Abstract
Protein homology is often limited to long structural segments that we have previously called modules. We describe here a suite of programs used to catalog the whole set of modules present in microbial proteomes. First, the Darwin AllAll program detects homologous segments using thresholds for evolutionary distance and alignment length, and another program classifies these modules. After assembling these homologous modules in families, we further group families which are related by a chain of neighboring unrelated homologous modules. With the automatic analysis of these groups of families sharing homologous modules in independent multimodular proteins, one can split into their component parts many fused modules and/or deduce by logic more distant modules. All detected and inferred modules are reassembled in refined families. These two last steps are made by a unique program. Eventually, the soundness of the data obtained by this experimental approach is checked using independent tests. To illustrate this modular approach, we compared four proteobacterial proteomes (Campylobacter jejuni, Escherichia coli, Haemophilus influenzae, and Helicobacter pylori). It appears that this method might retrieve from present-day proteins many of the modules which can help to trace back ancient events of gene duplication and/or fusion.
There is now a wide consensus that proteins display a modular structure. In our working model (Riley and Labedan 1997; Labedan and Riley 1999), we considered a “module” a long (mean size 220 residues) structural segment of homology found in prokaryotic proteins. We introduced this concept of modular structure because we feel it is crucial to uncover the mechanisms that have shaped either protein evolution (gene ancestry) or chromosome evolution of microorganisms. Accordingly, we proposed that a great deal of protein history can be easily traced back through a simple working model based on two well known gene events, duplication and fusion (Labedan and Riley 1999). In our model, the alternate use of these two processes would have been sufficient to progressively create the immense majority of present-day proteins. Gene duplication (for a review, see Ohno 1970) allows more and more specialized functions when the obtained copies, kept in the same genome for a while (paralogous genes according to the Fitch 1970 nomenclature), will have progressively diverged over time. Moreover, a protein may evolve not only by gene duplication and divergence of the copies but also by fusion in various combinations of genes encoding these copies, at different moments of their differentiation.
If our working model is pertinent, comprehension of protein evolution requires a census of all events of gene duplication and gene fusion. Until now, we identified modules through a combination of semiautomatic and manual methods. With the present deluge of data released by the whole-genome sequencing programs, it is clear that we need a new experimental approach. Accordingly, we have devised a suite of automatic programs that allows us to detect in a few steps the whole set of modules which constitute the proteome of any organism for which the complete sequence is available. We have also added a new, salient step: Distant modules may be deduced by logic from the analysis of groups of families of multimodular proteins which share at least one homologous module. Such an experimental approach allows us to identify more and better defined ancestral modules and to propose plausible scenarios which trace the building of present-day proteins from the duplication and fusion of genes encoding various ancient modules.
RESULTS
Identifying All Homologous Proteins
To assess the homologous relationships among proteins belonging to either the same proteome (paralogy) or proteomes of various organisms (orthology), we set up a strategy based in part on previous results.
The evolutionary distance separating two proteins deriving from a common ancestor and displaying significant sequence similarities is given in PAM units, a PAM unit being defined as the number of accepted point mutations per 100 residues separating two sequences (Dayhoff et al. 1978; Schwartz and Dayhoff 1978). The frequency with which any particular pair of (mutated) amino acids occur at a given position in two properly aligned homologous proteins can be used as a PAM score to evaluate the evolutionary distance separating the two proteins. It has been shown (Dayhoff et al. 1978) and frequently confirmed (see, e.g., Feng et al. 1985; Gonnet et al. 1992) that for many comparisons, the best scoring systems to detect distant homologous proteins correspond to PAM 250 scores.
Using a rationale based on information theory, Altschul (1991, 1993) further showed that, to be statistically significant, an alignment of sequences separated by a distance of 250 PAM units must be longer than 83 residues. Therefore, to define the significance of sequence similarities in terms of putative homology between distant proteins, we adopted the following two limits: Any sequence alignment must extend for at least 80 residues, and have a PAM distance less than 250 PAM units.
The DARWIN (data analysis and retrieval with indexed nucleotide/peptide sequences) package of programs (Gonnet et al. 1992; Benner et al. 1993; Gonnet and Hallett 1997) remarkably fits our theoretical approach and experimental needs. This package of programs (available at http://cbrg.inf.ethz.ch/Darwin/index.html) tries to distinguish biologically meaningful from chance similarities using a maximum likelihood approach. We previously adapted the DARWIN AllAll program to build a set of procedures (De Rosa and Labedan 1998) allowing one to collect, in one step, all of the significant matches displaying an alignment length greater than 80 residues and a PAM distance below 250.
Tables 1 and 2 summarize the results obtained in applying this approach to the comparison of proteins encoded by four proteobacterial genomes: Campylobacter jejuni, Escherichia coli, Haemophilus influenzae, and Helicobacter pylori. Table 1 shows a matrix of the 36661 pairs of homologous modules found in the AllAll output. The diagonal of the matrix gives the number of paralogous pairs for each bacterium, with the other figures corresponding to the different sets of orthologous pairs for each pair of organisms. From these data, we can compute the respective number of paralogs for each organism (Table 2). The relative proportion of paralogous proteins is found to be markedly larger in E. coli than in the three other proteobacteria.
Table 1.
Numbers of Pairs of Homologous Modules Detected During an Exhaustive Protein Comparison Using the Adapted DARWIN AllAll Program
| E. coli | C. jejuni | H. influenzae | He. pylori | |
|---|---|---|---|---|
| E. coli | 9173 | — | — | — |
| C. jejuni | 5319 | 1706 | — | — |
| H. influenzae | 6791 | 2684 | 1321 | — |
| He. pylori | 3751 | 2672 | 1939 | 1305 |
Table 2.
Percentages of Paralogous Proteins in the Analyzed Species
| Species | Total number of proteinsa | Number of paralogs | Percent |
|---|---|---|---|
| E. coli | 4094 | 2487 | 60.7 |
| C. jejuni | 1533 | 758 | 49.4 |
| He. pylori | 1457 | 646 | 44.3 |
| H. influenzae | 1618 | 653 | 40.3 |
Uniquely proteins longer than 79 residues.
Identifying and Collecting Homologous Modules
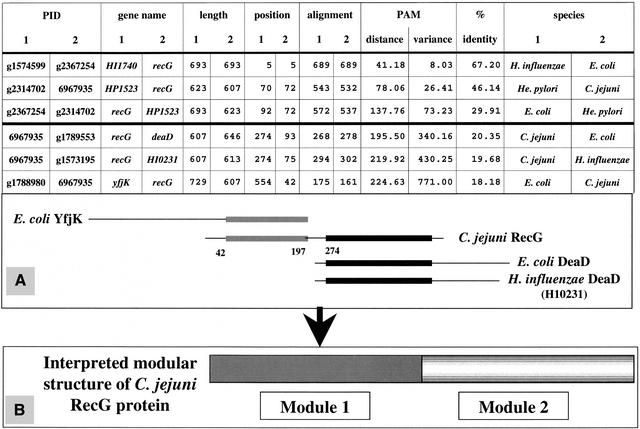
The output of the DARWIN AllAll comparisons was designed to exhibit the length of the structural segments of homology detected in the sequence of each partner of pairs of homologous proteins (see Methods and legend to Fig. 1). The three first lines of the table in Figure 1 show the full alignment of different orthologs of the ATP-dependent DNA helicase RecG: pairs H. influenzae–E. coli, H. pylori–C. jejuni, and E. coli–H. pylori, respectively. Note that one ortholog has been found for each species, but, among the six expected (n(n−1)/2) pairs, only three have been detected. The missing pairs are most probably below our thresholds, illustrating that our data underestimate the actual number of homologs. The next three lines of the table in Figure 1 show various cases of partial alignment which are summarized in the scheme shown in box A. The N-terminal half of C. jejuni protein RecG is homologous to a C-terminal segment of the unknown E. coli protein YfjK, and its C-terminal half is pairing with the N-terminal segment of the inducible ATP-independent RNA helicase DeaD encoded by E. coli and by H. influenzae (gene HI0231), respectively. Box B of Figure 1 shows how we interpret this experimental result in terms of modular structure. It appears that the RecG protein encoded by C. jejuni is formed of two modules.
Figure 1.
Detecting modules in an exhaustive comparison of a protein database. The table illustrates how the output of the DARWIN AllAll procedure was designed to exhibit essential information on the sequence alignment for each pair of homologous proteins. (A) How the data of the last three lines of the table can be interpreted; the homologous structural segments present in the pairing proteins are shown as boxes (black or gray) on the primary structure (line) of each protein. (B) How we interpret the presence of these structural segments in terms of protein multimodularity.
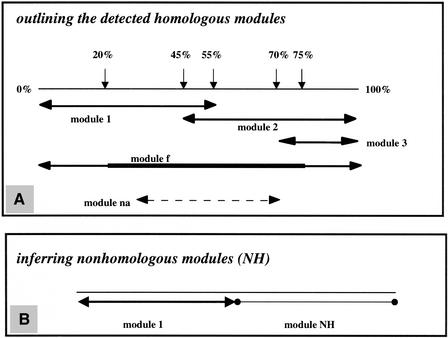
To extract automatically every module found in any pair of homologous proteins using the informations in the AllAll output, we wrote a second program, “Module,” based on the following rationale. Since our objective is to delineate the two or three major segments underlying protein structure, we adopted the following criteria (Fig. 2A). We will call module_1 any segment beginning at a residue located between 0% and 20% of the total length of the protein and having an alignment extending no longer than 55% of this total length. If the segment beginning at a residue located between 0% and 20% is extending more than 75% of the total length of the protein (corresponding to a full alignment) it will be called module_f (fusion module). We will call module _2 any segment beginning at a residue located after 45% of the total length of the protein, and in the case of large proteins we further define as module_3 any segment beginning at a residue located after 70% of the total length. At any rate, the minimal length of a module must be longer than 25% of the total protein length. Any alignment different from any of these criteria will be called a non-assigned module, module _na. Finally, a segment without homolog in any protein of the set of proteomes under study will be defined as a nonhomologous (NH) module (Fig. 2B).
Figure 2.
Delineating the different modules structuring homologous proteins. To define the nature of the different structural segments of homology, we imposed a series of thresholds on the positions at which these segments begin and finish in terms of percent of the total length of each protein (A). (B) How nonhomologous modules are inferred.
Numbering Gene Duplication Events
Once identified, the homologous modules detected by the AllAll and Module programs may be clustered in families in order to number the putative ancestral genes which gave birth to these families. The program “Families”, written in C language, allows us to assemble families of modules using a transitive approach (see Methods for details).
When comparing the four proteobacteria, 12,545 modules were detected and clustered in families using two complementary approaches. (1) The whole set of modules formed a total of 2565 families. (2) For each pair of species, we can obtain families of paralogous and orthologous modules, as summarized in Table 3. Here, for the sake of simplicity, we will call orthologs the homologous modules belonging to two different species, and paralogs the homologous modules belonging to the same species (Fitch 1970, 2000). For example, in the case of the pair E. coli/C. jejuni, we have 1020 families made of 5527 E. coli paralogous modules, 334 families made of 1662 C. jejuni paralogous modules, and 1156 families made of 4678 E. coli/C. jejuni orthologous modules. Table 3 confirms that there are far more homologous segments in E. coli than in the three other proteobacteria.
Table 3.
The Families of Paralogous and Orthologous Modules for Each Pair of Species
| E. coli | C. jejuni | H. influenzae | He. pylori | |
|---|---|---|---|---|
| E. coli | 1020 families | 1156 families | 1419 families | 1036 families |
| 5527 modules | 4678 modules | 5548 modules | 3833 modules | |
| C. jejuni | 334 families | 898 families | 952 families | |
| 1662 modules | 2860 modules | 3327 modules | ||
| H. influenzae | 334 families | 772 families | ||
| 1098 modules | 2303 modules | |||
| He. pylori | 301 families | |||
| 1235 modules |
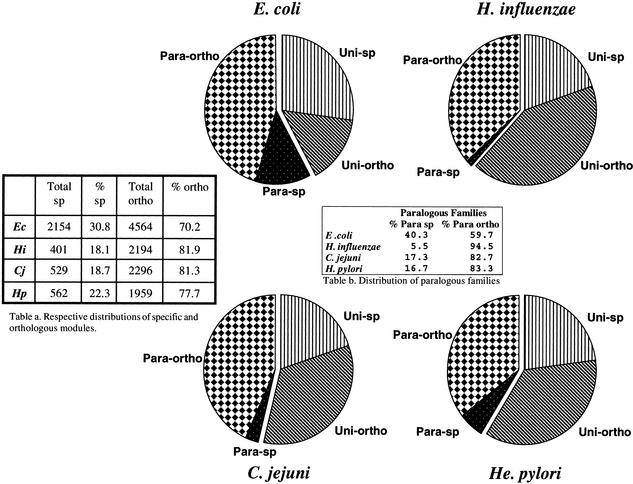
Integration of these different figures allowed us to distribute the whole sets of proteins belonging to these four proteobacteria in four different categories, as shown in Figure 3. The first two categories correspond to proteins which are found only in one species (sp) and which either have a paralog (para-sp) or are unique to their species (uni-sp). The last two categories correspond to proteins found in more than one organism and which either have a paralog (para-ortho) or are unique to their species (uni-ortho). Because the homology is defined at the module level, we further categorized the multimodular proteins according to the information obtained for each module. For example, a bimodular protein made of an orthologous module and a paralogous module will be classified as para-ortho.
Figure 3.
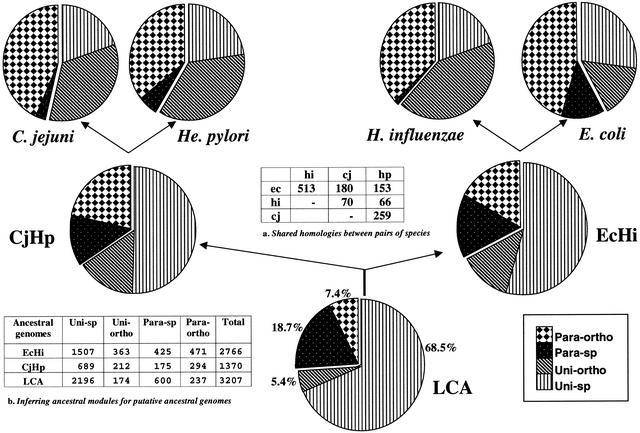
Calculating the respective proportions of each class of genes in various genomes. Data obtained through the intergenomic comparison of the four proteobacteria E. coli (Ec), H. influenzae (Hi), C. jejuni (Cj), and H. pylori (Hp) are give as pie charts and summarized in two tables. (Table a) The respective percent of genes specific to each organism and of the set of orthologs. (Table b) The respective percent of para-sp and para-ortho for the paralogous families. Uni-sp: genes unique to a species without any homolog. Uni-ortho: genes unique to a species with at least one ortholog. Para-sp: paralogous genes without any homolog in another species. Para-ortho: paralogous genes with at least one ortholog.
Again, it can be seen that proteins are distributed differently in E. coli than in the three other smaller proteomes. In particular, there are far more orphans, both paralogous and unique ones, in E. coli than in the three other bacteria (see respective numbers in Tables a and b of Fig. 3). As already suggested, many of the homologs unique to E. coli may have been lost to pathogenesis during the evolution of the three other proteobacteria (De Rosa and Labedan 1998). Moreover, the set of E. coli paralogous orphans (para-sp) split into as many as 411 families (40.3% of the total, see Table b), indicating that the occurrence of a large level of functional differentiation is probably necessary to the way of life of this more complex bacterium. In contrast, there is a very limited number of paralogous families specific to each of the other three organisms (Fig. 3, Table b).
Studying Gene Fusion Events
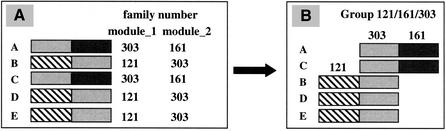
Multimodular proteins were further analyzed in order to disclose the relationships among the different families to which their adjacent modules belong. As shown in Figure 4A, a module belonging to family 303 may have fused either with a module belonging to family 161 (proteins A and C) or with a module belonging to family 121 (proteins B, D, and E). Thus, families 303, 121, and 161 may be grouped to reflect that they are connected by a chain of neighboring unrelated homologous modules (Fig. 4B).
Figure 4.
Analysis of multimodular proteins to trace back the events of gene fusion. A list of various multimodular proteins sharing homologous modules (e.g., modules belonging to family 303) which have different unrelated neighbors (belonging to either family 121 or family 161).
Two programs were created to automate the grouping of families. First, pairs of families of adjacent segments were identified by using the program “prep_group”, which scans the whole list of multimodular proteins for which family numbers of neighboring modules are indicated. This will create a list of pairs of families to which the program “Families” is further applied to group the families. A family without a relationship with any other family will be referred to as unique herein. Each group of families was further analyzed to trace back the timing of duplication versus fusion events. For example, in the simple case shown in Figure 4B, we can suppose that an ancient gene coding an ancestral module 303 has duplicated and each copy has further fused, one with an ancient gene coding the ancestral module 161 and the other with an ancient gene coding the ancestral module 121. Then, both fused ancestral genes, namely 303–161 and 121–303, would have further duplicated to give the five present-day proteins.
Inferring Distant Modules in Multimodular Proteins
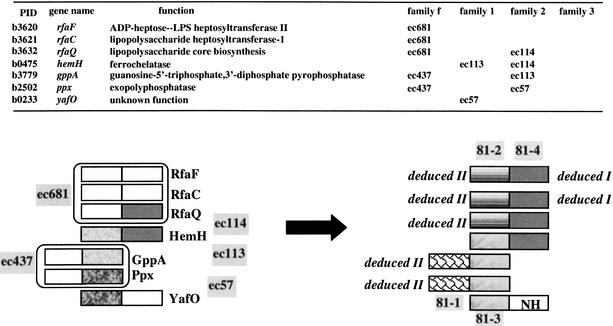
Detailed analysis of these groups of families revealed that, in many instances, we can deduce by logic modules which were either too distant to have been detected previously in the exhaustive Darwin AllAll analysis or were included in fusion modules (module_f). Figure 5 illustrates such an approach in the case of the E. coli paralogous group 81. Proteins RfaF, RfaC, and RfaQ which are required for biosynthesis of the lipopolysaccharide core are homologous along their full length (family ec681). Since RfaQ contains a module_2 homologous to the module_2 of the ferrochelatase HemH (family ec114), we can infer that RfaF and RfaC must also have a distant homologous module_2. We designate such inferred modules deduced modules of type I (in short, deduced I). Moreover, since RfaF, RfaC, and RfaQ are homologous along their full length, we can further infer that their module_1s are homologous. We will call this second type of inferred modules deduced modules of type II (in short, deduced II). Now, the ferrochelatase HemH has its module_1 homologous to the module_2 of the guanosine-5′-triphosphate,3′-diphosphate pyrophosphatase GppA (family ec113). Because GppA aligns along its full length to the exopolyphosphatase Ppx (family ec437), we will infer that the two module_1s of GppA and Ppx are deduced II modules. Furthermore, because module_2 of Ppx is also homologous to module_1 of YafO (family ec57), we can infer that they are homologous to module_1 of HemH and module_2 of GppA. Thus, in total, these seven proteins appear to be made of 13 homologous modules belonging to four refined families, hereafterdubbed 81–1, 81–2, 81–3, and 81–4, and a nonhomologous module NH. During this process of detection of distantly related modules, five module_fs (belonging to families ec681 and ec437) have been replaced by their elementary modules.
Figure 5.
Deducing supplementary modules in groups of families of detected modules. In the upper part of the figure, a list of proteins belonging to group 81 made of five paralogous families of E. coli proteins are listed with the nature of their modules and family number. Below, the different steps of the deduction approach are dissected with replacement of fused modules by their elementary units and clustering of distant modules in refined families (e.g., 81-3).
Therefore, the most plausible (and parsimonious) scenario describing the history of these seven present-day proteins could be as follows: (1) The ancient genes encoding the ancestral modules 81–2 and 81–4 have fused and the fusion product duplicated at least twice to give proteins RfaF, RfaC, and RfaQ; (2) the ancient gene encoding the ancestral module 81–3 has duplicated at least twice and the different copies have fused either with the ancient gene encoding an ancestral module 81–4 to give HemH, or with a unique module to give YafO, or with the ancient gene encoding the ancestral module 81–1, and this fusion product 81–1/81–3 has duplicated at least once to give GppA and Ppx proteins.
Detecting All Modules and Grouping Them in Refined Families
The approach detailed in Figure 5 has been automated by writing in C++ a program, SortClust, which performs the following tasks. First, “Module sorter” allows to identify and collect the deduced modules of type I and those of type II and the deduced NH modules for all homologous proteins present in families belonging to a group. To do that, the script “Module sorter” analyzes a table (similar to that shown at the top of Fig. 5) in which for each homologous protein are successively listed information on its PID number and the respective number of the family to which each AllAll-detected module belongs. The program SortClust is also able to resolve two specific problems: Internal duplications, and the treatment of modules which are found homologous to splittable modules. In a second step, the script “Module cluster” will assemble all of these better-defined homologous modules into new refined families.
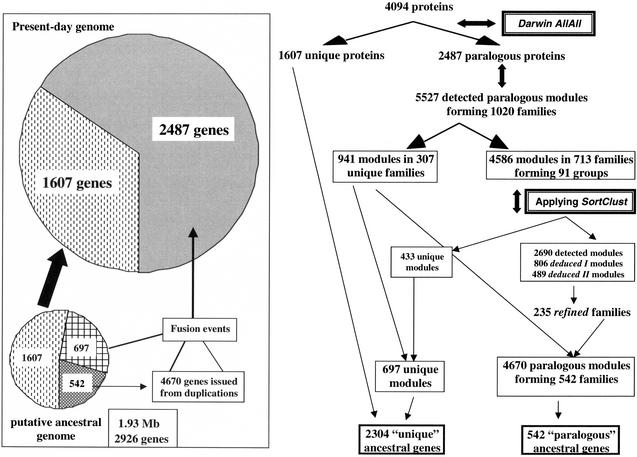
Figure 6 illustrates the use and interest of the SortClust program. For the sake of simplicity, we have detailed only the treatment of the E. coli set of paralogous modules. The 5527 paralogous modules (Table 3) detected by the Darwin AllAll program form 1020 families which can be separated into 307 unique families (941 modules) and 713 families (4586 modules) which can be clustered in 91 groups of families connected by a chain of neighboring unrelated homologous modules. When applying SortClust to these 713 families, 1896 of their 4586 detected modules can be reinterpreted as 806 deduced modules of type I and 489 deduced modules of type II. The 2690 remaining detected modules and the 1295 deduced ones may been assembled in only 235 refined families. Moreover, SortClust identified 433 unique modules. If we add the 264 nonhomologous modules, the 413 module_fs, and the 264 modules belonging to the 307 unique families, we get a total of 697 unique modules and 4670 paralogous modules forming 542 families. From these data, we can infer that the 2487 paralogous present-day proteins are coded by genes which descend from ancestral genes issued from the duplication of 542 genes, followed by the fusion of these more or less differentiated copies either between themselves or with the 697 genes coding unique modules. Thus, the ancestral genome would have been made of three categories of genes: 1607 genes at the origin of the present-day unique proteins, 542 at the origin of the present-day 4670 paralogous modules, and 697 at the origin of the present-day modules which fused in various combinations with 4670 paralogous modules to create the 2487 paralogous present-day proteins (box in Figure 6). Such an “ancestral” genome would contain 2926 genes, with a putative size of 1.93 Mb if we assume that the mean size (220 residues) of present-day modules is mirroring that of the product encoded by these “ancestral” genes. Thus, such an approach does not go too far into the past, because the size of this putative ancestor is larger than that of the three modern pathogens C. jejuni (1.64), H. pylori (1.67), and H. influenzae (1.83), and amounts to as much as 41.6% of the present-day E. coli genome.
Figure 6.
Summary of the flux of information gathered to number the putative ancestral genes of the E. coli genome deduced by our suite of programs. Right: How our different programs concur to determine the different types of modules (unique, detected, and deduced) and to assemble the homologs in families. Left: Illustration of how a putative ancestral genome may be proposed in terms of size and distribution of gene classes.
Checking the Soundness of the Obtained Data
The data obtained as a result of this suite of programs are based in part on imposed assumptions, mainly the assignation of module range (step 3 of our suite of programs) and the inferring of distant modules (step 6). In particular, because this inference step is made without any threshold on the PAM distance, it requires a posteriori stringent controls. Therefore, to check the validity of these assumptions and that of the obtained data, we examined in detail each refined family for its multiple alignment and genealogical tree.
The amino acid sequence of detected modules was computed by the program Module using the data obtained with the AllAll program (see above). That of deduced modules was inferred from the multimodular protein on the basis of the module size (see Methods). Any inferred module which either cannot be multiply aligned with the other members of its refined family or is found too distant (lower PAM distance to the "closest" family member higher than 250) was discarded.
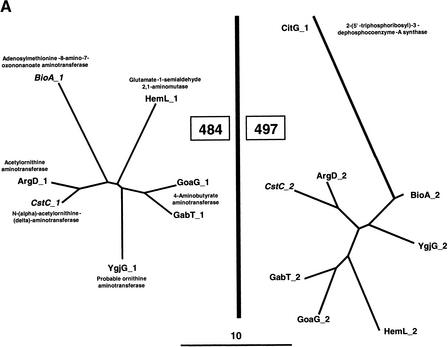
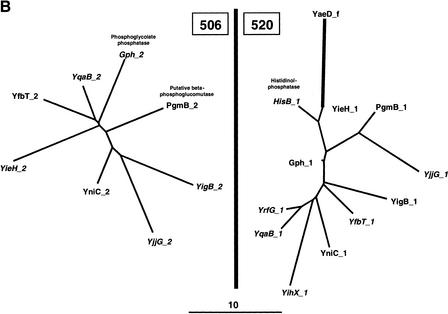
The refined families which escaped this first filter were further examined for their tree topology. Two types of dubious topology were found. Figure 7A shows an example where a deduced module displays a too faint homology. The module_1 and module_ 2 of seven aminotransferases form families 484 and 497, respectively. However, 497 contains also another module, CitG_1, which has an apparently unrelated function and which is very distant from the other members of this family. Because there is arguably doubt about such a remote homology, we preferred to exclude such cases. Figure 7B shows another arguable location of a deduced module. Again, we have a set of bimodular proteins whose function is mainly unknown, where module_2 forms family 506 and module_1 forms family 520, respectively. In family 520, the module YaeD_f seems to branch directly on module YieH_1. This unexpected, apparently aberrant topology, makes the assignation of YaeD to family 520 highly controversial. Accordingly, we decided to remove any module displaying such an aberrant position.
Figure 7.
Examples of genealogical trees of refined families displaying a suspect topology. The evolutionary relationships between detected (in italics) and deduced modules are shown for two pairs of grouped families. Branch lengths (in PAM units) are drawn to scale. (A) Families 484 and 497 belonging to group 7. (B) Families 506 and 520 belonging to group 79.
In total, 66 of the E. coli inferred modules were discarded after these two challenging steps. Thus, 95% of the modules we deduced by our approach seems sound.
DISCUSSION
The main objective of this article is to present a new experimental approach which tries to extract the maximum of information from completely sequenced microbial genomes in order to trace back the history of their genes. This automatic approach was designed to identify all modules in homologous proteins with the underlying idea that such retrieved modules are the signatures of ancient gene duplication and/or gene fusion events. Accordingly, we intentionally put aside many small regions of homology which correspond to what are often described as domains, motifs, or signatures according to their respective sizes. Such data are already nicely described in well known databases such as Prosite (Falquet et al. 2002), Prodom (Corpet et al. 2000), Domo (Gracy and Argos 1998), Pfam (Bateman et al. 2002), and SMART (Letunic et al. 2002). Our so-called modules often contained several such domains and/or motifs (not shown).
A Suite of Programs
Table 4 summarizes the suite of automatic programs that we have progressively created as a tool for a thorough analysis of a microbial proteome. Each of these programs, described in detail above, can be used independently. Several of these programs are based on the way we are translating the experimental data in the conceptual frame of our working model of gene ancestry. For example, the program Module is dependent on the way we are defining (Fig. 2) the different classes of structural segments of homology found previously by the Darwin AllAll program (Fig. 1) in terms of ancient events of gene duplication and gene fusion. Likewise, the program SortClust is built entirely on the way we are interpreting by logic the modular structure of proteins which belong to groups of families which are connected by a chain of neighboring unrelated homologous modules (Fig. 5). This approach, going back and forth between experimental data, then interpreting them to gain again new data through our suite of programs, appears to be acceptable only because we have introduced, at the end of this suite, a procedure to check the soundness of the obtained data. Interestingly, the vast majority of the inferred data responded correctly to the challenging criteria we used to define the reliable modules (e.g., >95% of the deduced E. coli modules seem sound).
Table 4.
The Suite of Home-Made Programs to Find Out All Modules in Homologous Proteins
| Main experimental step | Program | Language | Written by | |
|---|---|---|---|---|
| 1 → | Collecting protein dataset and translation in SGML language | sgml | C | K. Abou-Merhy |
| 2 → | Detecting homologous proteins | DARWIN AllAlla | Maple and C | R. De Rosa |
| 3 → | Detecting homologous modules | Module | Maple | H. Arfaoui |
| 4 → | Assembling homologous modules in families | Families | C | K. Abou-Merhy |
| 5 → | Inferring distant modules in multimodular proteins after grouping families and clustering them in new refined families | Module Sorter Module Cluster SortClust | C C C++ | S. Langevin S. Langevin I. Montaland |
| 6 → | Challenging deduced modules : checking evolutionary tree for each refined family | Disttree | C | A. Bazureau |
The AllAll program is part of the DARWIN suite of programs written by Gonnet et al. (1992).
To present this experimental approach as simply as possible, we have illustrated it with some of the results we obtained in the case of the comparison of four proteobacteria, but this strategy might work with as many microbial genomes as needed. The exhaustive comparison of the four proteomes corresponds simultaneously to (1) an intergenomic analysis (determination of all orthologous relationships between each combination of species), and (2) four intragenomic analyses (determination of all paralogous relationships for each proteobacterium). This allows the determination, for each analyzed protein, of its content in structural segments of homology. Thus, a protein (gene) may have partial, full, or mixed homology with another protein segment or may be unique. These different types of segments are viewed as modules in our working model because they all play a role in the mechanism of combinatorial construction of a gene from ready-made basic components. In our view, identifying a module is operationally equivalent to determining the ancestor to this gene segment.
Protein Classes
When comparing a set of several organisms, we can define four different classes of proteins for each genome. Firstly, inside the paralogous category, we can define the class of paralogs which have an ortholog in at least another species and the class of paralogs which are unique to a genome. Secondly, among the category of the genes which are unique to a species, we can define those which have an ortholog and those which are orphans. Figure 3 summarizes the main data obtained in the case of four proteobacterial genomes. It can be seen that the relative proportions of each protein class are rather different. The abundance of paralogous elements in E. coli compared to the three obligatory pathogens probably reflects the way of life of this bacterium, which is able to adapt to many different environmental conditions. In particular, we observe a significant proportion of genes specific to E. coli in both the unique and paralogous categories, these specific paralogs forming a strikingly high number of small families (varying in size from two to six members). Many of these paralogous small families code for putative transcriptional regulators, resistance to various substances (including antibiotics), or putative membrane proteins involved in various stages of transport of metallic ions and other rare environmental substances. These different functions may be important for survival of E. coli in adverse conditions.
Tracing Back Protein History
A large proportion of ancient events of gene duplication and gene fusion can be traced back when analyzing a unique proteome (Fig. 6). Here, the program SortClust plays a major role, because it allows one to resolve many fused modules in more elemental components and to detect distant homologies (see example, Fig. 5). In the case of E. coli, the number of putative ancestral modules decreased from 1020 (families detected in the AllAll output) to 542 (refined families). The number of ancestral genes can then be deduced for each genome from the determination of the unique proteins, unique modules, and families of paralogous modules. Figure 6 details the case of E. coli, allowing the reconstruction of a putative ancestral genome which would contain genes encoding unique modules detected or inferred in present-day proteins as well as the ancestors of the refined families of paralogous modules. Such an intragenomic analysis was helpful in determining that there are at least two classes of genes displaying very different behaviors: a majority of genes apparently never duplicated or, if they did, all extra copies were drastically eliminated; in contrast, a minority (18.5%) of genes did not stop to duplicate, with survival of the majority of the differentiated copies, leading to increasingly large families. The equilibrium between these two opposing tendencies may be achieved by some homeostatic mechanisms regulating the genome size (see, e.g., Mira et al. 2001). Then, many of the products deriving from this minor set of ancestral genes fused between them or with some unique genes in various combinations to increase the palette of available functions. These intragenomic data are thus helpful to disclose many of the ancient events which created present-day proteins, but they are of limited use for exploring the distant history of genomes.
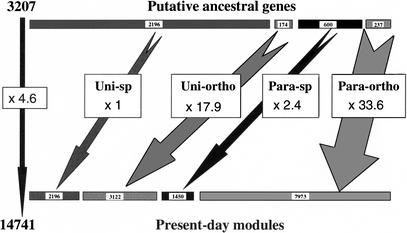
To go a step further back to more ancient ancestral genes, we turned to an intergenomic analysis. Figure 8 summarizes how we can use the whole set of data gathered when applying our suite of programs to the four proteobacteria. A comparison of the number of shared homologies between the different pairs of species shows that we have echi >> eccj > echp and cjhp >> cjec > hihp (table “a” in Fig. 8). Thus, we could tentatively reconstitute the gene distribution of both EcHi, putative last common ancestor (LCA) to the pair E. coli/H. influenzae, and CjHp, putative LCA to the pair C. jejuni/H. pylori. Although we started from different sets of class combinations (Fig. 3), the two ancestors EcHi and CjHp look strikingly alike in their respective inferred proportions of gene classes, with about half of the genes being unique to the species (54.5% for EcHi, 50.3% for CjHp), about one third being paralogous (32.4% for EcHi, 34.2% for CjHp), and the rest inferred as unique orthologs (13.1% for EcHi, 15.5% for CjHp). Thus, the LCA to these four proteobacteria would have been made of 3207 genes (table “b” in Fig. 8), which can be split again in two different categories: (1) 73.9% which never duplicated and which have been very frequently (68.5%) lost by one of the organisms after a speciation event, the rest (174) being the ancestors of the 3122 genes which form the 1301 families of orthologs found in present-day species; (2) 26.1% of the 3207 ancestral genes went through more or less frequent events of duplication with conservation of the differentiated copies. This small set of ancestral genes gave birth to either the 1450 members of the 600 paralogous families specific to each organism or the 7973 members of the 664 families of paralogous orthologs. Figure 9 shows another view of the process, which allowed the expansion from the 3207 ancestral genes to the 14,741 present-day modules, a 4.6-fold increase in the number of evolutionary units. The respective distributions of gene classes are given in respective percentages for both the ancestral genome LCA (upper bar, Fig. 9) and the four contemporary proteobacterial genomes (lower bar). The relative numbers of uni-ortho and especially para-ortho evolutionary units have exploded by a factor of near 18 (from 174 to 3122) and around 33.5 (from 237 to 7973), respectively. Such data are in strong support of the forecast hypothesis of Ohno (1970) that gene duplication is the main driving force to create new proteins/functions.
Figure 8.
Tentative reconstruction of the last common ancestor (LCA) to the four analyzed proteobacteria. Symbols in the pie charts are as in Fig. 3. Table a: The shared homologies between the different pairs of genomes of the four proteobacteria E. coli (ec), H. influenzae (hi), C. jejuni (cj), and H. pylori (hp). Table b: Summary of the respective figures for the four classes of genes found in the ancestors CjHp (ancestor to C. jejuni and H. pylori) and EcHi (ancestor to E. coli and H. influenzae) and in the LCA, respectively.
Figure 9.
From the putative ancestral genes to their progeny found in present-day proteins. Symbols used are as in Fig. 3. The upper bars give the distribution of the four gene classes in the genome of the putative LCA (gene content: 3207) with a size dependent on their relative percentage. The lower bars give the distribution of the contemporary modules for the same four gene classes (content in equivalent genes: 14,741) with the same size criteria. The boxed figures attached to the different arrows give the respective increase factor for each class as well as for the total (black arrow on the extreme left). See legend to Fig. 3 for the definition of the gene classes.
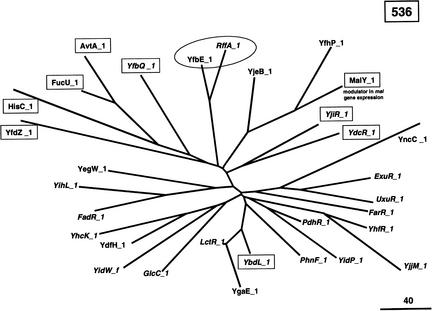
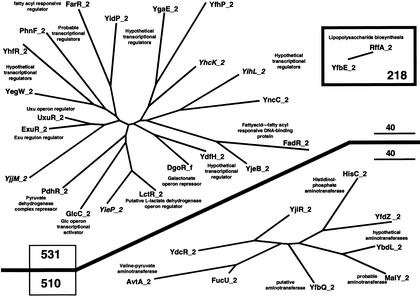
In this context, our experimental strategy seems pertinent to find out how a limited number of peculiar events of gene duplication and gene fusion helped to create such new proteins/functions. In particular, the thorough analysis of refined families belonging to the same group appears to be a powerful tool to trace back protein history. To illustrate this point, let us focus on the E. coli group 89 made of 63 paralogous modules belonging to 33 present-day proteins (Figs. 10, 11). This group 89 is made of four refined families of unequal size and contrasting groupings. The largest one, 536, is made of one module_f and thirty module_1s (Fig. 10). The corresponding module_2s are scattered in three different families (Fig. 11): a two-member family, 218, a nine-member family, 510, made predominantly of aminotransferases, and a larger family, 531, made mainly of transcriptional regulators. Thus, the most parsimonious scenario to explain the formation of these four refined families can be sketched as follows. Four ancestral genes, 536, 531, 510, and 218, fused in various combinations to create at least three ancestral functions. The fusion 536–218 led to a protein involved in lipopolysaccharide biosynthesis. The fusion 536–510 gave a gene encoding an ancestral aminotransferase, and the fusion 536–531 gave a gene encoding an ancestral transcriptional regulator. Then, two of these fusion products (536–510 and 536–531) duplicated once or several times to diverge in more or less large families of more specialized biochemical functions. Moreover, several of these bimodular proteins may be bifunctional, as it has been demonstrated for the recently crystallized MalY protein (Clausen et al. 2000). The presence of MalY_1 in family 536 and that of MalY_2 in family 510 appears consistent with the demonstration by Clausen et al. (2000) that MalY is capable of a direct protein-protein interaction with MalT, the central transcriptional activator of the maltose system and is composed of a pyridoxal 5′-phosphate-binding domain and a structural domain similar to aminotransferases.
Figure 10.
Genealogical tree of the refined family 536. The evolutionary relationships between detected (in italics) and deduced modules belonging to E. coli refined family 536 are shown. Branch lengths (in PAM units) are drawn to scale. Circled are the two module_1s whose module_2s belong to family 218. The module_1s whose module_2s belong to family 510 are boxed. The neighboring module_2s of the remaining module_1s belong to family 531.
Figure 11.
Genealogical trees of the refined families 510 and 531. The evolutionary relationships between detected (in italics) and deduced modules belonging to E. coli refined family 510 (upper part) and family 531 (lower part) are shown. Branch lengths (in PAM units) are drawn to scale. When known, the experimentally determined or putative function is indicated. The small family 218 is shown in a box.
Moreover, it may be underlined that the high level of information gained from the analysis of such refined families may also be very useful for functional annotation of modules of unknown proteins.
Conclusions
The enormous burst of data recently delivered by the whole-genome sequencing programs has radically changed the study of the mechanisms of protein evolution. Like other groups, we soon felt the need for new tools to undertake a global and automatic analysis of whole proteomes. This was the main incentive for our progressive building of appropriate programs to number—as far as we are able to detect them—the ancestral genes which were at the origin of the genes encoding proteins in present-day organisms. We have shown here that a large proportion of ancient events of gene duplication and gene fusion can be traced back when analyzing a proteome. Moreover, when comparing species, we could date at least some of these events of gene duplication and gene fusion with respect to the events of speciation and go further back to even more ancient ancestral genes. As we enlarge this approach to as many microorganisms as possible, we will be able to reach more and more ancient states for primordial genomes and their genes.
METHODS
Protein Sequences
The whole set of translated open reading frames obtained for completely sequenced genomes was imported from GenBank or EMBL databanks. We wrote a small program in C to extract and translate in SGML language the following information for each entry (each protein of each genome): a protein identification number, the species name, the gene name, the length of the protein, and its sequence (Table 4, step 1). This translation in SGML language is necessary for the search of homologies using the DARWIN program (Gonnet et al. 1992).
Comparing Protein Sequences
The DARWIN AllAll program was designed as a two-step procedure (Gonnet et al. 1992) to search a protein database to find the homologs using a maximum likelihood approach. We adapted the AllAll program to our own use (Table 4, step 2, see also De Rosa and Labedan 1998). First, the dynamic programming algorithm (Needleman and Wunsch 1970) is used with the PAM 250 matrix as a substitution score matrix (Schwartz and Dayhoff 1978). In this first step, the introduction of gaps is strictly regulated by penalties computed as a function of the PAM distance separating the two sequences (Benner et al. 1993). Then, in a second step, each alignment is refined using two other tools. The first tool tries to extend as far as possible the initial alignment using the Smith and Waterman (1981) algorithm. The second tool recalculates the initial substitution score matrix using the current data set of proteins in order to find the best matrix. This optimization process is monitored by computing the variance of the PAM distance, searching for the lowest value. When this lowest value cannot be decreased further, the alignment is registered as the optimal one for the two proteins studied.
We designed the output of the DARWIN AllAll analysis to exhibit essential information on the sequence alignments for each pair of homologous proteins (table in Fig. 1). First, information is given for each protein: the protein identification (PID number), the name of the gene which encodes it, and its length (number of residues). We then obtain information on the alignment itself, at which position it begins, and how long it is. Finally, information on homology is displayed: The value of the distance separating both proteins is computed in PAM units, followed by the variance of this evolutionary distance and the percent identity. Because the AllAll output file contains a significant number of “doubles” (duplicates), we added a small program in C which eliminates all duplicated pairs (program netto_mod written in C language by A. Bazureau, unpubl.).
It must be noted that such an exhaustive AllAll comparison is a time-consuming step. Typically, comparing two bacterial proteomes corresponding to about 5000 proteins requires about 20 h.
Registering All Modules
The cleaned list of pairs is then scanned by another program written in Maple to identify for each pair the nature of the modules and for each protein its modular structure (Table 4, step 3). The program Module successively completes two tasks: (1) During the AllAll analysis, it adds to each line of the DARWIN output (Fig. 1) information about the nature of the respective module from each protein involved in the found alignment according to the conditions outlined in Figure 2. (2) When the AllAll analysis is completed, it writes a file giving, for each homologous protein, the list of the modules found, the length, and the amino acid sequence of each module computed from the alignment data. When a module pairs with several homologs, the retained sequence is the longest alignment found.
Grouping Homologous Modules in Families
The program previously written in Caml language (De Rosa and Labedan 1998) has been replaced by a much faster one, written in C language (Table 4, step 4). This program, Families, automatically gathers into one family all homologous modules that are related by a chain of similarities, collecting all relatives of both members of each pair until no further pairwise relationship is found. This program further sorts the families by their size (number of members), then by alphanumeric order of PID numbers, which are themselves sorted within each family.
Genealogical Trees of Families
An evolutionary tree is derived for each family containing at least four members (after elimination of all nonassigned modules) using a script written in Maple to compute a PAM distance matrix and a matrix of the variances of each PAM distance. An adaptation of the program PhyloTree of the DARWIN package is then used to reconstruct a distance tree which is an approximation to a maximum likelihood tree (Gonnet et al. 1992). To automate these steps of family selection, translation in SGML language, and adaptation of PhyloTree program, the program prep_fam was written in C language by A. Bazureau (unpubl.).
Grouping Families
Another program, prep_group (written in C language by A. Bazureau, unpubl.), allows the user to identify all families which share homologous modules in multimodular proteins, as shown in Figures 4 and 5. The selected families are then clustered in groups by using again the program Families. Then, the list of all required information (see the table in Fig. 5) is automatically extracted for each protein belonging to these grouped families.
Inferring Distant Modules and Splitting Fusion Modules
The complete list of proteins containing information about their modules and family number is analyzed by the program SortClust (Table 4, step 5). SortClust consists of two procedures: Module Sorter compares multimodular proteins belonging to the same group of families to detect by logic distant modules. Module Cluster groups together all homologous modules, either detected or deduced, in new refined families. SortClust is also able to detect and deal with internal duplications and to treat complex cases where a module pairs with another splittable module. Depending on the complexity of the dataset to be analyzed, SortClust may require 1–2 h of computer time.
Acknowledgments
We gratefully acknowledge the invaluable help provided by the undergraduate students Karim Abou-Merhy, Hichem Arfaoui, and Adrien Bazureau in the development of several of the computer programs described herein.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
4 Corresponding author.
E-MAIL labedan@igmors.u-psud.fr; FAX 33 (0)1 6915-78 08.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.393902.
REFERENCES
- Altschul SF. Amino acid substitution matrices from an information theoretic perspective. J Mol Biol. 1991;219:555–565. doi: 10.1016/0022-2836(91)90193-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF. A protein alignment scoring system sensitive at all evolutionary distances J. Mol Evol. 1993;36:290–300. doi: 10.1007/BF00160485. [DOI] [PubMed] [Google Scholar]
- Benner SA, Cohen MA, Gonnet GH. Empirical and structural models for insertions and deletions in the divergent evolution of proteins. J Mol Biol. 1993;229:1065–1082. doi: 10.1006/jmbi.1993.1105. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T, Schlegel A, Peist R, Schneider E, Steegborn C, Chang Y-S, Haase A, Bourenkov GP, Bartunik HD, Boos W. X-ray structure of MalY from Escherichia coli: A pyridoxal 5′-phosphate-dependent enzyme acting as a modulator in mal gene expression. EMBO J. 2000;19:831–842. doi: 10.1093/emboj/19.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F, Servant F, Gouzy J, Kahn D. ProDom and ProDom-CG: Tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res. 2000;28:267–269. doi: 10.1093/nar/28.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC. A model for evolutionary change. In: Dayhoff MO, editor. Atlas of protein sequence and structure. Vol. 5 1978. , Suppl. 3, pp. 345–352. National Biomedical Research Foundation, Washington, D.C. [Google Scholar]
- De Rosa R, Labedan B. The evolutionary relationships between the two bacteria Escherichia coli and Haemophilus influenzae and their putative last common ancestor. Mol Biol Evol. 1998;15:17–27. doi: 10.1093/oxfordjournals.molbev.a025843. [DOI] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJA, Hofmann K, Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng DF, Johnson MS, Doolittle RF. Aligning amino acid sequences: Comparison of commonly used methods. J Mol Evol. 1985;21:112–125. doi: 10.1007/BF02100085. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- Fitch WM. Homology: A personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Gonnet, G. and Hallett, M. 1997. The DARWIN Manual. http://www.wr.inf.ethz.ch/personal/gonnet/DarwinManual/DarwinManual.html.
- Gonnet GH, Cohen MA, Benner SA. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- Gracy J, Argos P. DOMO: A new database of aligned protein domains. Trends Biochem Sci. 1998;12:495–497. doi: 10.1016/s0968-0004(98)01294-8. [DOI] [PubMed] [Google Scholar]
- Labedan B, Riley M. Genetic inventory: Escherichia coli as a window on ancestral proteins. In: Charlebois R, editor. Organization of the prokaryotic genome. 1999. , Ch. 17, pp. 311–329. ASM Press, Washington, D.C. [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Riley M, Labedan B. Protein evolution viewed through Escherichia coli protein sequences: Introducing the notion of a structural segment of homology, the module. J Mol Biol. 1997;268:857–868. doi: 10.1006/jmbi.1997.1003. [DOI] [PubMed] [Google Scholar]
- Schwartz RM, Dayhoff MO. Matrices for detecting distant relationships. In: Dayoff MO, editor. Atlas of Protein Sequence and Structure. Vol. 5 1978. , Suppl. 3, pp. 353–358. National Biomedical Research Foundation, Washington, D.C. [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]