the synthesis scale by a factor of one-quarter, it also reduces the cost to make each oligo. That is, if 384 oligos were synthesized on a 60-nmol scale, 1536 oligos would be synthesized on a 15-nmol scale. This is still significantly more than is needed for a number of applications, including PCR reactions, and is still slightly larger than that offered by some commercial oligo vendors.
METHODS
Many of the parts used to build both the reagent-delivery machine and the mask-making machine have been obtained from commercial distributors. A single computer (Dell XPS T500, 392.6 MB of RAM, 12.7 GB hard drive) running Microsoft Windows NT version 4.00.1381 was used to control both the reagent-delivery machine and the mask-making machine, with separate custom programs written in Microsoft Visual Basic 6.0. Twelve-volt 2- and 3-way Teflon valves rated at either 30 psi or 100 psi used to control reagent delivery in the reagent-delivery machine were supplied by Cole Parmer (Vernon Hills, IL). Vacuum flow in the reagent-delivery machine and air flow to the air cylinders (Norgren RLC00J-SAN-AA00, 250 max psi, Littleton, CO) in the mask-making machine were controlled using stainless steel three-way valves (MAC valves, 35 series, Wixom, MI). Computer control over all these valves is afforded by electromechanical relays (WRODC5) and controlling boards (PCIDIO) obtained from Cyberresearch (New Haven, CT). Each machine had its own variable DC power supply (Elecno Electronics, Wheeling, IL) to provide power to the valves. All of the linear motion tables were obtained from Lintech (Monrovia, CA). The linear motion tables used in the reagent-delivery machine are 20" (“x” table) and 8" (“y” table) ball-screw tables, and both are driven by Parker-Hannifin (Sunnyvale, CA) SM231 servomotors. A 20" ball-screw table driven by a Parker-Hannifin (Sunnyvale, CA) SM233 servomotor is used in the mask-making machine. Each table has its own dedicated Parker-Hannifin TQ-10 servo amplifier. All tables are controlled by a single Parker-Hannifin AT6450 4-axis controller. The air cylinder (BIMBA EF2 series, 1" stroke, 40mm bore) and pneumatic gripper (phd, 19090-2-001) of the mask-changing device were obtained from Shepherd Controls (Allen, TX). The 230-L tank of liquid Argon (to supply the inert atmosphere) and three Argon tanks (each with 249 ft3 of gas) that supply pressure to the acetonitrile drum, the various reagent bottles (deblock, dA, dG, dC, T, activator, cap A, cap B, and oxidizer), and the air cylinder and pneumatic gripper of the mask-changing device were obtained from BOC Gases (Murray Hill, NJ).
DNA synthesis reagents were obtained from a variety of sources. Acetonitrile (anhydrous), deblock, cap A, cap B, activator, and ammonium hydroxide (40% w/w) used to cleave oligos from the CPG were obtained from Fisher Scientific (Suwanee, GA). Five-gram bottles of phosphoramidite were obtained from Glen Research (Sterling, VA) and Annovis (Aston, PA). CPG was obtained from Applied Biosystems. The prepacked commercial columns used for 96-well synthesis were obtained from BioSearch (Novato, CA). Custom synthesis columns were made from pipette tips (Finntip 50) obtained from Fisher Scientific, and polyethylene frit material was obtained from Small Parts (Miami Lakes, FL).
Standard protocols were used to perform PCR reactions and MALDI analyses. The latter were performed on a PerSeptive Biosystems Voyager-DE PRO mass spectrometer (Foster City, CA).
Acknowledgments
This work was funded by the Defense Advanced Research Projects Agency (DARPA), grant number N65236-99-1-5426, to S.A.J. Thanks are also given to Vickie Seward for performing the PCR reactions shown in Figure 3.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Eric.Livesay@UTSouthwestern.edu; FAX (214) 648-4156.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.359002.
REFERENCES
- Dietrich R, Wirsching F, Opitz T, Schwienhorst A. Gene assembly based on blunt-ended double-stranded DNA-modules. Biotechnology Techniques. 1998;12:49–54. [Google Scholar]
- Functions, Cycles and Procedures, 1992, Models 392 and 394 DNA/RNA Synthesizers User's Manual, version A. Foster City, CA: Applied Biosystems.
- Gait MJ. Oligonucleotide synthesis: A practical approach. Chapters 1 and 4. New York, NY: Oxford University Press; 1984. [Google Scholar]
- Goforth S. The core of DNA synthesis. The Scientist. 2002;16:43. [Google Scholar]
- Hager, K., Fox, J., Gunthorpe, M., Lilley, K., and Yeung, A. 1999. Survey of current trends in DNA synthesis core facilities. http://www.abrf.org/JBT/1999/December99/dec99hager.html. [PMC free article] [PubMed]
- Ivanetich K, Niece R, Rohde M, Fowler E, Hayes T. Biotechnology core facilities: Trends and update. FASEB J. 1993;7:1109–1114. doi: 10.1096/fasebj.7.12.8375609. [DOI] [PubMed] [Google Scholar]
- Jaklevic J, Garner H, Miller G. Instrumentation for the Genome Project. Annu Rev Biomed Eng. 1999;1:649–678. doi: 10.1146/annurev.bioeng.1.1.649. [DOI] [PubMed] [Google Scholar]
- Kenneth B, Logsdon N, Anderson R, Esponosa-Lara J, Maldonado-Rodriguez R, Frost J., III Gene synthesis technology: Recent developments and future prospects. Biotechnol Appl Biochem. 1988;10:510–521. [PubMed] [Google Scholar]
- Lander E, Linton L, Birren B, Nusbaum C, Zody M, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lashkari D, Hunicke-Smith S, Norgern R, Davis R, Brennan T. An automated multiplex oligonucleotide synthesizer: Development of high-throughput, low-cost DNA synthesis. Proc Natl Acad Sci. 1995;92:7912–7915. doi: 10.1073/pnas.92.17.7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldrum D. Automation for genomics, part one: Preparation for sequencing. Genome Res. 2000a;10:1081–1092. doi: 10.1101/gr.101400. [DOI] [PubMed] [Google Scholar]
- Meldrum D. Automation for genomics, part two: Sequencers, microarrays, and future trends. Genome Res. 2000b;10:1288–1303. doi: 10.1101/gr.157400. [DOI] [PubMed] [Google Scholar]
- Niece R, Beach C, Cook R, Hathaway G, Williams K. State-of-the-art biomolecular core facilities: A comprehensive. FASEB J. 1991;5:2756–2760. doi: 10.1096/fasebj.5.13.1916100. [DOI] [PubMed] [Google Scholar]
- Pon R, Buck G, Niece R, Robertson M, Smith A, Spicer E. A survey of nucleic acid services in core laboratories. Biotechniques. 1994;17:526–534. [PubMed] [Google Scholar]
- Rayner S, Brignac S, Bumeister R, Belosludtsev Y, Ward T, Grant O, O'Brien K, Evans G, Garner H. Mermade: An oligodeoxyribonucleotide synthesizer for high throughput oligonucleotide production in dual 96-well plates. Genome Res. 1998;8:741–747. doi: 10.1101/gr.8.7.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T. RNA interference and small interfering RNAs. Chembiochem. 2001;2:239–245. doi: 10.1002/1439-7633(20010401)2:4<239::AID-CBIC239>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Venter J, Adams M, Myers E, Li P, Mural R, Sutton G, Smith H, Yandell M, Evans C, Holtet R, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Williams K, Niece R, Atherton D, Fowler A, Kutny R, Smith A. The size, operation and technical capabilities of protein and nucleic acid core facilities. FASEB J. 1988;2:3124–3130. doi: 10.1096/fasebj.2.15.3192042. [DOI] [PubMed] [Google Scholar]