Abstract
We have measured, by reverse transcription and real-time quantitative PCR, the steady-state levels of the mitochondrial and nuclear transcripts encoding several subunits of the human oxidative phosphorylation (OXPHOS) system, in different normal tissues (muscle, liver, trachea, and kidney) and in cultured cells (normal fibroblasts, 143B osteosarcoma cells, 143B206 ρ0 cells). Five mitochondrial transcripts and nine nuclear transcripts were assessed. The measured amounts of these OXPHOS transcripts in muscle samples corroborated data obtained by others using the serial analysis of gene expression (SAGE) method to appraise gene expression in the same type of tissue. Steady-state levels for all the transcripts were found to range over more than two orders of magnitude. Most of the time, the mitochondrial H-strand transcripts were present at higher levels than the nuclear transcripts. The mitochondrial L-strand transcript ND6 was usually present at a low level. Cultured 143B cells contained significantly reduced amounts of mitochondrial transcripts in comparison with the tissue samples. In 143B206 ρ0 cells, fully depleted of mitochondrial DNA, the levels of nuclear OXPHOS transcripts were not modified in comparison with the parental cells. This observation indicated that nuclear transcription is not coordinated with mitochondrial transcription. We also observed that in the different tissues and cells, there is a transcriptional coregulation of all the investigated nuclear genes. Nuclear OXPHOS gene expression seems to be finely regulated.
[The following individual kindly provided reagents, samples, or unpublished information as indicated in the paper: G. Attardi.]
Through the process of oxidative phosphorylation (OXPHOS), mitochondria provide most of the ATP to eukaryotic cells. Oxidative phosphorylation corresponds to the terminal stage of substrate oxidation in mitochondria, and is catalyzed by the OXPHOS system, which functionally associates the mitochondrial respiratory chain, composed of four complexes (complexes I, II, III, and IV), and ATP synthase (complex V) for ATP synthesis. The biogenesis of all these enzymatic complexes, except complex II, requires expression of genes located at the level of both the nuclear and the mitochondrial genomes. Mechanisms that putatively coordinate the expression of these genes are poorly understood at present.
Basically, gene expression may be regulated through both transcriptional and posttranscriptional mechanisms, but also in the specific case of the mitochondrial genes, through the number of mitochondrial DNA (mtDNA) copies in a cell. Intrinsically, modification of the number of mtDNA copies in a cell could allow concomitant variation in cellular concentration of mitochondrial mRNAs (Williams 1986; Kaufmann et al. 1996). However, this mechanism has not been supported by other data that indicate that the mtDNA copy number poorly contributes to the differences in OXPHOS subunit expression (Van den Bogert et al. 1993).
Mitochondrial DNA has only two different promoters involved in expression of the 13 structural genes, and transcription of mtDNA generates two large polycistronic mRNAs. As a consequence, mRNAs coding for 12 out of the 13 mitochondrial-encoded subunits are transcribed stoichiometrically. Only the ND6 gene is transcribed independently (for review, see Taanman 1999). Under specific inhibitory or pathological conditions, the levels of only a few of the mitochondrial transcripts changed (Collombet et al. 1997; Cantatore et al. 1998; Heddi et al. 1999). This indicated that modification of the steady-state level of mitochondrial transcripts was not always a simultaneously and proportionate event affecting all the 13 OXPHOS transcripts. For example, it has been reported that a chronic energetic stress on differentiated myotubes, using sodium azide to inhibit the cytochrome-c oxidase activity, resulted in an increase in the steady-state levels of only the CoII mRNA, as detected by Northern blot analyses. Under these conditions, mtDNA concentration and mRNA levels for other mitochondrial transcripts remained unaffected (Leary et al. 1998). These reported uncoordinated variations of individual transcripts are quite unexpected, keeping in mind that most of the mitochondrial genes are transcribed in a polycistronic way. Very likely posttranscriptional mechanisms, including mRNA stability, should be taken into account (Sbisà et al. 1992; Tullo et al. 1994).
As for the nuclear genes, transcriptional regulation is assumed to be achieved in trans by several transcription factors such as NRF1 or NRF2, Oxbox and Rebox, and Mt family factors (Evans and Scarpulla 1989; Suzuki et al. 1990; Chung et al. 1992). Some of these factors have also been suggested to interfere directly (Mt family) or indirectly (NRF1) with mtDNA replication and transcription (Suzuki et al. 1991; Scarpulla 1997). Interestingly, these data indicate that some transcription factors may act as nuclear signals to control mitochondrial gene expression. As a consequence, these factors may participate in a molecular dialogue between nucleus and mitochondria to coordinate expression of the different genes required for biogenesis of the OXPHOS system. Understanding this dialogue is, of course, of fundamental importance to elucidation of normal and pathological aspects of cellular energetics. Conversely, it can also be expected that signals from mitochondria could induce specific adaptations of nuclear expression. However, reports showing that cells fully depleted of mtDNA do not modify the level of expression of several nuclear-encoded mitochondrial proteins (Herzberg et al. 1993; Buchet and Godinot 1998; Procaccio et al. 1999) might indicate that mitochondria are not operating a feedback control on nuclear gene expression. In-depth characterization of the specific chemical mediators involved in the nucleus–mitochondria cross-talk remains to be performed.
To date, transcription analyses of the OXPHOS system have focused on the variations in expression, under different physiological, toxic, or pathological conditions, of the 13 mitochondrial genes or of a limited number of the nuclear genes coding for subunits of complexes II, III, and IV. A comprehensive appraisal of the status of the expression of genes coding for subunits that are parts of all the OXPHOS complexes has never been carried out. This is partly owing to the fact that characterization of the numerous cDNAs or genes coding for the complex I subunits has only recently been achieved (Loeffen et al. 1998; see MitoPick database, http://www-dsv.cea.fr/thema/MitoPick/Default.html).
We have used new technologies for transcriptome analysis by quantitative PCR to investigate the steady-state levels of several OXPHOS complex transcripts in human tissues and cultured cells. This original analysis revealed that nuclear and mitochondrial transcripts are present in a very large range of concentrations in the analyzed samples. The possibility of coregulation between the different genes was also investigated.
RESULTS
Comparison Between Quantitative RT-PCR and SAGE Method to Assess OXPHOS Transcript Amounts in Samples
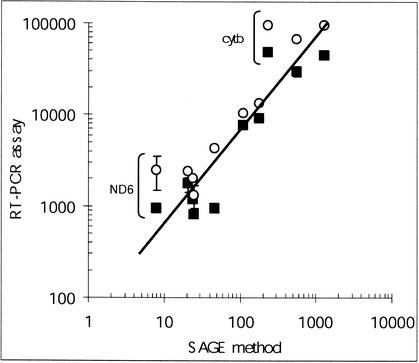
To ascertain whether the PCR-based quantitative assay of OXPHOS transcripts was a valuable approach, we compared quantitative data generated by our study and data from SAGE experiments carried out for large gene-profiling analyses. The SAGE method is a powerful technical approach to high-throughput assessment of the relative proportions of numerous transcripts in cellular samples. This technique is based on specific procedures for random concatenation and cloning of small cDNA fragments, followed by extensive sequence analysis as a final step for identification and quantitative determination of transcripts. SAGE data for muscle transcripts produced by Welle et al. (1999) allowed us to compare the SAGE method with our quantitative RT-PCR assay. These SAGE data, expressed as arbitrary hit numbers, and our data for muscular samples were plotted in Figure 1. Our RT-PCR data were assayed in skeletal muscle samples obtained from two different providers (Ambion and Clontech). It appeared that there was a fairly good correlation between the two technical approaches in assessing the concentration of most of the transcripts. As shown in Figure 1, in comparison to our technical approach, the SAGE method underestimated both the cytochrome-b and the ND6 mitochondrial transcripts. Some limitations in the SAGE technique, as discussed by Welle et al. (1999), might explain these underestimations.
Figure 1.
Relation between abundances of the muscular OXPHOS transcripts measured by quantitative RT-PCR and abundances of the corresponding cDNA tags assessed by the SAGE method. Quantitative data for skeletal muscle samples (from Ambion and Clontech providers) reported in Table 1 were plotted versus the quantitative data reported by Welle et al. (1999) for skeletal muscle by using the SAGE method. The SAGE data are available at the Rochester Muscle Database (http://www.urmc.rochester.edu/smd/crc/Swindex.html). (○) Sample from Ambion; (▪) sample from Clontech. Data corresponding to the cytochrome-b and the ND6 transcripts are labeled. Standard deviations were reported as vertical bars when exceeding the size of the symbols.
Steady-State Levels of Several Transcripts Coding for Subunits of the OXPHOS Complexes
In the course of screening for mutations by sequencing the cDNAs coding for the complex I subunits in pathological human samples, we found that cDNA synthesis by RT-PCR amplifications resulted in very different yields from transcript to transcript. This observation indicated that mitochondrial and nuclear transcripts that code for some of the complex I subunits might be present in disproportionate steady-state levels relative to each other. We decided specifically to investigate this point by assaying the steady-state levels of several complex I transcripts in different tissues. We also checked whether similar features could be observed with transcripts coding for subunits belonging to the other OXPHOS complexes (complexes II, III, IV, and V).
We set up and optimized quantitative RT-PCR experiments using a PCR-cycler instrument, which monitors PCR fragment concentrations in real time (Light Cycler, Roche Diagnostics). Quantitative accuracy and reproducibility were extensively checked by performing appropriate quality controls and by running standard samples made from titrated stock solutions of cDNA fragments, as described in Methods. We measured the steady-state levels of one nuclear transcript for complex II (sdhb subunit), one nuclear and one mitochondrial transcript for complexes III, IV, and V (Rieske subunit and cytb for complex III; CoIV and CoI for cytochrome-c oxidase; βATPase and ATPase8 for complex V, respectively), and finally, five nuclear and two mitochondrial transcripts for complex I (51 kD, 30 kD, 24 kD, MLRQ, MWFE, ND1, and ND6). Quantitative data were expressed as the ratio between the number of molecules of mRNA corresponding to a given OXPHOS subunit and the number of molecules of hypoxanthine phosphoribosyltransferase (HPRT) mRNA in the total RNA samples extracted from cultured cells or tissues. Average values corresponding to four different determinations are reported in Table 1.
Table 1.
Steady-State Levels of the OXPHOS Transcripts Measured in the Different Tissue and Cellular Total RNA Samples
| OXPHOS complexes | Transcripts | Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Muscle (Clontech) | Muscle (Ambion) | Liver | Kidney | Trachea | Control fibroblasts | 143B cels | 143B206ρ0 cells | ||
| Complex I | 24 kD (N) | 817 (±22) | 1310 (±368) | 408 (±4) | 111 (±12) | 70 (±3) | 552 (±91) | 434 (±43) | 505 (±73) |
| 30 kD (N) | 742 (±14) | 1464 (±333) | 206 (±5) | 124 (±9) | 79 (±8) | 658 (±218) | 366 (±49) | 326 (±17) | |
| 51 kD (N) | 513 (±57) | 762 (±287) | 273 (±111) | 174 (±9) | 94 (±9) | 451 (±7) | 342 (±82) | 268 (±182) | |
| MLRQ (N) | 7697 (±867) | 10,305 (±260) | 1362 (±181) | 1061 (±105) | 494 (±41) | 3138 (±820) | 1696 (±196) | 1474 (±620) | |
| MWFE (N) | 1170 (±23) | 2029 (±141) | 285 (±13) | 315 (±17) | 139 (±6) | 1051 (±373) | 482 (±120) | 672 (±277) | |
| ND1 (M) | 29,751 (±3219) | 66,150 (±6490) | 43,948 (±4946) | 17,509 (±1131) | 6745 (±615) | 3215 (±997) | 2405 (±293) | — | |
| ND6 (M) | 949 (±88) | 2487 (±1010) | 3678 (±852) | 1591 (±163) | 438 (±25) | 283 (±76) | 68 (±4) | — | |
| Complex II | sdhb (N) | 491 (±49) | 715 (±64) | 218 (±0) | 113 (±12) | 43 (±3) | 406 (±119) | 266 (±26) | 162 (±50) |
| Complex III | Rieske (N) | 1276 (±186) | 1833 (±42) | 184 (±33) | 171 (±13) | 108 (±9) | 792 (±278) | 460 (±43) | 506 (±108) |
| cytb (M) | 47,880 (±2876) | 95,111 (±8002) | 73,922 (±5483) | 34,840 (±128) | 10,927 (±848) | 9348 (±3014) | 1307 (±243) | — | |
| Complex IV | CoIV (N) | 949 (±13) | 4300 (±1297) | 291 (±49) | 253 (±32) | 223 (±10) | 2362 (±375) | 1220 (±189) | 1260 (±268) |
| Col (M) | 9143 (±254) | 13,365 (±758) | 10,821 (±87) | 11,170 (±2129) | 2362 (±167) | 6502 (±1266) | 1078 (±12) | — | |
| Complex V | β ATPase (N) | 1749 (±324) | 2391 (±260) | 310 (±72) | 336 (±21) | 165 (±11) | 1921 (±649) | 1060 (±19) | 928 (±113) |
| ATPase8 (M) | 45,024 (±5939) | 94,586 (±5878) | 60,118 (±4423) | 33,022 (±717) | 7325 (±646) | 12,913 (±2281) | 4358 (±4) | — | |
Transcripts were assayed by quantitative RT-PCR as described in Methods. Data are expressed as the ratio between the amount of each transcript of interest and the amount of HPRT transcript in the sample. Values are expressed as mean (± SD) corresponding to at least four different determinations. (N) Nuclear-gene transcript; (M) mitochondrial-gene transcript.
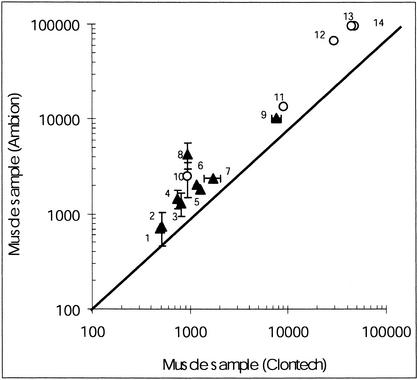
As a quality control for samples, we compared the amounts of OXPHOS transcripts measured in one muscle RNA extract from one provider (Ambion) with those measured in a muscle RNA extract obtained from another provider (Clontech). The data, plotted in Figure 2, showed that similar gene expression profiles were obtained with the two different muscle samples (symbols are close to the identity line). Similar plots were also obtained with data from RNA extracts prepared in the laboratory from fresh skeletal muscle biopsies (data not shown). From this plot, it was inferred that the quality of the muscular RNA extracts was consistent in all the different samples and nearly independent from their origins. This plot also revealed that the OXPHOS mRNA amounts ranged, from the least to most abundant transcript, over nearly two orders of magnitude in muscle extracts. The least abundant mRNA was the nuclear transcript coding for the complex II sdhb subunit, and the most abundant was the mitochondrial transcript that encodes the complex III cytb subunit.
Figure 2.
Comparison of the data obtained for two skeletal muscular samples. (▴) Nuclear transcripts coding for (1) sdhb; (2) 51 kD; (3) 24 kD; (4) 30 kD; (5) Rieske; (6) MWFE; (7) βATPase; (8) CoIV; (9) MLRQ. (○) Mitochondrial transcripts coding for (10) ND6; (11) CoI; (12) ND1; (13) ATPase8; (14) cytb. The line of identity was drawn. Statistical fluctuations of the values were reported as horizontal and vertical bars when they exceeded the size of the symbols.
A multisample comparison was then performed by plotting the steady-state levels of the OXPHOS transcripts in various tissues versus the steady-state concentrations of the equivalent transcripts in cultured 143B cells. This plot revealed that the assessed transcripts could be grossly distributed in three different sets (Fig. 3). A first set consisted only of the mitochondrial transcript coding for the ND6 subunit of complex I, which was found to be the least abundant transcript among the investigated transcripts of the 143B extracts. A second set combined all the nuclear-encoded transcripts, and the third one was composed of the mitochondrial transcripts (other than ND6). In 143B cells, the amounts of OXPHOS transcripts also ranged over nearly two orders of magnitude (Fig. 3). This plot indicated that there was no strict correlation between the amounts of the OXPHOS transcripts from the 143B cells and those from the various tissues. In fact, as shown in Figure 3, on average, most of the tissues contained higher concentrations of mitochondrial transcripts than did 143B cells.
Figure 3.
Comparison between abundances of the OXPHOS transcripts in the different tissues and those measured in cultured 143B cells. Open symbols: mitochondrial transcripts; closed symbols: nuclear transcripts. (⋄) Skeletal muscle; (▵) liver; (□) trachea; (○) kidney. Data corresponding to the ND6 transcript are indicated. Statistical fluctuations of the values were reported as horizontal and vertical bars.
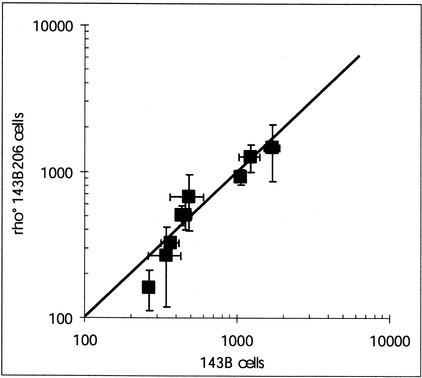
OXPHOS nuclear transcripts were also assayed in mtDNA-depleted cells (143B206 ρ0 cells). The data were plotted versus the amounts of the same nuclear transcripts in the 143B cells (Fig. 4). This plot clearly showed that the 143B cells and the daughter 143B206 ρ0 cells contained nearly identical concentrations of nuclear OXPHOS mRNA.
Figure 4.
Correlation between the steady-state levels of the nuclear OXPHOS transcripts in 143B206 ρ0 cells versus the parental 143B cells. The line of identity was drawn. Statistical fluctuations of the values were reported as horizontal and vertical bars.
Intracomplex Variations of OXPHOS Transcripts
To make some appraisal of the stoichiometries between the different transcripts, it was first of interest to study the ratio between the steady-state level of one mitochondrial transcript and the level of one nuclear transcript, both of them coding for subunits of one specific OXPHOS complex. The values for three different ratios concerning complex I and one ratio for complexes III, IV, and V are reported in Figure 5. This analysis illustrates the fact that there were large variations between the different ratios (see, e.g., the ND6/MLRQ and the ND1/51 kD ratios). In cultured cells (Fig. 5), the mitochondrial transcripts were found either in substoichiometric amounts compared with the nuclear transcripts (ND6 vs. MLRQ), nearly stoichiometric amounts (ND1 vs. MLRQ and CoI vs. CoIV), or in excess (cytb vs. Rieske, ATPase8 vs. βATPase, and ND1 vs. 51 kD). In the specific case of complex I, it is worth noting that assembly of a functional complex that presumably contains stoichiometric amounts of subunits involves ∼700 molecules of the ND1 transcript per 100 molecules of the 51-kD transcript and only 4 molecules of the ND6 transcript per 100 molecules of the MLRQ transcript.
Figure 5.
Ratios between mitochondrial and nuclear transcripts coding for subunits of separate OXPHOS complexes in 143B cells and muscle. (1) ND6/MLRQ; (2) ND1/MLRQ; (3) ND1/51 kD; (4) CoI/CoIV; (5) cytb/Rieske; (6) ATPase8/βATPase. (White bars) 143B cells; (black bars) muscle. Standard deviations were reported as thin vertical bars.
From the shape of Figure 3, it was deduced that cultured 143B cells contained fewer mitochondrial transcripts than tissue cells. Consistent with this conclusion, the mitochondrial-to-nuclear transcript ratios were lower than those measured in muscle (Fig. 5). On average, the ratios were nearly 5 to 10 times lower in cultured 143B cells than in tissues (see Table 1). Lower mitochondrial-to-nuclear transcript ratios in cells in comparison to tissues were also observed for cultured normal fibroblasts. It can be hypothesized that this fact may result from diminished amounts of mtDNA and/or numbers of mitochondria in these cells, correlated with an accompanying reduction in the level of mitochondrial transcripts. Accordingly, we checked whether the cellular concentration of mtDNA was decreased in cultured cells. To compare samples, we measured the respective values of the mitochondrial DNA/nuclear DNA ratio (mtDNA/nDNA). These values were found to be similar in cultured control fibroblasts and in tissues (data not shown). Consequently, the decrease in the intracomplex ratios between mitochondrial and nuclear transcripts in cultured cells in comparison to tissue samples (as evidenced in Fig. 3) was not the result of a decrease in mtDNA content in the cells.
Intercomplex Variations in OXPHOS Transcripts
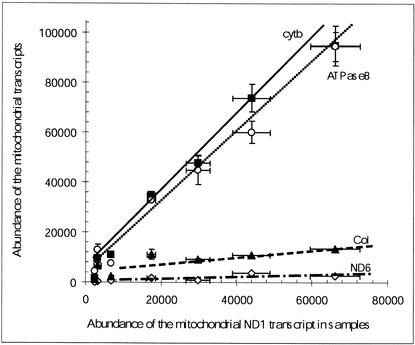
We also studied the correlations between the levels of the transcripts that code for subunits of different OXPHOS complexes (inter-OXPHOS complex variations in transcript abundances). Such an analysis could help to reveal any possible tissue-specific changes in the expression control of the genes of the OXPHOS complexes. To study the intercomplex variations in the levels of the transcripts of mitochondrial origin, we plotted the amounts of the different mitochondrial transcripts versus the amounts of the ND1 transcripts. As shown in Figure 6, there was a good relationship between the amount of transcript coding for ND1 and the amount of all the other measured mitochondrial transcripts in the different samples. The slopes of the lines in the graph were indicative of the respective stoichiometries of the mitochondrial transcripts. It can then be estimated that, in mitochondria, there were nearly 150 molecules of transcripts coding for cytb or ATPase8 per 100 molecules of transcripts coding for ND1 in all the investigated tissues and cells. The transcripts for CoI and ND6 were present in substoichiometric amounts compared with the ND1 transcript, with ∼20 and only 5 molecules of transcripts per 100 molecules of the ND1 transcript, respectively.
Figure 6.
Correlation between the abundances of the mitochondrial transcripts and the abundances of the ND1 transcript in the different tissues and cells. (⋄) ND6; (▴) CoI; (○) ATPase8; (▪) cytb. Fitted line equations are the following: ND6 versus ND1: y = 0.044x + 156; CoI versus ND1: y = 0.184x + 3254; ATPase8 versus ND1: y = 1.371x + 2423; cytb versus ND1: y = 1.399x + 1197. Statistical fluctuations of the values were reported as horizontal and vertical bars.
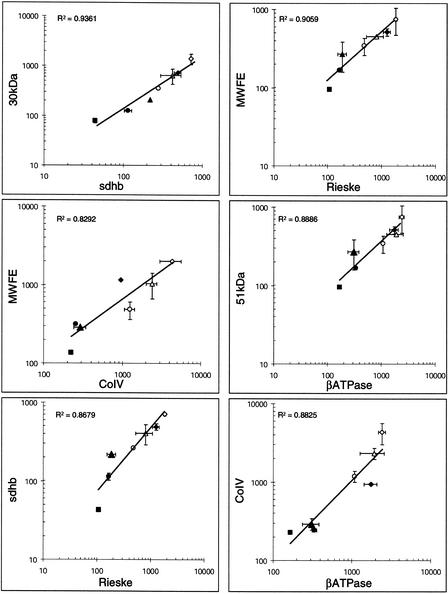
Intercomplex variations in the transcripts of nuclear origin were evidenced by plotting the measured amounts of one nuclear transcript for a given complex versus the amount of another nuclear transcript coding for a subunit of a different complex. The plots are shown in Figure 7. They illustrate the variations in the levels of transcripts coding for complex I subunits versus transcripts coding for a subunit of either complex II (30 kD vs. sdhb), complex III (51 kD vs. Rieske), complex IV (MWFE vs. CoIV), or complex V (51 kD vs. βATPase). They also show variations in the levels of transcripts coding for complex II subunit versus complex III subunit (sdhb vs. Rieske) and variations in the level of transcripts coding for complex IV subunit versus complex V subunit (CoIV vs. βATPase). In these plots, data are scattered over more than one order of magnitude and closely fit linear relations. This means that the relative proportions of the different transcripts coding for the subunits of different OXPHOS complexes are kept nearly constant in all the various investigated tissues and cultured cell samples. These observations indicated that in the tissues and cultured cells, the concentrations of all the nuclear transcripts are tightly coregulated. The slopes of the straight lines that fit the data in all the plots indicated the averaged ratios between the steady-state levels of the nuclear transcripts. These ratios ranged between 1 and 2 for most of the transcripts. This indicated that in the different samples most of the nuclear OXPHOS transcripts were in nearly stoichiometric amounts. The highest value of 15 was found for the ratio between the most abundant transcript (MLRQ) and the least abundant transcript (sdhb).
Figure 7.
Correlation between the abundances of the nuclear OXPHOS transcripts in the different tissues and cells. Correlation coefficients (R2) are given in each panel. (⋄) Muscle Ambion; (♦) muscle Clontech; (▴) liver; (●) kidney; (▪) trachea; (○) 143B cells; (▵) control fibroblasts. Statistical fluctuations of the values were reported as horizontal and vertical bars.
DISCUSSION
The mammalian mitochondrial OXPHOS system is multienzymatic and comprises five different multimeric complexes. The functional assembly of the OXPHOS system requires that at least 80 different genes, located in two different genomes (∼70 nuclear and 13 mitochondrial genes coding for subunits), must be expressed in a presumably coherent way. This raises the challenge of studying both the individual control mechanisms for mitochondrial and nuclear gene expression and the cross-talk between nuclear and mitochondrial genomes. Literature data mostly relate to analyses of the variations in the expression of OXPHOS genes under stressed or pathological conditions. However, a prerequisite for subsequent analyses is a comprehensive appraisal of the normal steady-state levels of all the transcripts coding for subunits that are parts of the five multimeric OXPHOS complexes.
We investigated the cellular steady-state levels of several transcripts coding for subunits of the five different complexes of the human OXPHOS system, both in various tissues and in cultured cells. We addressed the problem by performing carefully controlled quantitative RT-PCR experiments. We quantitatively analyzed a total of 14 different transcripts of both mitochondrial and nuclear origins.
Our data showed that all the investigated mitochondrial and nuclear transcripts coding for the subunits of the human mitochondrial OXPHOS system were in a large range of steady-state concentrations in tissues and cultured cells (about two orders of magnitude). This large variation in steady-state levels of OXPHOS transcripts evidenced by quantitative RT-PCR was similarly demonstrated by the data of Welle et al. (1999) recorded in SAGE experiments on human muscle samples. The amounts of most of the mitochondrial transcripts exceeded those of the nuclear transcripts. This last observation is in agreement with the data of Van den Bogert et al. (1993), who assessed some of the OXPHOS transcripts by Northern blot analysis, and also with data from SAGE experiments (Welle et al. 1999). Literature data are scarce concerning the respective steady-state amounts of all the mitochondrial transcripts under normal physiological conditions. Reported values indicated that mitochondrial H-strand transcripts are nearly equistoichiometric (Gelfand and Attardi 1981; Cantatore et al. 1984). Our results show that the highest value measured for the ratio between these individual transcripts was close to 7. L-Strand ND6 transcripts were much less abundant than the other mitochondrial transcripts, probably because they have a shorter half-life than H-strand transcripts (Gelfand and Attardi 1981).
Assuming that a waste-free assembly process of the OXPHOS components is probably required for efficient cellular bioenergetic production, several explanations can be put forward to rationalize the large range of steady-state levels of the OXPHOS transcripts. It can be hypothesized that the final amount of expressed subunits is not strictly correlated with the amount of the individual transcripts. Instead, the rates of subunit synthesis in the cells may be finely tuned for each OXPHOS complex simply by individual adjustment of the translation efficiency of all the various transcripts. Iron-mediated translation regulation of the 75-kD subunit of complex I, as recently evidenced by Lin et al. (2001), could support this hypothesis.
Control of the steady-state levels of functional and ready-to-assemble subunits of the OXPHOS complexes may combine sophisticated processes regulating not only the translation rate of transcripts, but also all the protein-processing steps (mitochondrial import, folding, ligand binding, etc.) and the overall subunit stability. In the case of complex I, this could be true for subunits whose degradation rate may be reduced after binding of cofactors like iron–sulfur clusters, ubiquinone, or FMN. In support of this point, it has been shown that in vitro proteolysis of the 39-kD subunit of the bovine enzyme is prevented by NADPH, a newly discovered ligand for complex I (Yamaguchi et al. 1998; Schulte et al. 1999). Moreover, some specific features of the complex I subunits may largely contribute to limiting the rate of both the mitochondrial import and the degradation steps. This might be relevant for the import of the MLRQ and MWFE subunits that have no consensual mitochondrial import signals and contain candidate membrane-spanning hydrophobic domains located close to their N termini. Increased steady-state levels of the MLRQ transcript may allow higher expression of this hydrophobic protein and, as a consequence, may improve its rate of mitochondrial import.
Excess of mitochondrial transcripts over the nuclear transcripts could compensate for poor intramitochondrial translation efficiency. It has been proposed that inefficient intramitochondrial translation is a consequence of the fact that mitochondria contain low levels of mito-ribosomes in comparison with the mRNA concentration, and that the structural features of mitochondrial mRNA, with its short 5′-untranslated regions, could be a rate-limiting factor for initiation of translation (Cantatore et al. 1987; Garstka et al. 1994; Taanman 1999). However, this hypothesis does not fit very well with the low concentration of the ND6 transcript of complex I observed in the different tissues and cells.
Regardless of the fact that the mitochondrial and nuclear transcripts are present at different steady-state levels in the cells, the equivalent genes may be either independently or coordinately expressed. Support for a transcriptional coordination between nuclear and mitochondrial genes has been provided by numerous reports (Hood et al. 1989; Gagnon et al. 1991; Van den Bogert et al. 1993; Connor et al. 1996; Heddi et al. 1999). However, data from analysis of changes in mitochondrial transcription in response to thyroid hormones (Enriquez et al. 1999) as well as data from reporter gene activity (Sewards et al. 1994) have indicated that mitochondrial and nuclear transcription systems operate independently.
The data presented in Figures 3 and 4 support the idea that the nuclear OXPHOS genes are transcribed independently of mitochondrial OXPHOS genes. This autonomous nuclear transcription is indicated by the fact that whereas steady-state levels of mitochondrial transcripts differed (high in tissues, reduced in cultured cells or nil in ρ0 cells), the steady-state levels of OXPHOS nuclear transcripts remained nearly unchanged. This may indicate that neither the cellular mtDNA concentration nor the mitochondrial RNA concentration plays a role in a putative message delivery from mitochondria to nucleus transcription machinery. These observations are consistent with data from studies of the nuclear OXPHOS gene expression in cells with deficient mitochondria (see below). In these studies, investigated models consisted either of cells depleted of mtDNA or of cells severely impaired in mitochondrial protein synthesis because of a pathological nuclear mutation or blockade by an inhibitor. Gene expression was monitored either directly at the level of the transcript concentration or indirectly at the level of the protein synthesis or enzymatic activity. For example, several nuclear-encoded complex I subunits were immuno-detected at a nearly normal concentration in mitochondria from 143B206 ρ0 cells (Procaccio et al. 1999). Normal intramitochondrial accumulation of the complex II subunits was also observed in Chinese hamster mutant cell lines with a defect in mitochondrial protein synthesis (Au and Scheffler 1997). Enzymatic activity of complex II was also normal or slightly decreased in 143B206 ρ0 cells or SFT.12 ρ0 transformant fibroblasts, two different types of human cells fully depleted of mitochondrial DNA (Tiranti et al. 1995; Procaccio et al. 1999). Immunochemical approaches also revealed that complex II subunits and some nuclear-encoded complex IV subunits are still present in mitochondria from normal fibroblasts depleted of mtDNA by ethidium bromide treatment or from patients' fibroblasts spontaneously exhibiting mtDNA depletion (Marusich et al. 1997). Expression of several complex IV subunits was also observed in cultured human myoblasts depleted of mtDNA (Herzberg et al. 1993). Finally, synthesis and assembly of nuclear-encoded subunits of complex V were demonstrated in cells treated with mitochondrial protein synthesis inhibitor or in ρ0 cells (Nijtmans et al. 1995; Buchet and Godinot 1998). Our work showing that nine nuclear genes coding for major subunits of the five OXPHOS complexes are normally transcribed in cells fully depleted of mtDNA is consistent with the above reports. However, the results of Li et al. (1995) partly disagree with ours and support the transcriptional coordination model. These investigators found that in ρ0 cells, the steady-state amount of transcript coding for the βATPase measured by Northern blot analyses was unchanged in comparison to control cells, but that the amounts of CoIV and CoVIaL transcripts were increased by 60% and 100%. respectively. Extended analysis of the variation in expression of all the OXPHOS nuclear genes under experimental conditions that allow restriction of mitochondrial gene expression will help to reveal any mitochondrial control of nuclear expression.
More experiments are clearly necessary if we are to understand fully the discrepancies between all the available results. It remains possible that the mitochondrial and nuclear genes could be coordinately transcribed under normal conditions. However, this coordination might be disrupted under extraphysiological conditions (DNA-depleted mitochondria or functionally impaired mitochondria) or by various stimuli.
Another very attractive point of the present report is the demonstration of an intercomplex and independent regulation of the transcription of the mitochondrial and nuclear OXPHOS genes. Coregulation of the expression of the mitochondrial genes can evidently result from the polycistronic transcription of the mitochondrial genes. Our study indicated that in tissues and cultured cells, the steady-state stoichiometries between all the mitochondrial transcripts are kept constant whatever the mitochondrial transcription activity. More surprisingly, the ratios between the investigated nuclear transcripts coding for subunits from the five different OXPHOS complexes were found to be identical in all the analyzed samples. Constant proportions of the enzyme of the glycolytic pathway were noticed, at the level of the enzyme activity, in a variety of tissues several years ago. This was also reported for several enzymes of the citric acid cycle and the respiratory chain (for review, see Pette and Dolken 1975; Rustin et al. 1994). Here we demonstrate for the first time that in the case of the subunits that are part of all the OXPHOS complexes, transcription coregulation is a major component for maintaining a constant proportion of these complexes among tissues and cells. The existence of a tightly coupled expression of the OXPHOS nuclear genes may indicate that the cells avoid a wasteful process for generation of the OXPHOS system. This coregulation indicates that the functional expression of the OXPHOS genes is under the control of a complicated process. At the molecular level, we can hypothesize that specific transcription factors are controlling the promoters of the nuclear OXPHOS genes to coordinate the transcription of these genes and consequently the expression level of the subunits. Analysis of the promoter activity of all the nuclear OXPHOS genes under normal or pathological conditions may help to discover the mechanisms and key factors that are responsible for this transcriptional coregulation of the nuclear genes.
METHODS
Cell Lines and Culture Conditions
Fibroblasts from forearm skin explants were obtained from healthy individuals, with informed consent. The 143B (TK−) osteosarcoma cell line (CRL 8303) was obtained from the American Type Culture Collection (Rockville, MD, USA) and the daughter 143B206 ρ0 cell line, completely depleted of mtDNA by long-term exposure to ethidium bromide, was kindly provided by G. Attardi (Caltech, Pasadena, CA). All the cells were cultured in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum in a 5% CO2 atmosphere at 37°C. For 143B and 143B206 ρ0 cells only, the growth medium was also complemented with 100 μg/mL 5-bromo-2‘-deoxyuridine (BrdU), 100 μg/mL pyruvate, and 50 μg/mL uridine.
RNA and DNA Samples
Total RNA was extracted from cultured cells, harvested at a nonconfluent state, by using the RNeasy kit from QIAGEN. Commercially available total human RNA samples from skeletal muscle were from Clontech and Ambion, and those from liver, trachea, and kidney were from Clontech. Total DNA was prepared from cultured cells and from liver biopsies as described by Sambrook et al. (1989).
RT-PCR and mRNA Quantification
Before reverse transcription, all types of RNA samples (5 μg samples) were freed of any contaminating DNA by treatment with an RNase-free DNase I (Life Technologies). After digestion (30°C, 15 min), DNAse I was denatured by heating (65°C, 10 min). The quality of the resulting RNA samples was carefully checked by assessment of the presence of mtDNA, taken as an indicator for any remaining DNA contamination. This was achieved by performing PCR experiments on 0.1 μg of the treated samples with specific primers for amplification of several mitochondrial genes. Resulting PCR products were analyzed by electrophoresis on agarose gels. Only RNA samples showing no trace at all of amplified signals were used in subsequent experiments.
Reverse transcription was conducted on 3-μg aliquots of RNA samples at 42°C for 50 min using a poly(T) oligonucleotide as a primer for the MMLV reverse polymerase (Invitrogen). Resulting single-stranded cDNA was used as the starting material for quantitative assay of the transcripts by real-time PCR (see below).
Design of primer pairs used for cDNA amplifications was based on two criteria: the size of the generated fragments (200–300 bp) and the annealing temperatures (usually in the range 55°–60°C). Whenever possible couples of primers were also chosen for their matching to cDNA domains resulting from transcription of two different exons from the genes of interest. Combination of both the strategy for primer selection and the DNase treatment of RNA samples ensured that amplification of PCR fragments was not owing to genomic DNA amplification. The quality and specificity of the resulting amplified PCR fragments were also carefully checked. First, amplified fragments were electrophoresed on agarose gels to confirm their expected sizes, and also to check the homogeneity of these fragments. Second, PCR fragments were sequenced, and their identity was ascertained by comparison with sequences in databases (data in MitoPick database, http://www-dsv.cea.fr/thema/MitoPick/Default.html; for the complex I cDNAs and in GenBank for the other amplified cDNAs coding for all the other OXPHOS subunits).
Quantitative PCR experiments, carried out to assay mRNA concentrations were performed on a Light Cycler apparatus (Roche Diagnostics). Samples of the single-stranded cDNA, obtained as described above after the reverse transcription step, were used for individual amplification of cDNAs coding for OXPHOS subunits in capillary tubes with specific primer pairs. DNA polymerization was catalyzed by the Taq polymerase (Promega). SYBR Green dye was used as a reporter for detection according to protocols from Roche Diagnostics. Quantitative determinations of the samples were obtained by running standard samples under the same conditions. Standards for all the investigated cDNA fragments were made from purified samples of the individually cloned cDNA fragments. Concentrations of the standard stock solutions (calculated in nanomoles per microliter) were measured by UV absorption. All calculations were made with the Light Cycler software (v. 3, Roche Diagnostics). Final data were expressed as the ratio multiplied by 1000 between the amount of each transcript of interest versus the amount of HPRT transcript (a moderately expressed housekeeping gene, usually taken as a convenient reference) so as to allow comparison between tissue and cellular samples. After running the experiments, two types of quality controls were carried out. First, final products were submitted to a Light Cycler built-in renaturing–denaturing thermal program. Fusion curves obtained during this step gave confirmation of the homogeneity of the amplified fragments. Second, aliquots of all the amplified products were downloaded and electrophoresed on an agarose gel for visual inspection.
mtDNA-to-Nuclear-DNA Ratio Assay
Quantitative PCR protocols were also set up to allow measurements of both the mtDNA and nDNA concentrations in a given DNA sample purified from cultured cells or biopsies. For mtDNA, primer pairs were selected for individual amplification of the 12S rRNA gene. Selectivity of the primers was ascertained by checking that these primers were unable to amplify unspecific nDNA targets when using ρ0 cell DNA extracts, as a starting material. Nuclear DNA concentrations were measured by amplification of the 18S rRNA gene. Quantitative assays follow the same principle as described above for cDNA PCR quantitative analyses using the Light Cycler.
WEB SITE REFERENCES
http://www-dsv.cea.fr/thema/MitoPick/Default.html; MitoPick database.
http://www.urmc.rochester.edu/smd/crc/Swindex.html; Rochester Muscle Database.
Acknowledgments
We are extremely grateful to G. Attardi (Caltech, Pasadena, CA) for the gift of 143B206 ρ0 cells. We gratefully acknowledge the Laboratoire d’Immunochimie (ICH/DRDC/CEA-Grenoble) for technical support. This work was supported by grants from the Région Rhône-Alpes, the Université J. Fourier, and the Association Française contre les Myopathies.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL j.issartel@genomex.com; FAX (33) 4 56 38 11 00.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.194102.
REFERENCES
- Au HC, Scheffler IE. A respiration-deficient Chinese hamster cell line with a defect in mitochondrial protein synthesis: Rapid turnover of some mitochondrial transcripts. Somat Cell Mol Genet. 1997;23:27–35. doi: 10.1007/BF02679953. [DOI] [PubMed] [Google Scholar]
- Buchet K, Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted ρ0 cells. J Biol Chem. 1998;273:22983–22989. doi: 10.1074/jbc.273.36.22983. [DOI] [PubMed] [Google Scholar]
- Cantatore P, Gadaleta MN, Saccone C. Determination of some mitochondrial RNAs concentration in adult rat liver. Biochem Biophys Res Commun. 1984;118:284–291. doi: 10.1016/0006-291x(84)91098-2. [DOI] [PubMed] [Google Scholar]
- Cantatore P, Flagella Z, Fracasso F, Lezza AM, Gadaleta MN, de Montalvo A. Synthesis and turnover rates of four rat liver mitochondrial RNA species. FEBS Lett. 1987;213:144–148. doi: 10.1016/0014-5793(87)81480-1. [DOI] [PubMed] [Google Scholar]
- Cantatore P, Petruzzella V, Nicoletti C, Papadia F, Fracasso F, Rustin P, Gadaleta MN. Alteration of mitochondrial DNA and RNA level in human fibroblasts with impaired vitamin B12 coenzyme synthesis. FEBS Lett. 1998;432:173–178. doi: 10.1016/s0014-5793(98)00857-6. [DOI] [PubMed] [Google Scholar]
- Chung AB, Stepien G, Haraguchi Y, Li K, Wallace DC. Transcriptional control of nuclear genes for the mitochondrial muscle ADP/ATP translocator and the ATP synthase β subunit. Multiple factors interact with the OXBOX/REBOX promoter sequences. J Biol Chem. 1992;267:21154–21161. [PubMed] [Google Scholar]
- Collombet JM, Faure-Vigny H, Mandon G, Dumoulin R, Boissier S, Bernard A, Mousson B, Stepien G. Expression of oxidative phosphorylation genes in muscle cell cultures from patients with mitochondrial myopathies. Mol Cell Biochem. 1997;168:73–85. doi: 10.1023/a:1006830807107. [DOI] [PubMed] [Google Scholar]
- Connor MK, Takahashi M, Hood DA. Tissue-specific stability of nuclear- and mitochondrially encoded mRNAs. Arch Biochem Biophys. 1996;333:103–108. doi: 10.1006/abbi.1996.0369. [DOI] [PubMed] [Google Scholar]
- Enriquez JA, Fernandez-Silva P, Garrido-Perez N, Lopez-Perez MJ, Perez-Martos A, Montoya J. Direct regulation of mitochondrial RNA synthesis by thyroid hormone. Mol Cell Biol. 1999;19:657–670. doi: 10.1128/mcb.19.1.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J Biol Chem. 1989;264:14361–14368. [PubMed] [Google Scholar]
- Gagnon J, Kurowski TT, Wiesner RJ, Zak R. Correlations between a nuclear and a mitochondrial mRNA of cytochrome c oxidase subunits, enzymatic activity and total mRNA content, in rat tissues. Mol Cell Biochem. 1991;107:21–29. doi: 10.1007/BF02424572. [DOI] [PubMed] [Google Scholar]
- Garstka HL, Facke M, Escribano JR, Wiesner RJ. Stoichiometry of mitochondrial transcripts and regulation of gene expression by mitochondrial transcription factor A. Biochem Biophys Res Commun. 1994;200:619–626. doi: 10.1006/bbrc.1994.1493. [DOI] [PubMed] [Google Scholar]
- Gelfand R, Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: The mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981;1:497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddi A, Stepien G, Benke PJ, Wallace DC. Coordinate induction of energy gene expression in tissues of mitochondrial disease patients. J Biol Chem. 1999;274:22968–22976. doi: 10.1074/jbc.274.33.22968. [DOI] [PubMed] [Google Scholar]
- Herzberg NH, Middelkoop E, Adorf M, Dekker HL, Van Galen MJ, Van den Berg M, Bolhuis PA, Van den Bogert C. Mitochondria in cultured human muscle cells depleted of mitochondrial DNA. Eur J Cell Biol. 1993;61:400–408. [PubMed] [Google Scholar]
- Hood DA, Zak R, Pette D. Chronic stimulation of rat skeletal muscle induces coordinate increases in mitochondrial and nuclear mRNAs of cytochrome-c-oxidase subunits. Eur J Biochem. 1989;179:275–280. doi: 10.1111/j.1432-1033.1989.tb14551.x. [DOI] [PubMed] [Google Scholar]
- Kaufmann P, Koga Y, Shanske S, Hirano M, DiMauro S, King MP, Schon EA. Mitochondrial DNA and RNA processing in MELAS. Ann Neurol. 1996;40:172–180. doi: 10.1002/ana.410400208. [DOI] [PubMed] [Google Scholar]
- Leary SC, Battersby BJ, Hansford RG, Moyes CD. Interactions between bioenergetics and mitochondrial biogenesis. Biochim Biophys Acta. 1998;1365:522–530. doi: 10.1016/s0005-2728(98)00105-4. [DOI] [PubMed] [Google Scholar]
- Li K, Neufer PD, Williams RS. Nuclear responses to depletion of mitochondrial DNA in human cells. Am J Physiol. 1995;269:C1265–C1270. doi: 10.1152/ajpcell.1995.269.5.C1265. [DOI] [PubMed] [Google Scholar]
- Lin E, Graziano JH, Freyer GA. Regulation of the 75-kDa subunit of mitochondrial complex I by iron. J Biol Chem. 2001;276:27685–27692. doi: 10.1074/jbc.M100941200. [DOI] [PubMed] [Google Scholar]
- Loeffen JL, Triepels RH, van den Heuvel LP, Schuelke M, Buskens CA, Smeets RJ, Trijbels JM, Smeitink JA. cDNA of eight nuclear encoded subunits of NADH:ubiquinone oxidoreductase: Human complex I cDNA characterization completed. Biochem Biophys Res Commun. 1998;253:415–422. doi: 10.1006/bbrc.1998.9786. [DOI] [PubMed] [Google Scholar]
- Marusich MF, Robinson BH, Taanman JW, Kim SJ, Schillace R, Smith JL, Capaldi RA. Expression of mtDNA and nDNA encoded respiratory chain proteins in chemically and genetically-derived ρ0 human fibroblasts: A comparison of subunit proteins in normal fibroblasts treated with ethidium bromide and fibroblasts from a patient with mtDNA depletion syndrome. Biochim Biophys Acta. 1997;1362:145–159. doi: 10.1016/s0925-4439(97)00061-6. [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Spelbrink JN, Van Galen MJ, Zwaan M, Klement P, Van den Bogert C. Expression and fate of the nuclearly encoded subunits of cytochrome-c oxidase in cultured human cells depleted of mitochondrial gene products. Biochim Biophys Acta. 1995;1265:117–126. doi: 10.1016/0167-4889(94)00203-q. [DOI] [PubMed] [Google Scholar]
- Pette D, Dolken G. Some aspects of regulation of enzyme levels in muscle energy-supplying metabolism. Adv Enzyme Regul. 1975;13:355–377. doi: 10.1016/0065-2571(75)90025-4. [DOI] [PubMed] [Google Scholar]
- Procaccio V, Mousson B, Beugnot R, Duborjal H, Feillet F, Putet G, Pignot-Paintrand I, Lombès A, De Coo R, Smeets H, et al. Nuclear DNA origin of mitochondrial complex I deficiency in fatal infantile lactic acidosis evidenced by transnuclear complementation of cultured fibroblasts. J Clin Invest. 1999;104:83–92. doi: 10.1172/JCI6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, Gérard B, Rötig A, Saudubray JM, Munnich A. Reference charts for respiratory chain activities in human tissues. Clin Chim Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sbisà E, Tullo A, Nardelli M, Tanzariello F, Saccone C. Transcription mapping of the Ori L region reveals novel precursors of mature RNA species and antisense RNAs in rat mitochondrial genome. FEBS Lett. 1992;296:311–316. doi: 10.1016/0014-5793(92)80311-4. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory chain expression in mammalian cells. J Bioenerg Biomembr. 1997;29:109–119. doi: 10.1023/a:1022681828846. [DOI] [PubMed] [Google Scholar]
- Schulte U, Haupt V, Abelmann A, Fecke W, Brors B, Rasmussen T, Friedrich T, Weiss H. A reductase/isomerase subunit of mitochondrial NADH:ubiquinone oxidoreductase (complex I) carries an NADPH and is involved in the biogenesis of the complex. J Mol Biol. 1999;292:569–580. doi: 10.1006/jmbi.1999.3096. [DOI] [PubMed] [Google Scholar]
- Sewards R, Wiseman B, Jacobs HT. Apparent functional independence of the mitochondrial and nuclear transcription systems in cultured human cells. Mol Gen Genet. 1994;245:760–768. doi: 10.1007/BF00297283. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Hosokawa Y, Toda H, Nishikimi M, Ozawa T. Common protein-binding sites in the 5′-flanking regions of human genes for cytochrome c1 and ubiquinone-binding protein. J Biol Chem. 1990;265:8159–8163. [PubMed] [Google Scholar]
- Suzuki H, Hosokawa Y, Nishikimi M, Ozawa T. Existence of common homologous elements in the transcriptional regulatory regions of human nuclear genes and mitochondrial gene for the oxidative phosphorylation system. J Biol Chem. 1991;266:2333–2338. [PubMed] [Google Scholar]
- Taanman JW. The mitochondrial genome: Structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- Tiranti V, Munaro M, Sandona D, Lamantea E, Rimoldi M, DiDonato S, Bisson R, Zeviani M. Nuclear DNA origin of cytochrome c oxidase deficiency in Leigh's syndrome: Genetic evidence based on patient's-derived ρ0 transformants. Hum Mol Genet. 1995;4:2017–2023. doi: 10.1093/hmg/4.11.2017. [DOI] [PubMed] [Google Scholar]
- Tullo A, Tanzariello F, D'Erchia AM, Nardelli M, Papeo PA, Sbisà E, Saccone C. Transcription of rat mitochondrial NADH-dehydrogenase subunits. Presence of antisense and precursor RNA species. FEBS Lett. 1994;354:30–36. doi: 10.1016/0014-5793(94)01080-3. [DOI] [PubMed] [Google Scholar]
- Van den Bogert C, De Vries H, Holtrop M, Muus P, Dekker HL, Van Galen M J, Bolhuis PA, Taanman JW. Regulation of the expression of mitochondrial proteins: Relationship between mtDNA copy number and cytochrome-c oxidase activity in human cells and tissues. Biochim Biophys Acta. 1993;1144:177–183. doi: 10.1016/0005-2728(93)90170-k. [DOI] [PubMed] [Google Scholar]
- Welle S, Bhatt K, Thornton CA. Inventory of high-abundance mRNAs in skeletal muscle of normal men. Genome Res. 1999;9:506–513. [PMC free article] [PubMed] [Google Scholar]
- Williams RS. Mitochondrial gene expression in mammalian striated muscle. Evidence that variation in gene dosage is the major regulatory event. J Biol Chem. 1986;261:12390–12394. [PubMed] [Google Scholar]
- Yamaguchi M, Belogrudov GI, Hatefi Y. Mitochondrial NADH-ubiquinone oxidoreductase (Complex I). Effect of substrates on the fragmentation of subunits by trypsin. J Biol Chem. 1998;273:8094–8098. doi: 10.1074/jbc.273.14.8094. [DOI] [PubMed] [Google Scholar]