Abstract
Extreme pathways are a unique and minimal set of vectors that completely characterize the steady-state capabilities of genome-scale metabolic networks. A framework is provided to mathematically characterize extreme pathway length and to study how individual reactions participate in the extreme pathway structure of a network. The length of an extreme pathway is the number of reactions that comprise it. Reaction participation is the percentage of extreme pathways that utilize a given reaction. These properties were computed for the production of individual amino acids and protein production in Helicobacter pylori and individual amino acid production in Haemophilus influenzae. Reaction participation classifies the reactions into groups that are always, sometimes, or never utilized for the production of a target product. The utilized reactions can be further grouped into correlated subsets of reactions, some of which are non-obvious, and which may, in turn, suggest regulatory structure. The length of the extreme pathways did not correlate with product yield or chemical complexity. The distributions of extreme pathway lengths in H. pylori were also very different from those in H. influenzae, showing a distinct systemic difference between the two organisms, despite overall similar metabolic networks. Reaction participation and extreme pathway lengths thus serve to elucidate systemic biological features.
Biochemical pathways are thought of as functional units of metabolic networks. As such, the definitions and characterizations of metabolic pathways allow for a detailed analysis of robustness, physiological capabilities, and other systemic features of these complex reaction networks. These emergent properties necessitate the development of clear and mathematically precise definitions for a metabolic pathway. Mathematical and systemic definitions of metabolic pathways have been proposed (Mavrovouniotis and Stephanopoulos 1990; Liao et al. 1996; Karp et al. 1999; Schuster et al. 1999; Ouzounis and Karp 2000; Schilling et al. 2000).
Extreme pathways are mathematically derived vectors that can be used to characterize the phenotypic potential of a defined metabolic network (Schilling et al. 1999, 2000). Extreme pathway analysis has the following characteristics: (1) it generates a unique and minimal set of systemic pathways; (2) it describes all possible steady-state flux distributions that the network can achieve by non-negative linear combinations of the extreme pathways; and (3) it enables the determination of time-invariant, topological properties of the network. The calculation of extreme pathways is computationally challenging and for large networks, generates a tremendous amount of numerical data. (Schilling and Palsson 2000; Samatova et al. 2002). These challenges are being met and extreme pathway analysis has been performed at a genome scale for amino acid production in Haemophilus influenzae (Papin et al. 2002) and protein production in Helicobacter pylori (Price et al. 2002). With the ability to compute extreme pathways for large networks, it is necessary to develop methods to study the salient features of large sets of extreme pathways.
Extreme pathways can be characterized by their length and reaction participation. Extreme pathway length is defined as the number of reactions involved in an extreme pathway. Extreme pathways describe the conversion of substrates into products, while also creating all byproducts needed to maintain the systemic elemental balance and maintaining all cofactor pools at steady state. This characteristic distinguishes the definition of an extreme pathway from the traditional pathway definition of a linear chain of reactions (i.e., the series of reactions that connect a substrate to a product). This distinction is important because the extreme pathways account for all the reaction steps a network must use to complete the synthesis process. Therefore, extreme pathways can have multiple inputs and multiple outputs and are network properties. Consequently, extreme pathway length can also be characterized as the size or complexity of the corresponding flux distribution map. Another important characterization of extreme pathways is the reaction participation number, which is defined as the percentage of extreme pathways in which a given reaction participates. Reactions that participate in a large number of extreme pathways may represent good targets for regulation. Extreme pathway lengths and reaction participation numbers can thus be used to characterize the large-scale properties of metabolic networks. This study presents the first calculations of extreme pathway length and reaction participation for genome-scale metabolic networks, using the networks of H. pylori and H. influenzae as case studies.
Conceptual Framework
The new concepts introduced in this study require fairly explicit definition and explanation. In this section, we describe the conceptual framework for extreme pathway analysis and characterization before analyzing the H. pylori and H. influenzae genome-scale metabolic networks.
Extreme Pathways
The phenotypic capabilities of genome-scale metabolic networks can be characterized by a set of systemically independent and unique extreme pathways (Schilling et al. 2000). Extreme pathways correspond to steady-state flux distributions through a metabolic network. Thus, extreme pathways do not simply describe a linear set of reactions linking substrate to product, but instead, characterize the relative flux levels through all the reactions necessary to convert substrates to products, to balance all cofactor pools, and to secrete any byproducts needed to maintain the network in a homeostatic state. The sets of extreme pathways studied here lead to the synthesis of a target product, such as an individual amino acid or all the protein in a cell. Therefore, each extreme pathway in the set corresponds to a complete flux map that synthesizes the target product within the metabolic network.
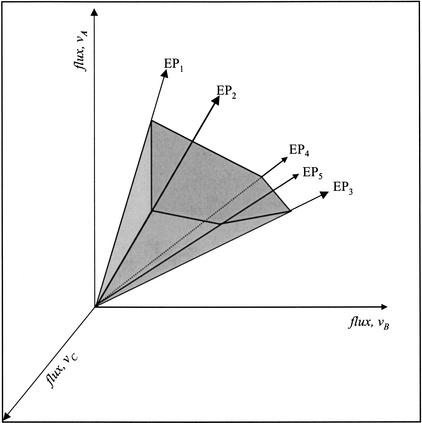
Extreme pathways are so named because they are the edges of a solution space and thus characterize the extreme functions of the network. The extreme pathways can be thought of as generating a convex cone in high-dimensional space, circumscribing all possible steady-state metabolic phenotypes (Fig. 1). All potential steady-state flux distributions through the network (hence, all metabolic phenotypes) are non-negative linear combinations of the extreme pathways. Consequently, the extreme pathways specify theoretical upper and lower bounds of the conversion of any substrates to any products.
Figure 1.
Schematic representation of a convex cone characterized by five extreme pathways. Extreme Pathways 1–5 (EP1, EP2, EP3, EP4, and EP5) circumscribe the solution space for the three fluxes indicated (vA, vB, vC). EP4 lies in the plane formed by fluxes vA and vB. Consequently, flux vC does not participate in that extreme pathway. EP3, EP4, and EP5 are all close and represent different uses of a network to achieve a similar overall result. All points within the convex cone can be described as a non-negative linear combination of the extreme pathways.
It should be noted that the extreme pathways are an irreducible, nonredundant subset of elementary modes (Pfeiffer et al. 1999; Schuster et al. 1999, 2000). Elementary modes for a given network are more numerous than the extreme pathways, but can all be represented by non-negative, linear combinations of the extreme pathways.
Pathway Length and Reaction Participation Matrices
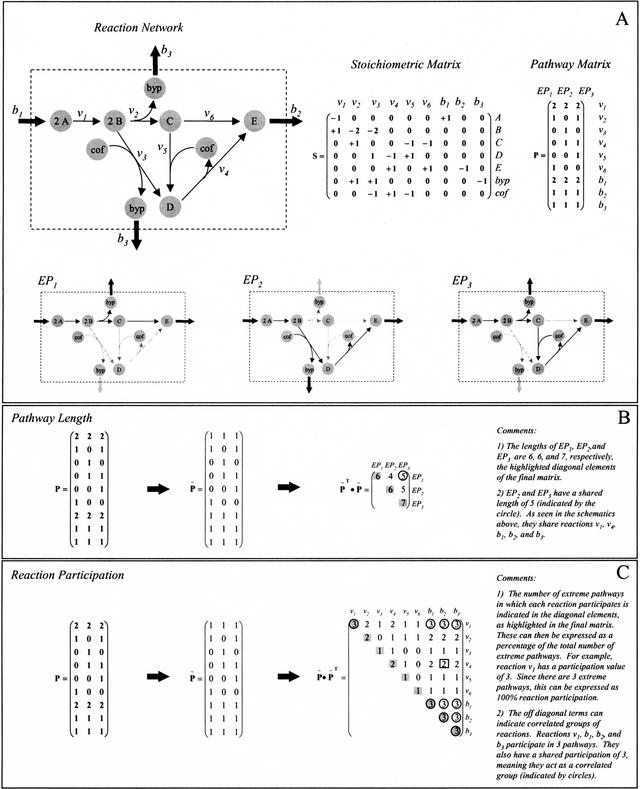
A matrix of extreme pathways can be formed in which each column is an extreme pathway and each row corresponds to a reaction in the network. The numerical value of the i,jth element corresponds to the relative flux level through the ith reaction in the jth extreme pathway. The Extreme Pathway Matrix is formed using all of the extreme pathways. A simple example reaction network and the corresponding Extreme Pathway Matrix are shown in Figure 2A. The Extreme Pathway Matrix shown in Figure 2A contains three extreme pathways. Each of these extreme pathways is displayed graphically in Figure 2A. EP1 and EP2 are not simply linear reaction chains, but instead, contain two outputs, E and the byproduct. Extreme pathways can have any number of inputs or outputs. EP3, like EP2, maintains cofactor pools at steady state. Each of the extreme pathways results in the production of the product E.
Figure 2.
Pathway Length and Reaction Participation Matrices for a simple network. (A) The example system with its corresponding stoichiometric and extreme pathway matrix. (A, bottom) Corresponding maps of the three extreme pathways. (B,C) The corresponding Length and Participation Matrices, as well as their respective properties. Note in A that although the metabolite byp only participates in one exchange flux, b3, there are two exchange flux arrows in the figure. However, both arrows correspond to the same single exchange flux.
The Pathway Length Matrix (PLM) is calculated directly from the Extreme Pathway Matrix, as shown in Figure 2B. The Extreme Pathway Matrix is first written in a binary form (P̃), in which each reaction is categorized as either used (1) or not used (0) within each extreme pathway. Then, the pathway length matrix, PLM, is computed by pre-multiplying the binary Extreme Pathway Matrix by its own transpose,
 |
1 |
resulting in a symmetric matrix. The PLM for the system in Figure 2A is shown in Figure 2B. The values along the diagonal of PLM correspond to the length of each extreme pathway. In the example system, the first value along the diagonal is 6, meaning that six reactions participate in EP1. A quick count of the reactions shown in EP1 (Fig. 2A) shows that there are six reactions participating in the first extreme pathway.
The off-diagonal terms of PLM are equally easy to interpret. They are the number of reactions that a pair of extreme pathways have in common. For example, notice the circled off-diagonal term in Figure 2B. This element is a comparison of EP3 (the column) and EP1 (the row) and contains a value of 5. This means that EP1 and EP3 have five reactions in common. Upon examining EP1 and EP3 in Figure 2A, one can readily see that the five reactions shared are b1, v1, v2, b2, and b3. Thus, the off-diagonal terms of the pathway length matrix are the reactions common to the two pathways being compared at each element of the matrix.
The Reaction Participation Matrix (RPM) is also calculated directly from the binary form of the Extreme Pathway Matrix. The Reaction Participation Matrix is calculated by post-multiplying the binary Extreme Pathway Matrix by its own transpose,
 |
2 |
also forming a symmetric matrix. The RPM was calculated for the example system, as shown in Figure 2C. The diagonal terms in RPM refer to the number of pathways in which the given reaction participates. For example, the first diagonal term, corresponding to reaction v1, has a value of 3. This means that reaction v1 participates in all three extreme pathways. An examination of EP1, EP2, and EP3 in Figure 2A shows that reaction v1, which converts A → B, is in fact utilized in all three extreme pathways. The values in the Reaction Participation Matrix can be characterized as percentages of the total number of extreme pathways. To accomplish this, the entire matrix, RPM, is normalized to the total number of extreme pathways, three in the example case. Thus, the first diagonal element would correspond to 100% reaction participation, as reaction v1 was utilized in all three extreme pathways.
The off-diagonal terms refer to the number of extreme pathways that contain both of the corresponding reactions. For example, notice the off-diagonal element boxed in Figure 2C containing a value of 2 (or 2/3, 67%). This element refers to the number of pathways that contain both reaction b2 (the column) and reaction v4 (the row). Upon examining the extreme pathways in Figure 2A, one can see that both of these reactions are utilized in EP2 and EP3, whereas only b1 is utilized in EP1. The circled elements in Figure 2C show reaction pairs that participate in exactly the same extreme pathways. In this particular case, each of these reaction pairs participates together in all of the extreme pathways. Thus, reactions v1, b1, b2, and b3 are always present. They form a reaction group, meaning that if one of them is utilized, the others must also be utilized.
This section has provided a simple method to study mathematically derived formulations describing extreme pathway length and reaction participation. The Pathway Length and Reaction Participation Matrices contain the minor and major product moments (Horst 1965) of the binary Extreme Pathway Matrix, respectively. The Pathway Length and Reaction Participation characterization of the Extreme Pathway Matrix can be utilized together to examine the integrated properties of large-scale networks.
RESULTS
The metabolic network for H. influenzae used in this study contained 461 reactions and 367 metabolites. The metabolic network for H. pylori used in this study contained 381 reactions and 332 metabolites. The reconstruction of metabolic networks has been reviewed previously (Covert et al. 2001).
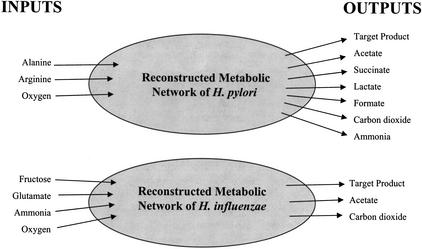
Reaction Participation and Pathway Length Matrices were calculated from various Extreme Pathway Matrices for the H. influenzae and H. pylori networks described previously (Papin et al. 2002; Price et al. 2002). Statistical analyses of these matrices for various data sets in H. influenzae and H. pylori were performed and the results are presented below. The Extreme Pathway Matrices for the following data sets were evaluated: (1) the individual production of the nonessential amino acids in H. influenzae, (2) the individual production of the nonessential amino acids in H. pylori, and (3) the production of the set of nonessential amino acids in H. pylori. Allowed inputs and outputs to the two genome-scale metabolic networks are shown in Figure 3.
Figure 3.
Illustration of the input and output constraints for the genome-scale metabolic networks in this study. The utilized inputs in H. influenzae and H. pylori are listed at left. More inputs were allowed to comprise previously defined minimal medium. However, only the inputs listed above were utilized by the metabolic networks for the synthesis of the specified products. The allowed outputs are listed at right. The target products for H. pylori included the nonessential amino acids as well as the simultaneously produced set of nonessential amino acids in both E. coli and equimolar ratios. The target products for H. influenzae included the nonessential amino acids.
Reaction Participation
The reaction participation values were calculated for the three data sets listed above. The percentages of extreme pathways in which each reaction participated were calculated and rank ordered as shown in Figure 4. The shape of each of the curves in Figure 4 shows three distinct regions as follows: (1) an initial flat portion of the curve representing the reactions that are utilized in all extreme pathways; (2) a region in which the reactions are sometimes, but not always, used in the extreme pathways that produce the target product; and (3) a final region containing those reactions that are never used in an extreme pathway that produces the target product. The second region contains the reactions that can be used for synthesis of the target product, but are not always necessary. Thus, this region represents reactions used in various alternate routes for the synthesis of the target product.
Figure 4.
Reaction participation values for all of the data sets calculated in this study. (A,B) The spectrum of reaction participation values, with a shortened x-axis so as to highlight the non-zero reaction participation values. Note the three regions in A and B as follows: (1) set of reactions that participate in all of the extreme pathways; (2) reactions that participate in varying amounts of extreme pathways; and (3) reactions that do not participate in any of the extreme pathways. Also note that the ordering of the reaction number is different for each data set so that the reaction number in each set corresponds to a different reaction.
The numbers of essential and utilized reactions for the analyzed data sets are summarized in Table 1. A comparison of the size of essential reaction sets for the production of amino acids with equivalent central metabolic precursors yielded an interesting result. The aromatic amino acids (which are all derived from phosphoenolpyruvate and erythrose 4-phosphate) in H. influenzae and H. pylori have the highest number of reactions that are in 100% of their respective extreme pathways (with the exception of histidine). Although the aromatic amino acids cluster together with regard to the number of essential reactions, not all amino acids with similar central metabolic precursors are similarly grouped. For example, the amino acids that are derived from oxaloacetate (aspartic acid, asparagine, methionine, threonine, lysine, and isoleucine) do not appear to group together according to the number of essential reactions.
Table 1.
Number of Reactions Involved in the Production of the Indicated Target Product
| H. pylori Target product | Essential reactions | Utilized reactions |
|---|---|---|
| Tryptophan | 32 | 105 |
| Tyrosine | 28 | 101 |
| Cysteine | 25 | 102 |
| Glycine | 22 | 97 |
| Lysine | 22 | 102 |
| Serine | 16 | 91 |
| Threonine | 14 | 96 |
| Asparagine | 13 | 91 |
| Aspartic Acid | 12 | 91 |
| Proline | 10 | 91 |
| Glutamic Acid | 7 | 91 |
| Glutamine | 6 | 91 |
| Equimolar Amino Acids | 85 | 140 |
| E. coli Ratio Amino Acids | 85 | 140 |
| H. influenzae Target product | Essential reactions | Utilized reactions |
| Histidine | 51 | 112 |
| Tryptophan | 41 | 108 |
| Phenylalanine | 36 | 108 |
| Tyrosine | 36 | 108 |
| Methionine | 34 | 106 |
| Isoleucine | 31 | 108 |
| Lysine | 31 | 108 |
| Glycine | 29 | 82 |
| Threonine | 26 | 103 |
| Asparagine | 25 | 98 |
| Serine | 25 | 97 |
| Leucine | 23 | 105 |
| Aspartic Acid | 22 | 97 |
| Glutamine | 21 | 102 |
| Proline | 18 | 103 |
| Valine | 17 | 102 |
| Alanine | 12 | 99 |
See Fig. 3 for the indicated network inputs and outputs. Essential reactions refers to the number of reactions that were used in every extreme pathway (region I in Fig. 4). Utilized reactions refers to the number of reactions that were used at least once in the set of extreme pathways for the production of the associated product (region II in Fig. 4). The individual amino acids are sorted in descending order according to the number of essential reactions. Equimolar amino acids refers to the set of amino acids in equimolar ratios. E. coli ratio amino acids refers to the set of amino acids in ratios analogous to those seen in E. coli biomass.
Correlation of Reaction Participation Values; Definition of Reaction Subsets
The choices of which reactions can be used in a particular extreme pathway are not independent. The off-diagonal elements of the Reaction Participation Matrix can be used to determine subsets of reactions that always appear together across all of the extreme pathways. One obvious reaction subset is the set of all reactions that must appear in every extreme pathway. Other reaction subsets can be nonobvious.
The reaction subsets are shown in Tables 2 and 3 for lysine production in H. influenzae and H. pylori, respectively. The reactions in these reaction subsets must be either all present or all absent in any extreme pathway. Because these enzymes must operate together in all steady states, it seems likely that these enzymes would be coexpressed (Pfeiffer et al. 1999; Schilling and Palsson 2000). Thus, these reaction subsets provide groups of enzymes that may be coregulated. Tables 2 and 3 are related to the data shown in Figure 4. The reactions listed in the first group of both Tables 2 and 3 correspond to the first region of their respective curves in Figure 4, indicating that these reactions were always used, whereas the rest of the reaction groups shown in Tables 2 and 3 correspond to the reactions in the drop-off region of the respective curves in Figure 4. Note that Tables 2 and 3 do not list reactions that are never used.
Table 2.
Reaction Subsets for Lysine Synthesis in H. influenzae
| 1 | THRA1, ASD, DAPA, DAPB, DAPD, DAPC, DAPF, LYSA, GDHA, ASPC2, FBA, TPIA, GAPA, PGK, GPMA, ENO, SUCCD, PPC, FRUTR, FRUK, PTA, ACKA, CO2TR, ACTR, NH3TR, ACxt, CO2xt, FRUxt, NH3xt, LYS |
| 2 | SERA, SERC, SERB |
| 3 | GLYA, GCV, FMT, FOLD1, FOLD2 |
| 4 | ASNA, ANSB, ADK1 |
| 5 | ASPA, FUMC |
| 6 | PGI1, ZWF, GND, RPIA, RPE, TALB, TKTA1, TKTA2, PGL |
| 7 | ACCABCD, FABD, FABH, FABB |
| 8 | DGKA, PAPHTSE |
| 9 | GLMS, NAGB |
| 10 | PYRG, NDK2, NDK3, CDD1, CMKB2, CMKB3, USHA6 |
| 11 | NDK1, UDK |
| 12 | NDK5, TMK2, DUT |
| 13 | TDK1, USHA2 |
| 14 | TDK2, USHA1 |
| 15 | GLGC, GLGA, GLGP |
| 16 | GLPA, GPSA |
| 17 | CYDA, O2TR, O2xt |
| 18 | MGSA, GLOA, GLOB |
Each of the reactions subsets above are correlated in the extreme pathways that correspond to lysine synthesis in H. influenzae. For example, in every extreme pathway, the reactions TDK2 and USHA1 (group 14) are either used or not used together. Group I contains the reactions that are always utilized (region I in Fig. 4) and Groups 2–18 above are in the variable region (region II in Fig. 4)
Table 3.
Reaction Subsets for Lysine Synthesis in H. pylori
| 1 | ASPB1, METL1, ASD, DAPA, DAPB, DAPD, DAPC, DAPE, DAPF, LYSA, MAEB, MDH, OOR, ALATP, SUCTP, FRDO, ATPA, NH3TP, ALAxt, NH3xt, SUCCxt, LYS |
| 2 | SDAA, SERA, SERC, SERB |
| 3 | PUTA1, PROC |
| 4 | PUTA2, ORNTRSN, ROCF, ARGTP, UREASE, ARGxt |
| 5 | DADA, ALR |
| 6 | ASNA, ANSB |
| 7 | PGI, FBP, FBA, TPI, GAP, PGK, PGL |
| 8 | PGM, ENO, PPSA |
| 9 | GND, RPI, RPE, TAL, TKTA1, TKTA2 |
| 10 | EDD, EDA |
| 11 | GLTA, ACNB, ICD |
| 12 | POR, FLDO |
| 13 | CDSA, CDH, CMKA, NDK3 |
| 14 | GUAB, GUAA, GUAC |
| 15 | NDK5, TMK2, DUT |
| 16 | PTA, ACKA |
| 17 | ACTP, ACxt |
| 18 | PROTP1, NATP |
| 19 | LACTP, DLD, LACxt |
| 20 | BC1O, O2TP, O2xt |
| 21 | CO2TP, CO2xt |
Each of the reactions subsets are correlated in the extreme pathways that correspond to lysine production in H. pylori. For example, in every extreme pathway, the reactions SDAA, SERA, SERC, SERB (group 2) are either used together or not used at all.
From detailed analysis of these reaction subsets, some interesting characteristics emerge. First, the pentose phosphate reactions in H. influenzae lysine synthesis (group 6 in Table 2) and the serine synthesis reactions (group 2 in Table 2) form obvious groups. A nonobvious group for lysine synthesis in H. influenzae consists of the reactions in group 4 of Table 2 as follows: (1)ASNA, ASP + ATP + NH3 → ASN + AMP + PPI; (2) ANSB, ASN → ASP + NH3; (3) ADK1, ATP + AMP ↔ 2 ADP, with metabolites ASP – L-aspartate, ASN – L-asparagine, ATP – adenosine triphosphate, AMP – adenosine monophosphate, ADP – adenosine diphosphate, NH3 – ammonia, and PPI – diphosphate. Reaction ASNA is catalyzed by the enzyme aspartate-ammonia ligase; reaction ANSB is catalyzed by the enzyme L-asparaginase II; and reaction ADK1 is catalyzed by the enzyme adenylate kinase. The first two reactions interconvert asparagine and aspartate (albeit with different byproducts and cofactors). However, the reaction ADK1 is not a reaction that would obviously be correlated to ASNA and ANSB. Initially, this reaction might be grouped with nucleotide synthesis reactions, or some other energy-associated reactions. It is also interesting to note that no other reaction correlates with this subset (i.e., another reaction involved in nucleotide synthesis or energy production).
For lysine synthesis in H. pylori, there were other nonobvious reaction subsets. Group 13 consists of the following reactions: (1) CDSA, PA + CTP → CDPDG + PPI; (2) CDH, CDPDG → CMP + PA; (3) CMKA, CMP + ATP ↔ ADP + CDP; (4) NDK3, CDP + ATP ↔ CTP + ADP, with metabolites PA – phosphatidate, CTP – cytidine triphosphate, CDPDG – CDP-diacylglycerol, PPI – diphosphate, CMP – cytidine monophosphate, CDP – cytidine diphosphate ATP – adenosine triphosphate, and ADP – adenosine diphosphate. It is interesting to note that two reactions (CDSA and CDH) associated with phospholipid and fatty acid metabolism were correlated with two reactions (CMKA and NDK3) associated with nucleotide synthesis. Another interesting grouping in H. pylori lysine synthesis was seen in groups 7 and 8. Both subsets contain reactions associated with glycolysis, and yet, the reactions do not group together. This result ocurrs because the metabolite 3-phosphoglycerate (3PG) can be used in the reaction SERA; thus, 3PG produced by the reaction PGK need not be consumed in the reaction PGM. Rather, 3PG can be siphoned off from glycolysis and consumed by the reaction SERA. Consequently, PGK and PGM are not in the same reaction subset for lysine synthesis in H. pylori.
Extreme Pathway Length
The extreme pathway lengths were calculated for the three data sets described above. Figures 5 and 6 show the histograms of extreme pathway lengths for the production of the amino acids in H. pylori and H. influenzae, respectively. Table 4 presents a summary of the statistical properties for the extreme pathway length distributions shown in Figures 5 and 6.
Figure 5.
Extreme pathway length distributions in H. pylori. The x-axis in each of the figures represents the length of an extreme pathway. The y-axis in each of the figures represents the number of extreme pathways at the corresponding length.
Figure 6.
Extreme pathway length distributions in H. influenzae. The x-axis in each of the figures represents the length of an extreme pathway. The y-axis in each of the figures represents the number of extreme pathways at the corresponding length. Tyrosine and phenylalanine have the exact same pathway length distributions.
Table 4.
Summary of the Statistical Analyses of Extreme Pathway Lengths
| H. pylori Target product | Number of EPs | Pathway length | |||
|---|---|---|---|---|---|
| average | maximum | minimum | coefficient of variation | ||
| Asparagine | 340 | 44 | 54 | 28 | 15% |
| Aspartic Acid | 491 | 43 | 52 | 24 | 14% |
| Cysteine | 1022 | 59 | 71 | 45 | 10% |
| Glutamine | 315 | 41 | 53 | 23 | 18% |
| Glutamic Acid | 493 | 41 | 53 | 25 | 17% |
| Glycine | 377 | 51 | 60 | 38 | 10% |
| Lysine | 611 | 54 | 66 | 39 | 12% |
| Proline | 867 | 43 | 56 | 15 | 16% |
| Serine | 355 | 45 | 54 | 33 | 12% |
| Threonine | 469 | 48 | 60 | 31 | 14% |
| Tryptophan | 1958 | 64 | 73 | 51 | 6% |
| Tyrosine | 1008 | 58 | 68 | 44 | 7% |
| Equimolar Amino Acids | 6032 | 106 | 112 | 99 | 2% |
| E. coli Ratio Amino Acids | 5553 | 106 | 112 | 99 | 2% |
| H. influenzae Target product | Number of EPs | Pathway length | |||
| average | maximum | minimum | coefficient of variation | ||
| Alanine | 1739 | 36 | 49 | 18 | 10% |
| Asparagine | 445 | 39 | 52 | 29 | 13% |
| Aspartic Acid | 690 | 35 | 49 | 27 | 14% |
| Glutamine | 690 | 37 | 46 | 28 | 11% |
| Glycine | 456 | 39 | 48 | 35 | 7% |
| Histidine | 1507 | 65 | 74 | 61 | 3% |
| Isoleucine | 1480 | 47 | 61 | 37 | 9% |
| Leucine | 3884 | 42 | 55 | 31 | 10% |
| Lysine | 1168 | 47 | 61 | 37 | 9% |
| Methionine | 1343 | 48 | 63 | 40 | 8% |
| Phenylalanine | 1758 | 51 | 64 | 43 | 7% |
| Proline | 2624 | 38 | 51 | 25 | 11% |
| Serine | 690 | 37 | 50 | 30 | 10% |
| Threonine | 1318 | 42 | 55 | 32 | 10% |
| Tryptophan | 3540 | 58 | 69 | 49 | 6% |
| Tyrosine | 1758 | 51 | 64 | 43 | 7% |
| Valine | 1739 | 39 | 52 | 23 | 9% |
The coefficient of variation is the standard deviation normalized to the average (expressed as a percent). Equimolar amino acids refers to the set of amino acids in equimolar ratios. E. coli ratio amino acids refers to the set of amino acids in ratios analogous to those seen in E. coli biomass. EPs, extreme pathways.
Extreme Pathway Length Distribution
The histograms of extreme pathway lengths for the production of each of the nonessential amino acids in H. pylori are shown in Figure 5. The distributions in extreme pathway length corresponding to the production of each of the amino acids are very diverse. However, a few common features exist among these distributions. One striking characteristic shown in Figure 5 is that many of the distributions have more than one peak. Thus, it seems that there are often multiple common extreme pathway lengths around which deviations can be made.
The histograms for the extreme pathway lengths for the production of each of the amino acids in H. influenzae are shown in Figure 6. The extreme pathway length distribution of valine and alanine are essentially identical, except that the extreme pathway lengths of valine are shifted. It can be seen that for all of the longer pathways, it takes three extra reaction steps to make valine instead of alanine. However, for the shorter extreme pathways, it actually takes five extra reaction steps to make valine instead of alanine. Thus, the number of extra reaction steps needed to make valine instead of alanine depends upon the set in which the active extreme pathways lie.
It is instructive to compare the extreme pathway length distributions for the same products between H. pylori and H. influenzae. For example, a comparison of aspartic acid production in H. pylori and H. influenzae reveals a distribution that is roughly reversed between the two organisms. A similar pattern is seen with tryptophan and tyrosine. The extreme pathway length distributions for asparagine are similar for the two organisms, whereas many distributions of the other amino acids are quite different.
The extreme pathway length distributions for the simultaneous production of the set of nonessential amino acids in H. pylori is shown in Figure 5, both for equimolar ratios and for ratios corresponding to the amino acid composition in Escherichia coli. The range of pathway lengths was quite small, varying between 99 and 112 for both sets with a coefficient of variation of 2%. The shape of the distributions for both compositions was very similar.
Correlation to Product Yield and Molecule Complexity
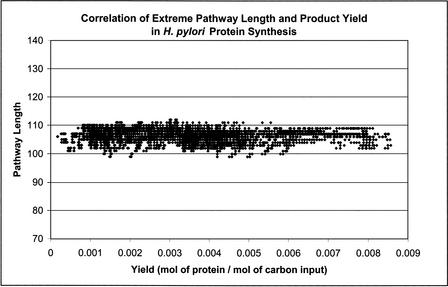
The yield of the target product (defined as output flux per unit carbon input flux) was plotted against the pathway length for all data sets in H. pylori and H. influenzae, with a representative set shown in Figure 7. In all cases, there was a very poor correlation between the yield of the target product (amino acids, ribonucleotides) and the length of the extreme pathway. However, for proline synthesis in H. pylori and alanine and valine synthesis in H. influenzae, the maximum yield pathways were also the pathways with the shortest length (data not shown). There was no obvious correlation between the yield and the pathway length for the other extreme pathways. Thus, finding the shortest extreme pathway between substrate and product did not correlate to finding the extreme pathway of highest yield.
Figure 7.
Correlation of extreme pathway length and yield (mol of protein/mol of carbon input) in H. pylori protein synthesis. There was essentially zero correlation between target product yield and extreme pathway length in all of the data sets evaluated in this study, as illustrated in this figure.
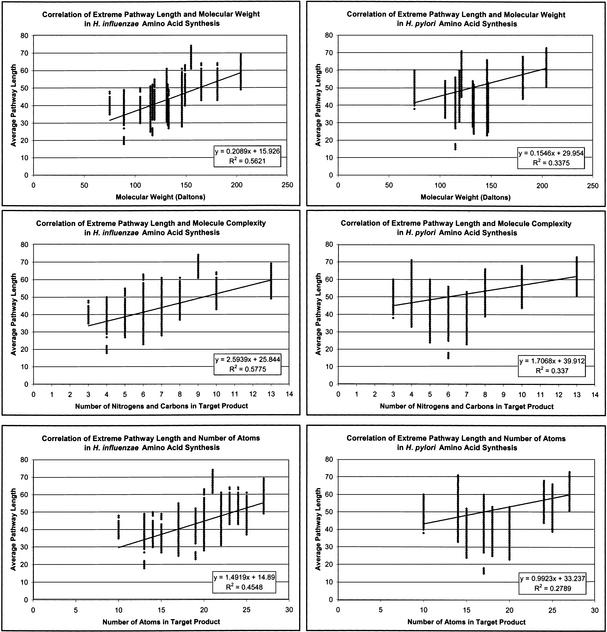
The extreme pathways were also evaluated to determine whether extreme pathway length could be correlated with the complexity of the molecule being synthesized. Molecular complexity was defined by (1) the number of carbon and nitrogen atoms in the molecule, (2) the total number of atoms in the molecule, and (3) the molecular weight of the molecule. The average pathway length was plotted against these characterizations of molecular complexity (Fig. 8). A line was fitted to the data to determine the degree of linear correlation between the two variables. The resultant R2 values for the three cases are indicated on the plots (with P values << 0.001 for the slopes and intercepts in all three cases). Thus, there appears to be a weak correlation between extreme pathway length and the chemical complexity of the target product.
Figure 8.
Correlation of extreme pathway length with molecule complexity. The molecule complexity is characterized by the number of carbons and nitrogens, the total number of atoms, and the molecular weight of the target product. The line that best fits the data is indicated, along with its corresponding equation and R2 value. All of the weak linear correlations are statistically significant (P-value << 0.001).
DISCUSSION
This study presents mathematically precise definitions of extreme pathway length and reaction participation, and their evaluation for genome-scale metabolic networks. These extreme pathway characterizations help to study emergent properties of metabolic networks. From the Reaction Participation and Pathway Length Matrices, several key results were obtained as follows: (1) the reaction participation values demonstrated the computation of essential reaction subsets without which the network would be incapable of synthesizing the product of interest; (2) the off-diagonal terms of the Reaction Participation Matrix elucidated sets of reactions that were systemically correlated; (3) the minimal extreme pathway length indicated the minimum number of reactions necessary to synthesize a given product, including a set of variable reactions in addition to the essential reaction set; (4) the extreme pathway lengths were not correlated with the product yield and were poorly correlated with different measures of molecular complexity of the target product; and (5) there were distinct extreme pathway length distribution differences between the two organisms studied. Thus, Reaction Participation and Pathway Length Matrices can be used to clearly and quantitatively enumerate emergent properties of defined metabolic networks.
The Reaction Participation Matrix allowed the definition of essential reaction sets. The determination of these essential reaction sets could enable the identification of the organism's weaknesses. A knockout of any one of these reactions would result in the complete crippling of the synthesis capability for the corresponding product. For lysine synthesis in H. influenzae, there were 31 reactions in all of the extreme pathways forming the core set of reactions that must be present in lysine synthesis. However, there were 37 reactions in the shortest extreme pathway, showing that at least 6 additional reactions were needed to complete a pathway in addition to the core set of reactions. For the synthesis of the equimolar amino acid set in H. pylori, there were 85 reactions in all of the extreme pathways, with a length of 99 reactions for the shortest extreme pathway. The difference between the size of the essential reaction set and the minimum and maximum pathway lengths represents a certain degree of redundancy in the network, an ability of the metabolic network to make a selection in how the product is synthesized. Identifying the reactions in this variable group can indicate where an organism's robustness resides.
An analysis of the off-diagonal values of the Pathway Length and Reaction Participation Matrices led to interesting characterizations. The off-diagonal terms of the Pathway Length Matrix indicated the number of reactions that a pair of extreme pathways has in common, in other words, their shared length. Many core reactions are used in the synthesis of most products. From the Reaction Participation Matrix, we can determine the number of pathways that use both of a given pair of reactions. The shared participation in part indicates the correlation between reactions for a given metabolic network under specified conditions. Interestingly, nonobvious reaction subsets were found. Whereas obvious reaction subsets (e.g., pentose phosphate reactions) indicate an expected functional connection under the specified conditions, less-obvious groups indicate a functional connection that goes beyond traditional classifications, as discussed above. Subsequently, these less-obvious reaction subsets could correspond to genes that are transcriptionally coregulated (Pfeiffer et al. 1999; Schilling and Palsson 2000). This study presents the first analysis of reaction correlation from the full complement of metabolic genes. If the groups are not transcriptionally coregulated, it would be interesting to investigate why the reactions in a subset are functionally coregulated, perhaps serving to better understand physiological behavior, objectives, or adaptive pressures.
Interestingly, the length of an extreme pathway was found to be uncorrelated with the yield values for all cases studied herein. This result suggests that a simple visual inspection of a metabolic network cannot readily identify optimal pathways for the production of a given product. For example, simply identifying the shortest pathway from reactant to product is not necessarily the pathway of maximum yield. There could be multiple routes that combine carbon lost as a byproduct in one reaction step and reincorporated in another reaction step to produce the highest yield for a given product. Furthermore, various measures of molecular complexity were also not strongly correlated with the target product yield. This result further supports the observation that metabolic networks are so inherently interconnected and complex that integrative analytical approaches are needed to elucidate systemic characterizations.
In summary, the Pathway Length and Reaction Participation Matrices are presented herein as unambiguous and mathematically derived characteristics of metabolic networks. Extreme pathways represent a unique and independent set of vectors whose non-negative linear combinations characterize all possible steady-state phenotypes. As such, the Pathway Length and Reaction Participation Matrices serve as unique and independent characterizations of the metabolic network. These characterizations have elucidated emergent systemic properties of reconstructed genome-scale metabolic networks in H. influenzae and H. pylori. As modern queries of biological systems expand in complexity, emergent systemic properties will become more difficult to parse out of the raw data. Extreme Pathway Matrices and their resultant characterizations may become an important method for the analysis necessary to determine these systemic properties.
METHODS
Genome-Scale Maps
The in silico models for H. pylori (Schilling et al. 2002) and H. influenzae (Edwards and Palsson 1999; Schilling and Palsson 2000) were reconstructed previously using established methodologies (Covert et al. 2001). The reactions used in these in silico models can be found at http://gcrg.ucsd.edu/downloads. These in silico strains were constrained to minimal medium requirements and defined exchange fluxes. Figure 3 illustrates schematically the systemic input and output constraints utilized in this study.
Extreme Metabolic Pathways and Convex Analysis
The Extreme Pathway Matrices utilized in the present study were calculated as described previously for H. influenzae (Papin et al. 2002) and H. pylori (Price et al. 2002). Briefly, Extreme Pathway Matrices are derived directly from the stoichiometric matrix describing the known metabolic network of an organism. The m × n stoichiometric matrix, S, of a reconstructed metabolic network includes all metabolites (m rows) and all corresponding metabolic reactions and transport processes (n columns). The flux represents the amount of mass moving through the associated reaction. An exchange flux corresponds to a flux across the system boundary. An internal flux corresponds to a reaction within the system.
A metabolic network can be constrained by implementing simple thermodynamic principles regarding the irreversibility of reactions. With reversible reactions decomposed into their respective forward and reverse directions, all internal fluxes are constrained to be non-negative, as expressed in Equation 3.
 |
3 |
in which v is a vector of all the internal fluxes of the metabolic network.
The stoichiometric constraints of a metabolic system (conservation of mass) at steady state can be described by Equation 4, in which S is the m × n stoichiometric matrix described above. The fluxes through each of the n corresponding reactions of the stoichiometric matrix are represented by the vector, v.
 |
4 |
A convex basis is constructed to span all solutions to Equation 4, subject to the inequality constraints from Equation 3, so that pathways do not use fluxes opposite the direction of an irreversible reaction. The vectors forming this convex basis are the extreme pathways (pi). From this convex basis, a cone can be generated to circumscribe all allowable solutions. Every point within the cone can be written as a non-negative combination of the extreme pathways (Equation 5). Thus, this cone circumscribes all valid solutions to Equation 4.
 |
5 |
Any point in the interior of this cone represents a valid steady-state set of flux values for the metabolic network and corresponds to a particular metabolic phenotype. A more detailed description of the theory behind extreme pathway analysis and a description of the algorithm used to calculate the pathways can be found elsewhere (Schilling et al. 2000). For the purposes of this study, the flux corresponding to the target product was constrained to be positive.
Statistical Calculations
All statistical calculations and plots were generated with Statistica (Statsoft) and Excel (Microsoft) software.
Reaction Subset Calculations
The correlated reaction groups were determined by inspecting the off-diagonal terms of the Reaction Participation Matrix. A MATLAB program was used to scan through all of the off-diagonal terms of the reaction participation matrix. Reaction i and reaction j were placed into the same subset whenever element i,j was equal to both element i,i and element j,j. These subsets of two were then joined whenever they had elements in common, until no more groupings could be made and each reaction was a member of no more than one group. For example, if the elements i,i, and j,j each had a value of 30, reactions i and j would participate in 30 extreme pathways. If the element i,j had a value of 30, then the reactions i and j would participate together in 30 extreme pathways. Subsequently, when the elements i,i, j,j, and i,j have equivalent values, then the corresponding reactions always appear together.
Acknowledgments
The authors acknowledge the helpful comments and suggestions from Dr. Nagiza Samatova, Markus Covert, Iman Famili, Jennifer Reed, and Sharon Wiback. Financial support for this work was provided by grants from the National Institutes of Health (GM57089), the National Science Foundation (BES 98-14092, MCB 98-73384, and BES 01-20363), and the Whitaker Foundation (Graduate Research Fellowship to J.P.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL palsson@ucsd.edu; FAX (858) 822-3120.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.327702.
REFERENCES
- Covert MW, Schilling CH, Famili I, Edwards JS, Goryanin II, Selkov E, Palsson BO. Metabolic modeling of microbial strains in silico. Trends Biochem Sci. 2001;26:179–186. doi: 10.1016/s0968-0004(00)01754-0. [DOI] [PubMed] [Google Scholar]
- Edwards JS, Palsson BO. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J Biol Chem. 1999;274:17410–17416. doi: 10.1074/jbc.274.25.17410. [DOI] [PubMed] [Google Scholar]
- Horst P. Factor analysis of data matrices. New York: Holt Rinehart and Winston; 1965. [Google Scholar]
- Karp PD, Krummenacker M, Paley S, Wagg J. Integrated pathway-genome databases and their role in drug discovery. Trends Biotech. 1999;17:275–281. doi: 10.1016/s0167-7799(99)01316-5. [DOI] [PubMed] [Google Scholar]
- Liao JC, Hou SY, Chao YP. Pathway analysis, engineering and physiological considerations for redirecting central metabolism. Biotech Bioengr. 1996;52:129–140. doi: 10.1002/(SICI)1097-0290(19961005)52:1<129::AID-BIT13>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Mavrovouniotis ML, Stephanopoulos G. Computer-aided synthesis of biochemical pathways. Biotech Bioengr. 1990;36:1119–1132. doi: 10.1002/bit.260361107. [DOI] [PubMed] [Google Scholar]
- Ouzounis CA, Karp PD. Global properties of the metabolic map of Escherichia coli. Genome Res. 2000;10:568–576. doi: 10.1101/gr.10.4.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin JA, Price ND, Edwards JS, Palsson BO. The genome-scale metabolic extreme pathway structure in Haemophilus influenzae shows significant network redundancy. J Theor Biol. 2002;215:67–82. doi: 10.1006/jtbi.2001.2499. [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Sanchez-Valdenebro I, Nuno JC, Montero F, Schuster S. METATOOL: For studying metabolic networks. Bioinformatics. 1999;15:251–257. doi: 10.1093/bioinformatics/15.3.251. [DOI] [PubMed] [Google Scholar]
- Price ND, Papin JA, Palsson BO. Determination of redundancy and systems properties of Helicobacter pylori's metabolic network using genome-scale extreme pathway analysis. Genome Res. 2002;12:760–769. doi: 10.1101/gr.218002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatova, N.F., Geist, A., Ostrouchov, G., and Melechko, A.V. 2002. Parallel out-of-core algorithm for genome-scale enumeration of metabolic systematic pathways. Proc. First IEEE Workshop on High Performance Computat. Biol. (HiCOMB2002), Ft. Lauderdale, FL.
- Schilling CH, Palsson BO. Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis. J Theor Biol. 2000;203:249–283. doi: 10.1006/jtbi.2000.1088. [DOI] [PubMed] [Google Scholar]
- Schilling CH, Schuster S, Palsson BO, Heinrich R. Metabolic pathway analysis: Basic concepts and scientific applications in the post-genomic era. Biotech Prog. 1999;15:296–303. doi: 10.1021/bp990048k. [DOI] [PubMed] [Google Scholar]
- Schilling CH, Letscher D, Palsson BO. Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. J Theor Biol. 2000;203:229–248. doi: 10.1006/jtbi.2000.1073. [DOI] [PubMed] [Google Scholar]
- Schilling CH, Covert MW, Famili I, Church GM, Edwards JS, Palsson BO. Genome-scale metabolic model of Heliicobacter pylori 26695. J Bacteriol. 2002;184:4582–4593. doi: 10.1128/JB.184.16.4582-4593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S, Dandekar T, Fell DA. Detection of elementary flux modes in biochemical networks: A promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 1999;17:53–60. doi: 10.1016/s0167-7799(98)01290-6. [DOI] [PubMed] [Google Scholar]
- Schuster S, Fell DA, Dandekar T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat Biotechnol. 2000;18:326–332. doi: 10.1038/73786. [DOI] [PubMed] [Google Scholar]