Abstract
The regulation of transcription and subsequent gene splicing are crucial to correct gene expression. Although a number of regulatory sequences involved in both processes are known, it is not clear how general their functions are in the genomic context, nor how the regulatory regions are distributed throughout the genome. Here we study the distribution of known mutagenic elements within human introns and exons to deduce the properties of regions essential for splicing and transcription. We show that intronic splicing regulators are generally found close to the splice sites, but may be found as far as 200 nucleotides away from the splice junctions. Similarly, sequences important for splicing may be located as far as 125 nucleotides away from the junctions, within exons. We characterize several types of simple repetitive sequences and low-complexity regions that are overrepresented close to both intron ends and are likely to play important roles in the splicing process. We show that the first introns within most genes play a particularly important regulatory role that is most likely, however, to be involved in transcription control. We also study the distribution of two known regulatory motifs, the GGG trinucleotide and the CpG dinucleotide, and deduce their respective importance to splicing and transcription regulation.
The protein coding functions of genes have long been considered their most important attributes. However, the correct functioning of the encoded protein depends critically on the proper spatial and temporal expression of the gene, followed by appropriate splicing of the transcribed pre-mRNA. It is becoming clear that, not only changes in the coding sequence, but also mutations affecting transcription regulation (Klesert et al. 1997) and the splicing machinery (Ars et al. 2000; Blencowe 2000; Cartegni et al. 2002) are responsible for human genetic disease. Here, we focus on investigating the distribution and characterization of regions responsible for splicing regulation. However, since the physical range of splicing regulators overlaps that of transcriptional regulators, we also infer several properties of regions involved in transcription control.
Exon-intron boundaries are defined by simple rules (Kent and Zahler 2000); an intron begins with a splice acceptor, the AG dinucleotide, and ends with a splice donor, the GT dinucleotide. There are very few exceptions to this rule. However, such dinucleotides are abundant within each gene. Additional information is necessary for accurate splicing. One piece of information is provided by the presence of a branch site. This is a defined 7-nucleotide intronic sequence, usually at 15–40 nucleotides from the 3′ end of the intron. However, the branch site alone is not sufficient for correct gene splicing, even in short introns (Lim and Burge 2001).
Additional splicing control elements are known to exist. They may be involved in recognition of the appropriate splice site or in the suppression or enhancement of certain splice site usage, particularly in alternative splicing cases. It is difficult to make a clear distinction between splicing recognition and regulation, since an element may serve as a constitutive splicing enhancer/suppressor or promote alternative splicing, depending on the expression pattern of the gene and the availability of trans-acting associated factors. It has been estimated that a large proportion—estimates range from 35% to 59% (Mironov et al. 1999; Brett et al. 2000; Lander et al. 2001)—of all human genes are alternatively spliced. Hence, we will refer to all regions necessary for correct splicing as splicing control elements (SCEs).
Relatively little is known about the nature of SCEs. Generally, they do not obey defined consensus rules and are known to consist of diverse sequences (Fairbrother et al. 2002). Although some common SCEs have been observed, for example the GGG triplet (McCullough and Berget 1997; Carlo et al. 2000; Brudno et al. 2001), purine-rich elements (McCullough and Schuler 1997; Hastings et al. 2001), or polypyrimidine tracts commonly present in the 3′ intronic regions (Hastings and Krainer 2001), most newly discovered elements tend to consist of entirely new motifs, rather than belong to known groups (Blencowe 2000). Even less is known about the potential location of SCEs. They are most likely present in the vicinity of a splice site but have also been found as far away as 150 bp (Zheng et al. 2000; Rowen et al. 2002). They may be located within the intron or exon, and at their respective 5′ or 3′ ends (McCullough and Schuler 1997).
Although many gene promoter elements are well characterized and consist of several known essential motifs, such as the TATA box, the CAAT box, and numerous transcription factor binding sites located in the immediate proximity of the transcription initiation site, our knowledge of promoter regions is also far from complete (Suzuki et al. 2002). In particular, enhancer elements, which are often essential for correct gene expression, may be located at various distances throughout the gene and also belong to diverse classes of elements.
In this work, we use the distribution of known mutagenic elements in the human genome to deduce which regions are likely to be involved in transcription and splicing control. Deleterious mutations may be caused by any of the following: point mutations, insertions and deletions, insertions of transposable repetitive elements (Smit 1996), and microsatellite expansion or contraction. We use the complete human genome sequence and its annotation (Lander et al. 2001; Kent et al 2002), along with the single nucleotide polymorphism (SNP) database (Sachidanandam et al. 2001) to determine the distribution of SNPs, interspersed repeats, and simple microsatellite repeats in human genetic elements.
In general, deleterious events in regions important to gene expression and splicing regulation will be removed from the population by natural selection and will therefore be underrepresented in the available databases. Thus, since regulatory elements are likely to be present close to the splice sites, we expect to see fewer SNPs and repetitive elements close to the ends of introns than towards the middle of introns. In the case of simple repetitive elements, we also expect that some types of repeats should actually be overrepresented in the proximity of splice sites, because many SCEs are known to consist of repeated sequences (McCullough and Berget 1997; Jensen et al. 2000). Thus, we are able to determine the spatial distribution of regulatory elements within both introns and exons, and provide further insight as to the nature of such regulatory elements. We then look at the distribution of some known regulatory motifs (such as the GGG trinucleotide and the CpG dinucleotide) within genes and investigate their potential functions as splicing and transcription regulators.
RESULTS
Distribution of SNPs in Exons
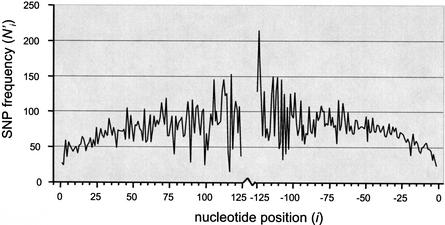
The distribution of SNPs in human coding exons is shown graphically in Figure 1. As expected, SNPs are underrepresented near the exon-intron boundaries, and the adjusted frequency N‘i increases with the distance from the boundary. This suggests that exonic SCEs are typically present near the exon-intron boundaries, but may be located (with decreasing probability) as far as 125 nucleotides into the exon. It is impossible to extend the analysis further into the exons, because there are relatively few large exons, and the adjusted frequency count becomes highly stochastic as a result of a decreased sample size.
Figure 1.
Distribution of SNPs in exons. The graph shows the frequency N‘i adjusted for uneven distribution of exon lengths (see Methods). Distances from the 5′ and 3′ exon ends are indicated as positive and negative values, respectively. The increasing trend in the adjusted frequency N‘i with distance from the exon-intron boundaries implies that the probability of finding an SCE decreases with distance from the splice site but is still nonzero at distances of more than 125 bp.
We noted that the frequency of SNPs in Figure 1 seems to be lower at the 5′ half of exons than in the 3′ half. To test this observation, we looked at all exons flanked by two introns (and therefore likely to contain regulatory elements at both ends), and, for each coding SNP, we determined whether the SNP is closest to the 5′ or the 3′ edge. In this analysis, 4684 SNPs were located closer to the 5′ end of an exon and 4855 were closer to 3′. However, this difference is not significant (χ2 = 3.07, P = 0.08).
Another prediction is that the ratio of nonsynonymous to synonymous substitutions in coding sequences should vary depending on whether the substitutions occur in regulatory or nonregulatory sequences. It can be shown (see Methods section) that the ratio of nonsynonymous/synonymous changes decreases as the density of regulatory regions increases. Hence, in view of the results from the spatial distribution of SNPs, we predicted that the ratio of nonsynonymous/synonymous changes should be highest towards the center of exons (where there are fewer regulatory sequences) and may also be higher at the 3′ than the 5′ ends of exons.
Table 1 compares the synonymous and nonsynonymous substitutions within subdivisions of exons. We can test the above hypotheses by collapsing appropriate cells of the table and performing χ2 tests on the resulting two-by-two tables. Hence, there is a greater proportion of nonsynonymous changes at the 5′ ends of exons than at the 3′ ends (χ2 = 6.37, P = 0.01). Furthermore, there is a greater proportion of nonsynonymous changes towards the center of exons than at the edges. This is true for comparing the first 125 positions with positions 126–250 from the 5′ end (χ2 = 5.03, P = 0.02) and from the 3′ end (the latter deviation is in the right direction, but is not significant, χ2 = 0.44, P = 0.51). There is also a higher proportion of nonsynonymous SNPs in single-exon genes (where we expect no splicing regulation) than in multiple-exon genes (χ2 = 8.07, P = 0.005). The last test confirms that we are accounting for SCEs, rather than general genetic control sites (e.g., transcription regulators).
Table 1.
Comparison of Frequencies of Synonymous and Nonsynonymous Changes at Different Exonic Locations
| Synonymous changes | Nonsynonymous changes | |
|---|---|---|
| Spliced genes | ||
| 5′ end | ||
| pos. 1–125 | 2596 | 2632 |
| pos. 126–250 | 384 | 460 |
| 3′ end | ||
| pos. 1–125 | 2525 | 2370 |
| pos. 126–250 | 242 | 242 |
| Single-exon genes | ||
| 5′ and 3′ ends | ||
| pos. 1–250 | 68 | 105 |
Distribution of SNPs in Introns
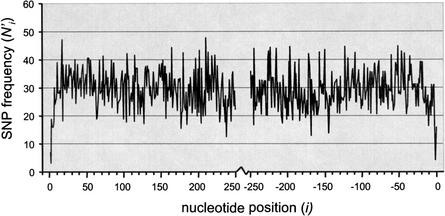
Figure 2 demonstrates, in agreement with expectations, that SNPs are underrepresented near splice sites at both intron ends. It is known that not only the splice donor and acceptor sites but also several adjacent bases follow consensus sequence rules (Lim and Burge 2001). Hence there is a very sharp rise in SNP density within the first few bases away from the boundary. Subsequently, the increasing trend stabilizes at a steady value at about 20 nucleotides from either end. We may speculate that because SCEs are often composed of low-complexity or repetitive sequences, they are not sensitive to disruption by point mutations. However, the trend may be caused largely by the nature of SNP ascertainment within and in the proximity of repetitive elements. In The SNP Consortium (TSC) discovery protocol, random sequence reads are discarded whenever they are composed largely of known repetitive elements (D. Altshuler, pers. comm.). Hence, the probability of SNP discovery decreases away from exon-intron boundaries, as the density of interspersed repeats increases. Because this ascertainment bias cannot be easily and accurately accounted for, we must limit ourselves to the one conclusion that is unaffected by the bias: At least the first and last 20 nucleotides of introns contain functional elements that are sensitive to disruption by single point mutations.
Figure 2.
Distribution of SNPs in introns. Point mutations certainly affect SCEs located within the first 20 bp from both ends. The leveling off of the distribution at farther distances may be an effect of SNP ascertainment problems in the proximity of repetitive elements.
Insertions of Interspersed Repeats in Introns
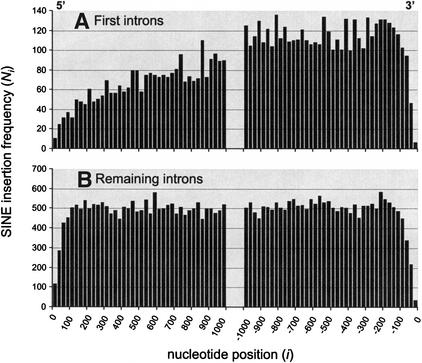
Interspersed transposable elements are a ubiquitous feature of mammalian genomes and are known to insert within genes (Makalowski 2000; Nekrutenko and Li 2001). The distribution of short interspersed nuclear elements (SINEs), averaged over all long human introns (>2500 bp), is shown in Figure 3. There is an obvious trend against insertions close to the splice sites. This is consistent with the expectation that SCEs are located close to the intron boundary, and their disruption is selected against. The frequency of insertions seems to reach a steady value at about 100–150 nucleotides away from the splice sites. In general, the distribution of SINE insertions is symmetrical with respect to the 5′/3′ intron subdivision (Fig. 3B). However, this is not true when only first introns of each gene are considered. Figure 3A shows that there are significantly more insertions in the 3′ portions of first introns that in the 5′ portions (a total of 5514 near 3′ vs. 2615 near 5′, χ2 = 1034, P < 10−10). This suggests that the 5′ ends of first introns are particularly functionally important. The increasing trend in SINE insertion frequency does not reach a steady value at 1000 nucleotides from the splice site, suggesting that important regulatory regions may be present even further away from the boundary. However, in view of the lack of such a trend in remaining introns, it is unlikely that those regions are involved in splicing regulation. Rather, the proximity to the start of transcription suggests that they could be responsible for the regulation of transcription.
Figure 3.
Distribution of SINEs in long human introns. The histogram has a 25-bp bin width; e.g., the first column at origin signifies the number of interspersed elements whose insertion points begin/end between 1 and 25 nucleotides from the 5′ splice site. Because SINE insertions are almost certainly mutagenic, the distribution corresponds to the inverse of the distribution of important regulatory regions.
Distribution of Microsatellite Repeats and Low-Complexity Regions in Introns
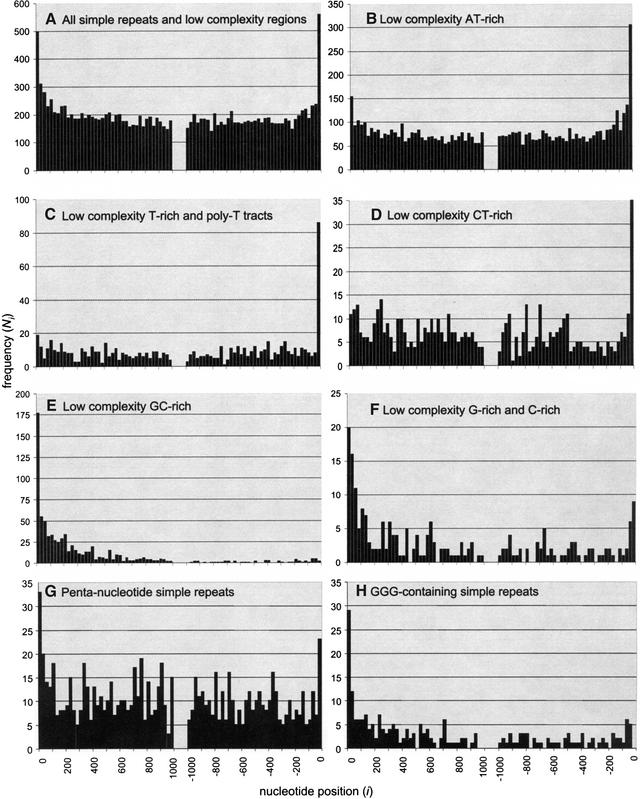
Microsatellites are short (1–7)-nucleotide simple tandem repeats, whereas low-complexity regions are composed of predominantly one or two kinds of nucleotides, but without any distinguishing repetitive pattern. The distribution of such simple repeats and low-complexity regions in introns clearly differs from that of interspersed repeats. In Figure 4A, both the 5′ and 3′ ends exhibit an elevated density of repetitive elements close to their respective boundaries. This suggests that many simple repetitive elements may actually be involved in splicing regulation. The remaining panels in Figure 4 show the distribution of the particular repetitive elements that actually contribute to the elevated density at the edges. Several types of low-complexity regions are overrepresented near the ends. AT- and T-rich regions seem to be mostly present disproportionately near the 3′ boundaries, yet they also show unexpected (but slightly lower) spikes very near the 5′ boundaries. A similar trend is seen in CT- and C-rich regions. Presumably, regions with elevated C and T content incorporate the polypyrimidine tracts ubiquitously present near the 3′ boundary. However, the polypyrimidine tracts are generally present directly adjacent to the splice site (less than 50 bp from the boundary), whereas the distribution of AT-rich low-complexity regions appears skewed up to about 300 bp towards the center of introns. This suggests that AT-rich elements may be involved in splicing regulation in the human genome, similarly to the role they play in plant genomes (McCullough and Schuler 1997). G- and C-rich regions seem to be equally overrepresented at both intron ends, implying that they play a more universal role as SCEs.
Figure 4.
Distribution of simple tandem repeats (1–7-nucleotide repeat unit) and low-complexity regions in introns. Panel (A) shows the combined distribution of all such regions, whereas the remaining panels identify the particular types of sequences that most significantly contribute to the overrepresentation near splice sites. Such sequences are most likely involved in splicing recognition and control.
Most simple tandem repeats are not overrepresented near intron boundaries. Thus, those repeats are not likely to be useful in splicing. Only poly-T tracts (Fig. 4C), and pentanucleotides [experimentally implicated in some regulatory elements, e.g., the Nova enhancer (Jensen et al. 2000)] have distributions distinctly skewed towards the edges. On closer inspection, most of the pentanucleotides responsible for this trend contain the GGG trinucleotide, known to be present in several experimentally defined SCEs (McCullough and Berget 1997; Lim and Burge 2001). In fact, all GGG-containing simple repeats (Fig. 4H) have such skewed distributions. The GGG motif, along with G-rich and GC-rich low-complexityregions, is particularly overrepresented near the 5′ intron boundaries and is thus likely to be mostly involved in splicing regulation at the 5′ ends.
Nucleotide Content Patterns in a Typical Gene
We conclude our analysis by studying a model gene, consisting of a 2000-bp promoter region, a 250-bp noncoding first exon, followed by a first intron, internal exon, internal intron, a terminal exon, and a terminator region. Although in reality introns are much longer than exons, we limit the analysis to the first 500 bp and last 500 bp of each intron, since those regions tend to demonstrate all of the unusual features present in introns. Each of the described genetic elements is in fact a composite average of all the known genes in the entire human genome (see the Methods section). Although this is not the most general representation of a human gene—an average gene consists of four introns (Deutsch and Long 1999)—we find that only the first introns and exons exhibit properties distinct from the others, and that combining all internal introns and exons is an accurate generalization. It is also worth noting that, since some mRNAs [possibly as many as 34% (Davuluri et al. 2000; Suzuki et al. 2002)] are incomplete at their 5′ ends, a significant proportion of what we believe to be first introns or exons may in reality be located further within the gene. Such misclassification introduces noise into our analysis but, as a result, our findings regarding the special properties of the promoter, first exon, and first intron (see below) are conservative—in a perfect genome annotation, the differences should be even more pronounced.
The most notable feature of genes is their elevated G+C content. This is expected, since genes are preferentially located in G+C rich isochores (Bernardi et al. 1985; Bernardi 2000). However, a closer look reveals that genes are not uniformly G+C-rich. We find that the G and C content increases monotonically within the promoter region, beginning to rise from the baseline of 42% G+C at a distance >2000 nucleotides from the start of transcription, to nearly 60% near the transcription start. The first noncoding exon is, on average, very G+C-rich (55–60%). The most G+C-rich element, however, is the first intron, with an average G+C content of up to 65% near the 5′ splice site, gradually decreasing towards the 3′ end. Internal exons exhibit a peak in G+C content at about 100–150 bp from either splice site, with a lower G+C content near the splice sites and towards the centers. Internal introns are characterized by elevated G+C percentage near the splice sites, approaching a baseline level at about 200 bp from each boundary The polypyrimidine tract extending to about 50 bp from the 3′ splice site constitutes a C-rich but G-poor region. Also, in general, the G content of introns is higher than the C content. Otherwise, the differences in relative G and C frequencies are minor. The terminal exon is unusual in that, at its 3′ end, the G+C content decreases towards the point of transcription termination. Finally, following the transcription end, G+C content increases to a steady value of about 45% at about 150 bp past the end of the transcript.
One should expect that such strong fluctuations of G+C content across various genetic features must be dictated by conditions necessary for the correct transcription (spatial and temporal), splicing, and possibly translation of genes. Thus, it is likely that G+C content should be associated with regulatory functions. However, it is important to distinguish whether the variation in the G+C content itself is sufficient to determine regulatory functions, such as transcription initiation, termination, and intron/exon definition, or whether it is the presence of specific G- and C-rich (or poor) regulatory elements, such as enhancers and repressors, that drives the local variation in G+C content.
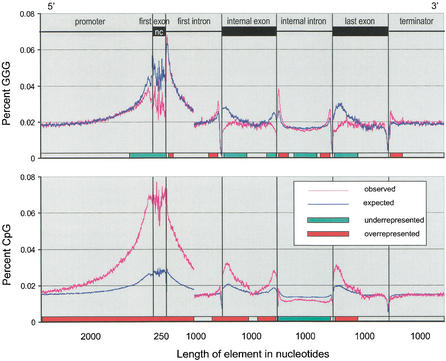
We answer this question by studying the distribution of two known regulatory motifs: the CpG dinucleotide [known to repress transcription when present in the methylated state (Siegfried et al. 1999)] and the GGG trinucleotide, which has been found to be overrepresented in short introns and to act as an intronic splicing enhancer (McCullough and Berget 1997). Figure 5 shows the observed and expected frequencies of the above motifs within the archetypal gene. The observed frequencies were determined using a 10-bp sliding window, whereas the expected frequencies were calculated using locally observed C and G frequencies, multiplied by a correction factor representing the relative overrepresentation of each motif in noncoding portions of the human genome (see Methods section). It is expected that in regions where either motif is responsible for regulatory functions, the overabundance of the motif will drive the local C and G contents; hence, the observed frequency of the regulatory motif should be in excess of the expected frequency. On the other hand, sequences not involved in regulation should be present at, or below, the expected frequencies.
Figure 5.
Observed and expected frequencies of common regulatory elements, the GGG trinucleotide and the CpG dinucleotide, in a typical human gene. The model gene contains information combined from all known genes in the entire human genome. The expected frequencies are calculated from local individual nucleotide frequencies, corrected for biases in occurrence of GGG and CpG in noncoding genomic regions. The slider bar at the bottom of each graph indicates the regions of relative over- and underrepresentation of each motif. Excess of a given motif over the expected frequency indicates a possible regulatory function.
The slider bar at the bottom of each graph in Figure 5 indicates regions of over- and underrepresentation of the two motifs. An excess of the GGG trinucleotide is observed in the first and last 200 bp of introns (excluding the 3′ polypyrimidine tract region, <50 bp from the 3′ splice site). This was suggested by earlier studies (Engelbrecht et al. 1992; Sirand-Pugnet et al. 1995; McCullough and Berget 1997) and our own analysis of repetitive sequences in introns. It is also present in excess within the 200 bp following the end of the transcription site. This has not been noted to date, and suggests that the GGG motif might function not only as an intronic splicing enhancer, but also may be involved in the termination of transcription.
On the other hand, GGG is underrepresented in the promoter region and exons, in particular near both 5′ and 3′ exon/intron boundaries. Hence, GGG is not likely to be involved in the regulation of transcription nor to be present in exonic splicing regulators.
Some equally interesting patterns are seen in the distribution of the CG dinucleotide. As expected, CpG content increases towards the transcription site. This reflects the methylation patterns near genes, since the unmethylated state of the CpG nucleotide is known to both drive transcription and prevent the mutational decay of the more commonly encountered methylated state. The large excess of CpG is also visible throughout leading noncoding exons and the 5′ part of the first intron. This suggests that CpG-dependent regulatory elements may be present as far as 2000 bp upstream from the start of transcription and also in the first exons (noncoding), and at least 1000 bp of the first introns (the expected/observed ratio approaches 1 at about 1000 bp from the 5′ splice site; data not shown on the graph). The excess of CpG is not seen in internal introns, implying a particular role for the leading intron.
Interestingly, a significant excess of CpG is also apparent in exons, particularly close to both exon/intron boundaries. This trend hints at a previously unnoted role for the CG dinucleotide as an exonic splicing regulator, in particular because the 3′ end of the last exon, which has no splice site, shows no excess of CpG.
In addition to the above two motifs, we also looked at the distribution of several other di- and trinucleotides within human genes. We found no remarkable trends, except for the distribution of the CCC triplet. Surprisingly, this sequence shows a pattern somewhat similar to the GGG triplet, with visible excess of CCC near both splice sites within introns. In addition, there is a slight excess of CCC within exons, up to 100 bp from both 5′ and 3′ exon/intron boundaries. This suggests a possible role for the CCC triplet in both intronic and exonic splicing regulation.
DISCUSSION
Exonic SCEs
The analysis of SNP distribution within exons shows that regions near the exon/intron boundaries are most likely to be involved in splicing regulation. This is indicated by both the frequency of SNPs, which increases away from the splice sites, as well as the ratio of nonsynonymous/synonymous substitutions, which is lowest close to the splice sites (indicating constraints other than protein coding). We note that the ratio of nonsynonymous/synonymous substitutions is higher in single exon genes (where no splicing regulation occurs), than in spliced genes, confirming that we are detecting constraints imposed by conservation of splicing control. We also note that the 5′ ends of exons are more likely to contain regulatory elements than the 3′ ends. The SNP analysis suggests that splicing control elements may be located at least as far as 125 bp (but possibly more) away from the splice site.
Our investigation of the spatial distribution of the GGG triplet and the CpG dinucleotide within genes suggests that the CpG dinucleotide may be involved in exonic splicing control. The frequency of CpG within exons shows a significant excess over the expected (Fig. 5). This excess is most pronounced at about 200 bp away from either splice site. We conclude that the putative CpG-dependent SCEs are not commonly present directly adjacent to the splice sites, where other types of control elements are likely to be necessary (these may include CCC-containing sequences, which we find to be overrepresented immediately adjacent to the exon/intron boundary). Again, the excess appears more extreme at the 5′ end than at the 3′ end, suggesting a greater relative importance of the 5′ ends of exons in splicing regulation.
CpG-containing elements have not been previously implicated in splicing regulation, but our analysis suggests that they may constitute a ubiquitous class of SCEs. It is interesting to note that terminal exons, which are not spliced at the 3′ end, exhibit an excess of CpG only at their 5′ end, whereas the 3′ end distribution follows the expectation. This supports the hypothesis that the overrepresentation of the CpG dinucleotide in exons is indeed associated with splicing control. However, we must note that we cannot rule out that such trends may also result from protein-coding constraints of exonic sequences, and that our finding needs empirical corroboration.
Intronic SCEs
The distribution of SNPs in introns (Fig. 2) implies that at least the first and last 20 bp of introns are important for the splicing process and sensitive to point mutation changes. This figure is likely to be a gross underestimate, because the SNP discovery frequency decreases in the vicinity of repetitive sequences, and the frequency of repetitive element insertions increases away from the splice sites. Therefore, we can use the information from the number of SINE insertions in introns to deduce the relative functional importance of different intronic regions. Because the frequency of SINE insertion reaches a steady-state value at about 150–200 bp from either splice site, we conclude that the first and last 150 bp of introns are likely to contain a significant number of elements required for the splicing process. It appears that the 25 bp directly adjacent to the 3′ splice site have the highest functional importance, containing only 30 SINE insertion points, compared to an average of 500 insertions per 25-bp segment in the interior of introns. This underscores the importance of the branch site and the polypyrimidine tract, usually found in the last 50 bp of introns, in splice site recognition.
At the same time, the SINE element distribution in Figure 3 reveals the unique functional importance of first introns, in particular their 5′ ends. The frequency of SINE insertions is markedly lower in the 5′ ends of first introns than in the 3′ ends, and it does not reach a steady level until over 1000 bp away from the splice site. Hence, the 5′ regions of first introns are likely to be particularly functionally important. It is possible that first introns are disproportionately significant for splicing processes, such as the assembly of the spliceosome. However, because of the proximity to the promoter region, we find it likely that the crucial function of first introns is linked to transcription regulation. In that case, 5′ regions of first introns are expected to harbor a high proportion of regulatory regions necessary for transcription, such as enhancer elements. While we cannot fully rule out either hypothesis, our investigation of the distribution of the CpG dinucleotide, whose importance to transcription is well documented (Lamb et al. 1991; Siegfried et al. 1999; Campanero et al. 2000), favors the transcription regulation alternative. CpG is highly overrepresented in the 5′ ends of first intron, but not in the 3′ end, nor in any part of the internal introns (Fig. 5).
The distribution of simple repeats and low-complexity regions is markedly different from that of SINEs (Fig. 3). The number of repetitive elements is highest in the proximity of splice sites. C-, T-, and CT-rich regions are mostly overrepresented directly adjacent to the 3′ splice sites and most likely constitute parts of polypirimidine tracts. However, G-, CG-, and AT-rich regions also show overabundance even at a distance > 200 bp from the splice sites. G-rich elements have been experimentally shown to act as splicing enhancers (Engelbrecht et al. 1992; Sirand-Pugnet et al. 1995; McCullough and Berget 1997). This analysis confirms the generality of this function. On the other hand, AT-rich elements are only known to act as splicing enhancers in plants (McCullough and Schuler 1997). Our result suggests that they may also be involved in splicing control in humans.
Of the simple microsatellite repeats, only some pentanucleotides and GGG-containing repeats appear overrepresented near the splice sites. Once again, the GGG motif has been frequently noted as an intronic splice enhancer, and our analysis confirms the generality of those findings. This observation also inspired the investigation of GGG distribution throughout human genes (Fig. 5).
The GGG trinucleotide was previously found to be overrepresented in short introns (McCullough and Berget 1997; Lim and Burge 2001). We find that this tendency is also observable in long introns, but limited to the first and last 200 bp. Although the excess of GGG is most pronounced in short introns, it is also present in long (>2000 bp) and very long (>5000 bp) introns (data not shown). Thus, we find that the GGG motif is a common intronic SCE, not limited solely to short introns.
Interestingly, we also find that the CCC triplet has a distribution similar to that of GGG; that is, CCC is overrepresented in the proximity of the splice sites, however not as significantly as the GGG motif. Since SCEs can act only by their presence on the transcribed strand, the two motifs are not simply reverse complements of each other, but must constitute distinct classes of control elements. The putative general function of CCC triplets as SCEs, suggested here, should be further explored empirically.
Transcription Control Elements
Although our study concerned mainly the investigation of regions involved in splicing control, we found that the range of splicing and transcription regulators cannot be fully decoupled. Hence we were able, and forced, to draw some interesting conclusions regarding the regulation of transcription.
The distribution of SINE insertions in the first introns was found to be markedly different than that in the internal introns. The low frequency of SINEs implied a particularly important function of the 5′ end of the first intron. In our subsequent investigation of GGG and CpG distributions, we observed that first introns also have unusually high frequencies of both of the above motifs. Furthermore, those high frequencies of occurrence are associated with a generally high G+C content of the first introns which appears to be driven by the excess of CpG.
The presence of the CpG dinucleotide is well known to be associated with transcribed genes (Cross and Bird 1995) and has been shown to be involved in transcription regulation. Methylation of CpG island represses gene expression; this is thought to occur through both the modification of chromatin structure, resulting in lack of accessibility of the gene and promoter region to the transcription machinery, and also the alteration of the binding sites of regulatory elements, such as enhancers (Siegfried et al. 1999; Campanero et al. 2000). Hence, we postulate that the high C+G and CpG content of first introns results mostly from the importance of those regions in transcription regulation.
The high CpG content is likely to be associated with lack of methylation. This probably serves dual functions: allowing transcription of the gene, and preventing mutational decay of the normally methylated cytosines. Our analysis shows that, generally, the first 2000 bp upstream of the transcription start, the first exon, and at least 1000 bp of first introns are likely to remain unmethylated and contain an unusually high proportion of the CpG nucleotide, and are probably involved in transcription regulation. The graphic representation of CpG frequency in Figure 5 is an interesting alternative to the somewhat arbitrary definition of CpG islands (Takai and Jones 2002) currently used in detecting gene-rich chromosomal regions.
We also note that the 3′ region of genes, directly following transcription termination, has an unusually high proportion of the GGG trinucleotide. Although transcription termination in eukaryotes is not yet fully understood, this finding may suggest that in a significant number of cases, termination of transcription in the human genome may occur by means of a G-rich stem-loop structure, similar to that commonly used by prokaryotes.
Summary
Our theoretical analysis provides early genomic guidelines regarding the distribution and nature of elements involved in gene splicing and expression. Briefly, we confirm the general importance of CpG dinucleotides in transcription control but also note their putative functions as exonic SCEs; we confirm the importance of the GGG triplet as an intronic SCE and also indicate its putative function in transcription termination; we note a possible role of the CCC trinucleotide as both an exonic and intronic SCE; we find that low-complexity regions in general are likely to harbor intronic SCEs; low-complexity AT-rich regions may constitute a common class of intronic SCEs presently noted only in plants; up to 2000 bp preceding the transcription start, the first exon (particularly if it is noncoding) and the 5′ portion (up to 1000 bp) of first introns are likely to be particularly important for transcription control; intronic SCEs are most likely to be present within the first and last 200 bp of introns; exonic SCEs are most common near exon/intron boundaries but may occur, with decreasing probability, at least as far as 125 bp away from splice junctions.
Of course, much more empirical work leading to the identification of regulatory elements needs to be done. However, our analysis should be a step forward for both the general understanding of regulatory functions and the identification of variants involved in genetically inherited diseases. We are becoming ever more aware that mutational changes other than those leading to amino-acid substitutions are often causes of genetic disorders (Klesert et al. 1997; Ars et al. 2000; Blencowe 2000). In this work, we hope to pave the road towards general categorization of such genetic changes.
METHODS
Databases
We used the August 2001 freeze of the University of Santa Cruz Golden Path human genome sequence, and the corresponding annotation database (http://genome.cse.ucsc.edu). The annotation includes the locations of known and predicted genes, SNPs, and different types of repetitive sequences, as determined by RepeatMasker (using the sensitive, -s setting, A.F. Smit, unpubl.; http://ftp.genome.washington.edu/RM/RepeatMasker.html). We used the known gene annotation track, originally obtained from the known gene database (RefSeq, NCBI, http://www.ncbi.nlm.nih.gov) and mapped to the genomic sequence by the University of California, Santa Cruz (Kent et al. 2002). In order to confirm the mapping process, we in silico-translated all the available genes, and discovered several inconsistencies. About 20% of the genes did not translate into the peptides contained in RefSeq. Hence, we used only the subset of confirmed correct genes (a total of 10,858, including known alternatively spliced variants) in further analyses.
SNPs in Exons
In this analysis we combined all the known human exons and determined the spatial distribution of SNPs. We determined the quantity Ni where i is the nucleotide position counting from the 5′ or 3′ end of each exon, and N is the total number of SNPs observed at that position, in the entire genome. Distances from the 5′ and 3′ exon ends are designated by positive and negative values, respectively. Ni needs to be adjusted to take into account the size distribution of exons. We define N‘i = Ni·v/vi where vi is the number of exons in the dataset that are at least 2i in length (and therefore actually do contain position i, and v = Σvi is the total number of exons in the dataset. Under the null hypothesis, that is, that SNPs occur randomly throughout exons, N‘i will have a uniform distribution. We considered positions 0 < | i| ≤ 125 nucleotides. In cases of short exons (<250 bp), the position was scored relative to the closest boundary. For example, an SNP at position 50 of a 200-bp exon is scored as 50 bp from the 5′ end, not 150 bp from the 3′end.
We limited the analysis to coding exons, since the overall frequency of SNPs in noncoding exons is expected to be much higher, in the absence of natural selection at the protein level, and may have a disproportionate effect on the observed results. We also excluded all genes that lack introns, since they are not expected to contain any SCEs. In the case of the first and last exon of each gene, which are flanked by only one intron, the position i was only measured from the edge adjacent to the intron, because the other end of the exon in not involved in the splicing process. Because we expect no spatial bias for SNP detection within exons, independent of the ascertainment method used, we used the entire SNP dataset for exon analysis—the SNP Consortium (TSC) and NIH tracks in the genome database—in order to utilize maximum sample size. However, similar trends in spatial distribution are obtained with various subsets of the data: random (TSC) SNPs, overlap SNPs, and SNPs from exon directed studies (data not shown).
It seems intuitive that the ratio, R, of nonsynonymous to synonymous amino acid substitutions should differ between regulatory and nonregulatory regions. Below we demonstrate this formally. Let pr denote the probability that a nucleotide substitution destroys an essential regulatory region and is therefore removed from population by natural selection. Similarly, let pp stand for the probability that a substitution destroys an essential protein and is removed from the population. Only nonsynonymous changes affect protein function, whereas all nucleotide substitutions are likely to affect regulatory regions, because the selection acts at the RNA level. Hence the expected observed frequency of nonsynonymous changes, Fns, is given by Fns = fns(1 − pr − pp), where fns is the frequency at which nonsynonymous changes occur in the absence of selection. Similarly, for synonymous changes, Fs = fs( − pr). The ratio, Fns/Fs is given by R = (fns/fs)·[(1 − pp)/(1 − pr)]. Because the frequencies at which synonymous and nonsynonymous mutations occur in the absence of selection are expected to be constant, R ∝ 1 − pp/(1 − pr). Thus the ratio of nonsynonymous/synonymous substitutions should be highest in nonregulatory regions (pr = 0) and lowest in regions involved in splicing regulation (pr > 0).
To calculate the above ratios, we in silico-translated all the RefSeq genes and determined whether or not the nucleotide substitution corresponding to each coding SNP in our dataset results in an amino-acid change.
SNPs and Repetitive Elements in Introns
In this analysis we combined all of the known human introns and determined the spatial distribution of SNPs and repetitive elements. The adjusted SNP distribution in introns was defined as N‘i, similarly to that in exons, as described above. In order to avoid a possible bias in SNP ascertainment in the proximity of coding sequences, we used only the randomly ascertained subset of SNPs, the SNP Consortium dataset (Sachidanandam et al. 2001), for this analysis.
The location of SINEs and simple repeats was quantified as follows. We obtained the start and end positions of repetitive elements, as determined by RepeatMasker from the Santa Cruz database. We measured the position of insertion, i, as the number of bases between the 5′ or 3′ end of an intron, and the nearest start/end of a repetitive element. We considered only long introns (size >2500 bp) and repetitive elements shorter than 500 bp (SINE and simple repeats) in order to avoid the bias of a priori reduced probability of insertion into small introns (∼500 bp), and also to avoid the overlap in measurement from the 5′ and 3′ ends of introns. Only insertion positions between 1 and 1000 bp from the ends were considered. Under the null hypothesis of random insertion, the insertion frequency Ni has a uniform distribution.
Model Gene Analysis
Our typical model gene consists of a 2-kbp promoter region, a first 250-bp noncoding exon, a 1-kbp first intron, one 1-kbp internal exon (representative of all internal exons), one 1-kbp internal intron (representative of all internal introns), and a terminal exon, followed by a 1000-bp terminator region. This model gene is actually a composite of all known genes and genetic elements in the human genome. We calculated specific nucleotide contents (e.g., C, G, CpG, etc.) of all of the genetic elements and averaged them to obtain a value representative of a typical human gene. For the introns and exons, the values are calculated for positions 1–500 bp from the 5′ end and 1–500 bp from the 3′ end. Because many introns and exons are longer than 500 bp, the curves for some of the nucleotide contents in Figure 5 are discontinuous at the mid-points of the genetic elements. The observed genome-wide average values were calculated as follows: The content of the nucleotide X at position i within a particular genetic element is given by the total number of nucleotides X at positions i within the entire genome, divided by the total number of elements containing the position i, that is, being at least 2i in length. A similar approach was used for polynucleotide content, but using a sliding window of the size of the polynucleotide, beginning at position i. In order to reduce stochasticity, the curves were subsequently smoothed by averaging within a window of 10 nucleotides.
To calculate the expected values for CpG and GGG motifs, we used local C and G content calculated above, corrected for the genome-wide biases in the occurrence of the above two motifs. The bias in CpG occurrence stems from hypermutation of the methylated dinucleotide and results in general underrepresentation of CpG. The cause for bias in GGG frequency is unknown, but results in a general excess of GGG triplets over the expected frequency. To determine this systematic average bias, we determined individual nucleotide and polynucleotide frequencies in noncoding regions found at a distance >50 kbp from known genes. The resulting correction factors,
fC fG/fCpG = 3.45, and fG3 /fGGG = 0.65, were then used to calculate the expected polynucleotide frequencies within genes, based on the locally measured G and C frequencies, under the null hypothesis of no functional significance.
WEB SITE REFERENCES
http://genome.cse.ucsc.edu/; human genome sequence and annotation.
http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker download site.
http://www.ncbi.nlm.nih.gov; SNP allele data.
Acknowledgments
This work was supported by grant HG00008 from the National Human Genome Research Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL majewski@complex.rockefeller.edu; FAX (212) 327-7996.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.606402.
REFERENCES
- Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. doi: 10.1016/s0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- Bernardi G, Olofsson B, Filipski J, Zerial M, Salinas J, Cuny G, Meunier-Rotival M, Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985;228:953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- Brett D, Hanke J, Lehmann G, Haase S, Delbruck S, Krueger S, Reich J, Bork P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29:2338–2348. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlo T, Sierra R, Berget SM. A 5′ splice site-proximal enhancer binds SF1 and activates exon bridging of a microexon. Mol Cell Biol. 2000;20:3988–3995. doi: 10.1128/mcb.20.11.3988-3995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Zhang MQ. CART classification of human 5′ UTR sequences. Genome Res. 2000;10:1807–1816. doi: 10.1101/gr.gr-1460r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch M, Long M. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 1999;27:3219–3228. doi: 10.1093/nar/27.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrecht J, Knudsen S, Brunak S. G+C-rich tract in 5′ end of human introns. J Mol Biol. 1992;227:108–113. doi: 10.1016/0022-2836(92)90685-d. [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Hastings M L, Krainer A R. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- Hastings ML, Wilson CM, Munroe SH. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. RNA. 2001;7:859–874. doi: 10.1017/s1355838201002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Zahler AM. Conservation, regulation, synteny, and introns in a large-scale C. briggsae-C. elegans genomic alignment. Genome Res. 2000;10:1115–1125. doi: 10.1101/gr.10.8.1115. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler AD. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klesert TR, Otten AD, Bird TD, Tapscott SJ. Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nat Genet. 1997;16:402–406. doi: 10.1038/ng0897-402. [DOI] [PubMed] [Google Scholar]
- Lamb BT, Satyamoorthy K, Li L, Solter D, Howe CC. CpG methylation of an endogenous retroviral enhancer inhibits transcription factor binding and activity. Gene Expr. 1991;1:185–196. [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski W. Genomic scrap yard: How genomes utilize all that junk. Gene. 2000;259:61–67. doi: 10.1016/s0378-1119(00)00436-4. [DOI] [PubMed] [Google Scholar]
- McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough AJ, Schuler MA. Intronic and exonic sequences modulate 5′ splice site selection in plant nuclei. Nucleic Acids Res. 1997;25:1071–1077. doi: 10.1093/nar/25.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, Fickett JW, Gelfand MS. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A, Li WH. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- Rowen L, Young J, Birditt B, Kaur A, Madan A, Philipps DL, Qin S, Minx P, Wilson RK, Hood L, et al. Analysis of the human neurexin genes: Alternative splicing and the generation of protein diversity. Genomics. 2002;79:587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi BZ, Cedar H. DNA methylation represses transcription in vivo. Nat Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- Sirand-Pugnet P, Durosay P, Brody E, Marie J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken β-tropomyosin pre-mRNA. Nucleic Acids Res. 1995;23:3501–3507. doi: 10.1093/nar/23.17.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Yamashita R, Nakai K, Sugano S. DBTSS: Database of human transcriptional start sites and full-length cDNAs. Nucleic Acids Res. 2002;30:328–331. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZM, Quintero J, Reid ES, Gocke C, Baker CC. Optimization of a weak 3′ splice site counteracts the function of a bovine papillomavirus type 1 exonic splicing suppressor in vitro and in vivo. J Virol. 2000;74:5902–5910. doi: 10.1128/jvi.74.13.5902-5910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]