Abstract
We have characterized a novel nuclease from the Kamchatka crab, designated duplex-specific nuclease (DSN). DSN displays a strong preference for cleaving double-stranded DNA and DNA in DNA-RNA hybrid duplexes, compared to single-stranded DNA. Moreover, the cleavage rate of short, perfectly matched DNA duplexes by this enzyme is essentially higher than that for nonperfectly matched duplexes of the same length. Thus, DSN differentiates between one-nucleotide variations in DNA. We developed a novel assay for single nucleotide polymorphism (SNP) detection based on this unique property, termed “duplex-specific nuclease preference” (DSNP). In this innovative assay, the DNA region containing the SNP site is amplified and the PCR product mixed with signal probes (FRET-labeled short sequence-specific oligonucleotides) and DSN. During incubation, only perfectly matched duplexes between the DNA template and signal probe are cleaved by DSN to generate sequence-specific fluorescence. The use of FRET-labeled signal probes coupled with the specificity of DSN presents a simple and efficient method for detecting SNPs. We have employed the DSNP assay for the typing of SNPs in methyltetrahydrofolate reductase, prothrombin and p53 genes on homozygous and heterozygous genomic DNA.
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank/EMBL/Date Bank under accession nos. AF520591. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: N.K. Yankovsky, A.V. Polyakov, and G.N. Rudenskaya.]
Nucleases are important analytical enzymes used widely in biomedical and biotechnological applications. Their application depends on the specificity and mode of action of a particular enzyme. Nucleases have been employed in the production of flavor enhancers, removal of nucleic acids, as therapeutic agents, and for the determination of nucleic acid structure, in particular, mutation typing (Reddy and Shankar 1993; Gite and Shankar 1995; Jasin 1996; Singwi and Joshi 2000; Williams 2001). Nucleases with novel and unusual properties may facilitate the development of advanced technologies in many areas of biotechnology and biomedicine.
Some nucleases show little sequence specificity but recognize certain structural features of their respective DNA substrates. These include enzymes with cleavage preference for single-stranded (ss) DNA, compared to double-stranded (ds) DNA (nucleases P1 and S1), those recognizing helical parameters such as groove width and flexibility (DNase I) or helical distortions caused by abasic sites (exonuclease III, HAP1), and enzymes specific for structures like flap DNA (5′-nucleases of eukaryotes, phage and eubacterial DNA polymerases) and four-way junctions (T4 endonuclease VII, RuvC; Suck 1997). DNase activity with cleavage preference for dsDNA has been reported in some marine invertebrates including sea-urchin and Kamchatka crab (Menzorova and Rasskazov 1981; Menzorova et al. 1993), but enzymes that cleave dsDNA in preference to ssDNA have not been cloned to date.
In this study, we characterized a novel nuclease from Kamchatka crab that displays considerable preference for DNA duplexes (dsDNA and DNA in DNA-RNA hybrids), compared to ssDNA. Moreover, this enzyme discriminates between perfectly and nonperfectly matched (up to one mismatch) short DNA duplexes. We applied this nuclease to single nucleotide polymorphism (SNP) typing.
SNP detection has considerable significance in association studies of complex diseases (Risch and Merikangas 1996; Brookes 1999; Cargill et al. 1999; Kruglyak 1999), pharmacogenetics (Housman and Ledley 1998; McCarthy and Hilfiker 2000; Roses 2001), population genetics (Hacia et al. 1999; Nielsena 2000), and physical mapping (Kruglyak 1997).
A number of different methods have been reported for the detection of single nucleotide variations (Landegren et al. 1998; Kwok 2000; Syvanen 2001). In general, current methods require preamplification of genomic DNA, usually by polymerase chain reaction (PCR), followed by SNP genotyping with an allele discrimination by one of four allele-specific reaction principals: hybridization, primer extension, oligonucleotide ligation, and cleavage of a flap probe. Each of these principals may be combined with either a homogeneous or solid-phase reaction format and a detection method such as fluorescence intensity, fluorescence polarization, or mass spectrometry. The specific advantages and disadvantages of each method have been covered in several reviews (Landegren et al. 1998; Kwok 2000; Syvanen 2001).
Although many methods of SNP detection exist, there is no single accepted technology of choice (Syvanen 2001). A suitable method for SNP detection must be accurate, adaptable for both solid-phase and homogenous reaction formats, and develop a robust and easily interpretable signal. These features lend the technology to automation.
In this study, we developed a novel assay for SNP detection that utilizes the unique properties of crab nuclease and fluorescence energy transfer tags, thus ensuring accurate and inexpensive discrimination between wild-type and mutated sequences. We have applied the DSNP assay to SNP analysis in p53, methyltetrahydrofolate reductase (MTHFR), prothrombin, and cytochrome C oxidase subunit I (COX1) genes on homo- and heterozygous DNA samples.
RESULTS
Cloning and Purification of a Kamchatka Crab Nuclease
Using degenerative primers corresponding to the most conserved amino acid sequences in DNA/RNA nonspecific nuclease family and SMART-RACE technology (Matz et al. 1999), we isolated a cDNA comprising full coding sequence of a novel nuclease from Kamchatka crab (Paralithodes camtschaticus).
The predicted crab protein shows 64% identity at the amino acid level with shrimp nuclease from Penaeus japonicus (Chou and Liao 1990; Wang et al. 2000). Analysis of the crab sequence by SMART software (Schultz et al. 2000) disclosed that the protein comprises a 27-aa signal peptide and a DNA/RNA nonspecific endonuclease domain (NUC domain). An alignment of the crab NUC domain with known domains of invertebrate, fungal, and bacterial proteins revealed that the crab domain contains a presumptive active site similar to the active site in shrimp nuclease (Wang et al. 2000).
The mature crab protein was expressed in E. coli. Although the recombinant protein was not active, it was used in rabbit immunization for polyclonal antibody generation. Nuclease from the Kamchatka crab hepatopancreas was purified by acetone precipitation, followed by column chromatography on DEAE Macro-Prep, Phenyl-Agarose, Hydroxyapatite, Heparin-Sepharose, and Sephadex G75 columns. Fractions from the purification steps were tested for both DNase activity and antibody binding. Purified crab nuclease was present as a single band of about 42 kD on SDS/PAGE.
Enzymatic Characteristics of Kamchatka Crab Nuclease
Similar to other members of the DNA/RNA nonspecific nuclease family, crab nuclease acquired its enzymatic activity in the presence of Mg2+ ions (7 mM required for optimal activity) and was inhibited by EDTA. The DNase activity of the purified Kamchatka crab nuclease was about 18,000 Kunitz units/mg protein, as determined with a modified Kunitz assay (Kunitz 1950; Liao 1974). The pH and temperature optima for activity were 7–8 and 55°–65°C, respectively. The nuclease was stable at a pH of greater than 6 and temperatures below 75°C.
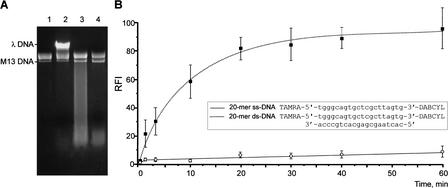
Surprisingly, crab nuclease exhibited strong cleavage preference for dsDNA substrates and little activity against ssDNA (Fig. 1). Moreover, although the Kamchatka crab nuclease was classified as part of a DNA/RNA nonspecific nuclease family, no significant cleavage activity on RNA substrates was observed. However, the nuclease effectively cleaved DNA molecules in DNA-RNA hybrid duplexes (data not shown). Based on these characteristics, we suggested the designation “duplex-specific nuclease” (DSN) for this enzyme.
Figure 1.
Determination of crab nuclease preference for specific structural features of DNA substrates. (A) Action of crab nuclease on ss phage M13 DNA and ds λ DNA. Lanes 1,2: negative controls, incubation without nuclease. Lane 1: phage M13 DNA alone; lane 2, mixture containing phage M13 DNA and λ DNA. Lanes 3,4: Digestion of phage M13 and λ DNA mixture by crab nuclease at 70°C for 1.5 min (lane 3) and 5 min (lane 4). For experimental details, see Methods. (B) Action of crab nuclease on synthetic ss and ds 20-mer DNA substrates. The cleavage reaction was performed as described in Methods at 35°C for different periods. Fluorescence intensity was measured at 570 nm (with excitation at 550 nm). The relative fluorescence increase in the oligonucleotide substrate, RFI, was defined as RFI = (Fi − Fo/Fmax − Fo) × 100%, where Fi is the fluorescence intensity of a substrate after incubation with nuclease, Fo is the substrate fluorescence in the absence of enzyme, and Fmax represents the fluorescence of 100% cleaved substrate. For kinetic graph construction, three identical experiments were performed, and the average values and standard deviations were plotted.
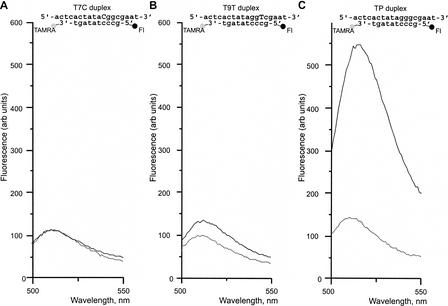
Analysis of DSN action on synthetic oligonucleotide substrates revealed that the enzyme discriminates between perfectly matched short DNA-DNA duplexes (8–12 bp) and duplexes of the same length with at least one mismatch (Fig. 2). The ability of DSN to distinguish between variations of one nucleotide in short DNA duplexes allowed the development of a novel efficient method of SNP detection, designated the DSN preference (DSNP) assay.
Figure 2.
Results of the DSN action on one mismatch-containing (A,B) and perfectly matched (C) DNA duplexes. Duplexes formed by 5-carboxyfluorescein (Fl)-5′-gccctatagt-3′-TAMRA signal probe and complementary targets were incubated with DSN at 35°C for 15 min. Emission spectra were obtained on the spectrofluorometer, with excitation at 480 nm. Dotted line, substrate fluorescence in the absence of enzyme; solid line, substrate fluorescence after incubation with DSN.
DSNP Assay Principle
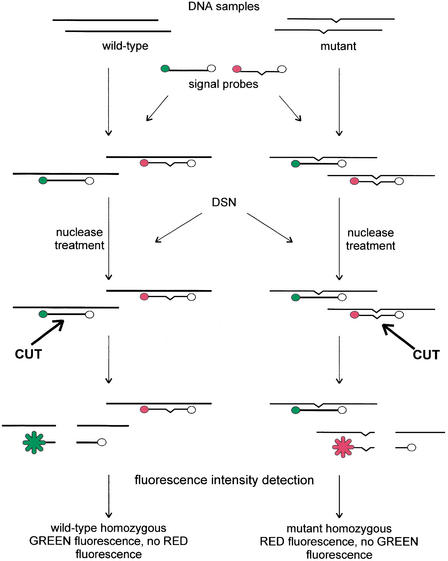
The principle of the method is depicted in Figure 3. Initially, the DNA fragment containing the SNP site of interest is amplified by PCR. Following amplification, an aliquot of the PCR reaction is mixed with two signal probes (usually 10-nt oligonucleotides) labeled with the fluorescence donor (at the 5′ end) and quencher (at the 3′ end). Each signal probe generates fluorescence at specific wavelengths after cleaving. The first oligonucleotide is complementary to the wild-type sequence, whereas the second is complementary to the mutant sequence. The mixture is incubated with DSN, during which the nuclease cleaves the PCR product to generate short DNA fragments that can effectively hybridize with signal probes. All perfectly matched duplexes generated by the DNA template and signal probes are cleaved by DSN to generate fluorescence.
Figure 3.
Scheme of the DSNP assay. The green circle represents the first fluorescent donor, the red circle the second fluorescent donor, and the open circle the fluorescent quencher.
To improve signal intensity, the reaction is performed in the presence of exonuclease-deficient Klenow fragment [KF(exo-)], which catalyzes strand displacement DNA synthesis by extension of the 3′-ends generated in the PCR fragment upon DSN nicking activity. Displaced DNA strands are involved in a genotyping reaction that results in a 5–20-fold increase in the specific fluorescent signal.
In the scheme described above, we employed two fluorescent dyes to distinguish between wild-type and mutant sequences in one tube. However, the assay may also be performed when both signal probes generate one type of fluorescence. In this case, wild-type and mutant sequence-specific signal probes are mixed with DNA substrates in two separate tubes.
Signal Probe Examination
To determine the generality of mismatch discrimination, we constructed a set of closely related 18-nt synthetic targets with single nucleotide variations along the 10-nt sequence and fluorescence resonance energy transfer (FRET)-labeled 10-nt signal probes capable of hybridizing with these targets to form perfect and one mismatch-containing duplexes (Table 1). Two types of signal probes were used, specifically, probes labeled with 5-carboxyfluorescein (Fl) (at the 5′ end) and TAMRA (at the 3′ end) and those labeled with Fl and DABCYL. All possible combinations between targets and signal probes were examined for cleavage by DSN.
Table 1.
Comparison of DSN Action on Perfect and Mismatched 10 bp DNA Duplexes
| Duplex Name | Target Structurea | ΔFm/ΔFp for signal probe: | |
|---|---|---|---|
| F1-5′-gccctatagt-3′TAMRA | F1-5′-gccctatagt-3′DABCYL | ||
| TP | 5′-actcactatagggcgaat-3′ | 1.00 | 1.00 |
| T1C | 5′-actcCctatagggcgaat-3′ | 0.04 | 0.48 |
| T1G | 5′-actcGctatagggcgaat-3′ | 0.04 | 0.25 |
| T1T | 5′-actcTctatagggcgaat-3′ | 0.04 | 0.42 |
| T2A | 5′-actcaAtatagggcgaat-3′ | 0.05 | 0.36 |
| T2G | 5′-actcaGtatagggcgaat-3′ | 0.02 | 0.46 |
| T2T | 5′-actcaTtatagggcgaat-3′ | 0.02 | 0.24 |
| T3A | 5′-actcacAatagggcgaat-3′ | 0.02 | 0.27 |
| T3C | 5′-actcacCatagggcgaat-3′ | 0.01 | 0.09 |
| T3G | 5′-actcacGatagggcgaat-3′ | 0.02 | 0.22 |
| T4C | 5′-actcactCtagggcgaat-3′ | 0.02 | 0.10 |
| T4G | 5′-actcactGtagggcgaat-3′ | 0.03 | 0.18 |
| T4T | 5′-actcactTtagggcgaat-3′ | 0.03 | 0.18 |
| T5A | 5′-actcactaAagggcgaat-3′ | 0.01 | 0.11 |
| T5C | 5′-actcactaCagggcgaat-3′ | 0.03 | 0.14 |
| T5G | 5′-actcactaGagggcgaat-3′ | 0.02 | 0.13 |
| T6C | 5′-actcactatCgggcgaat-3′ | 0.06 | 0.14 |
| T6G | 5′-actcactatGgggcgaat-3′ | 0.04 | 0.16 |
| T6T | 5′-actcactatTgggcgaat-3′ | 0.01 | 0.22 |
| T7A | 5′-actcactataAggcgaat-3′ | 0.03 | 0.07 |
| T7C | 5′-actcactataCggcgaat-3′ | 0.01 | 0.10 |
| T7T | 5′-actcactataTggcgaat-3′ | 0.06 | 0.09 |
| T8A | 5′-actcactatagAgcgaat-3′ | 0.04 | 0.06 |
| T8C | 5′-actcactatagCgcgaat-3′ | 0.05 | 0.01 |
| T8T | 5′-actcactatagTgcgaat-3′ | 0.06 | 0.05 |
| T9A | 5′-actcactataggAcgaat-3′ | 0.05 | 0.03 |
| T9C | 5′-actcactataggCcgaat-3′ | 0.08 | 0.03 |
| T9T | 5′-actcactataggTcgaat-3′ | 0.07 | 0.03 |
| T10A | 5′-actcactatagggAgaat-3′ | 0.03 | 0.03 |
| T10G | 5′-actcactatagggGgaat-3′ | 0.07 | 0.08 |
| T10T | 5′-actcactatagggTgaat-3′ | 0.05 | 0.15 |
Duplexes were generated from 10-mer signal probes and 18-mer synthetic targets. Fluorescence values, ΔF, were normalized according to the formula: ΔF = Fi-Fo, where Fi is the fluorescence of substrate after DSN treatment and Fo represents fluorescence at the start of the reaction. The ratio of DSN activity on mismatched (ΔFm) and perfect (ΔFp) duplexes was determined.
Sequence complementary to signal probes is underlined; mismatch position is marked by capital letter.
The observed fluorescence change for all mismatched duplexes was at least 10 times lower than that for perfectly matched duplexes in the experiments with the Fl-TAMRA-labeled probe (Table 1). No preference of DSN for specific mismatch positions was noted. In the case of the Fl-DABCYL-labeled probe, clear discrimination between perfect and nonperfect duplexes was observed if the variable nucleotide position was either at the 5′ end or the center of a signal probe (Table 1). Duplexes comprising mismatches near the 3′ end of the signal probe (T1–4) were cleaved by DSN with only 1.5–5 times less efficiency than perfect duplexes.
Thus, the effectiveness of the DSN-cleaving reaction depends on the fluorescent dye used in the signal probe. Nevertheless, most mismatches in target DNA were detectable with a common signal probe.
To determine the products of the DSN-cleaving reaction, several P32-labeled 10-mer signal probes were hybridized with 40-mer oligonucleotide targets and examined for cleavage by DSN. In each case, products of the cleavage reaction were subjected to polyacrylamide gel electrophoresis. We found that DSN cut the signal probes predominantly between fourth and fifth, and fifth and sixth nucleotides from the 5′-end. When the longer DNA duplexes were used as DSN substrates, cleavage reaction yielded products of 6 bp and shorter (data not shown).
Model Experiments
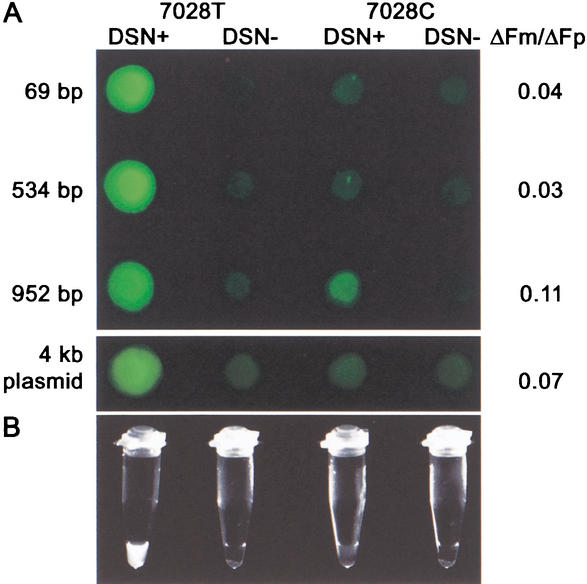
The DSNP assay was examined on several models, using wild-type and mutant DNA samples obtained from homozygous and heterozygous donors. We initially determined the size of PCR product suitable for the DSNP assay. PCR products of 952, 534, and 69 bp comprising C- or T- variants of the 7028 C-T mitochondrial polymorphous site (in COX1 gene) were generated and used with a T-variant specific signal probe in the DSNP assay. Clear results were obtained with all PCR products (see Fig. 4A). Moreover, we performed a specific DSNP assay reaction with plasmid DNA (about 4 kb) comprising a 69-bp COX1 fragment (Fig. 4A). From these studies, we conclude that the lengths of DNA targets may vary widely.
Figure 4.
Detection of 7028 C-T SNP in the COX1 gene with the DSNP assay. PCR fragments of 69, 534, and 952 bp comprising 7028 T or 7028 C variants, and pT-Adv plasmids with 69-bp fragment insertions were used in SNP typing with a T variant-specific signal probe. “DSN+”, reaction with DSN; “DSN−”, control samples, no enzyme. ΔFm/ΔFp was defined as in Table 1. (A) The multiwell PCR plate was photographed with an Olympus SZX12 fluorescent stereomicroscope equipped with a green filter. (B) PCR strips with DSNP results for 69-bp fragments were photographed with a MultiImage Light Cabinet (Alpha Innotech) under UV light.
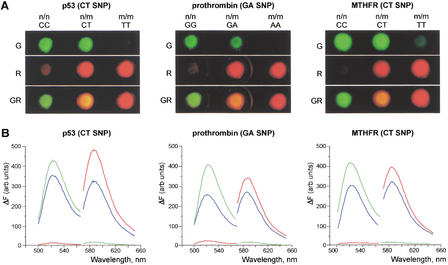
We further employed the DSNP assay with two fluorescent dyes to detect specific polymorphisms in genetic loci contributing to susceptibility to some diseases. We analyzed prothrombin 20210 G-to-A polymorphism associated with an increased risk of venous thrombosis and myocardial infarction (Poort et al. 1996), C677T polymorphism of the MTHFR gene associated with increased levels of total plasma homocysteine, a risk factor for coronary artery disease (Frosst et al. 1995), and p53 C309T polymorphism associated with carcinomas (Abarzua et al. 1995) in homozygous and heterozygous DNA samples (Fig. 5). All of the results obtained with the DSNP assay were confirmed by DNA sequencing.
Figure 5.
Analysis of p53 C309T, prothrombin 20210 G-to-A, and MTHFR C677T polymorphous sites in homo- and heterozygous DNA by the DSNP assay. (A) Photographs obtained on the fluorescent stereomicroscope equipped with green (G) and red (R) filters. GR, computer superposition of images obtained with green and red filters. n/n, homozygous DNA samples comprising wild-type sequence variant; n/m, heterozygous DNA samples; m/m, homozygous DNA samples comprising mutant sequence variant. (B) Normalized emission spectra of these samples obtained on a spectrofluorometer, with excitation at 480 nm (for green fluorescence) and 550 nm (for red fluorescence). Fluorescence values, ΔF, were normalized as described in Table 1. Green line, homozygous DNA samples comprising the wild-type sequence variant; red line, homozygous DNA samples comprising the mutant sequence; blue line, heterozygous DNA samples.
DISCUSSION
We cloned a novel crab nuclease enzyme (DSN) specific to DNA-containing duplexes. Based on sequence analysis, this enzyme was classified as part of the DNA/RNA nonspecific nuclease family. Typically, nonspecific nuclease family members cleave all types of nucleic acids with similar specificity for RNA, ss, and dsDNA substrates (Friedhoff et al. 1996; Ho et al. 1998; Meiss et al. 1998). Several nucleases of this family have either no or low RNase activity (Ho and Liao 1999; Wang et al. 2000), but no differences between cleavage activity against ss and dsDNA have been noted to date. Thus, DSN represents a novel type of nonspecific nuclease that displays no (or extremely low) RNase activity and a strong preference for cleaving dsDNA compared to ssDNA. Moreover, DSN is stable at a wide spectrum of pH values and temperatures, and insensitive to protease digestion (data not shown). These properties suggest that the protein is a good candidate for biotechnological use.
DSN cleavage activity against perfectly matched DNA duplexes exceeds its activity against nonperfectly matched duplexes of the same length. This DSN property is the most evident in short duplexes, in particular, those between short oligonucleotides (8–12 nt) and DNA substrates.
Based on this DSN characteristic, we developed a novel simple, robust, and automatable assay for SNP detection. There are several major advantages of the DSNP assay. Firstly, the DSNP assay works well with double-stranded DNA templates, specifically, unpurified PCR products. There are no separation, purification, or centrifugation steps. Currently, the assay takes about 1 h after PCR products are obtained.
Secondly, the DSNP assay does not require expensive instrumentation. Specific fluorescence signals produced in the assay may be visualized using standard laboratory equipment. For example, when green dye only is used for signal probe generation, the fluorescent signal is detectable, even under UV transillumination (Fig. 4B).
Thirdly, the assay has built-in internal controls. The fact that both alleles may be assayed in the same tube and the semiquantitative nature of fluorescence intensity changes make it relatively simple to identify PCR failures using this procedure.
Fourthly, it is possible to design PCR primers and probes for multiple different assays that may be performed well under one set of standardized reaction conditions. We found that the dependence of the DSNP assay on DNA template length is unimportant and the position of the blinking nucleotide may vary over a wide range. It allows the consistent selection of PCR primers and signal probes so that little or no optimization is necessary when a uniform set of criteria is employed to select primers and probes for different assays.
Finally, the DSNP assay is potentially suitable for a microarray format without the need for considerable modification. In this case, for each SNP to be detected, two signal probes are immobilized on the microarray. Signal probes are attached to the solid phase through a spacer followed by a 10-nt specific sequence, which differ in the positions corresponding to allelic distinction. Each probe is labeled with a fluorescent donor and a quencher at the ends of the specific sequence, so that a fluorescence signal is generated on a microarray after cleavage by DSN.
METHODS
DSN Cloning
Total RNA was isolated from crab hepatopancreas according to the protocol of Chomczynski and Sacchi (1987). First-strand cDNA synthesis and RACE procedures were performed using the SMART RACE cDNA Amplification Kit (Clontech) according to the manufacturer's instructions. The following degenerative primers were used for the 3′-RACE: 5′-cctcagtggca(g/a)gcttt(c/t)aat-3′ for the first PCR, and 5′-caggcctttaataatgg(t/a/g/c)aa(t/c)tgg-3′ for the nested PCR. Gene-specific primers (5′-ggccaggtctcgggtcgc-3′; 5′-gggtcgcgtattctaggta-3′, and 5′-ccattattgaaggcctgcca-3′) were used for 5′-RACE.
Sequence Analysis
Sequences were analyzed using BLAST software (http://www.ncbi.nlm.nih.gov/BLAST). Domain organization was verified with the Web-based SMART tool (http://smart.embl-heidelberg.de). Multiple sequence alignment was performed using the Clustal W version 1.8 software, following eye optimization.
Purification of DSN From Crab Hepatopancreas
Rabbit polyclonal antibodies were prepared against His-tagged mature DSN protein cloned into the pQE-30 vector (QIAGEN), produced in Escherichia coli, strain Ad 494 (Novagen), and purified by immobilized metal affinity chromatography using Talon Resin (Clontech).
Crab DSN nuclease was purified at 4°C. Fractions were subjected to testing for DNase activity and Western blotting (Harlow and Lane 1988) with rabbit polyclonal antibodies prepared against recombinant crab protein.
Fresh crab hepatopancreas was homogenized in two volumes of 100 mM Tris-HCl (pH 8.0) with 100 mM EDTA and centrifuged at 10,000 g for 30 min. The supernatant was diluted with 1.5 volumes of acetone, incubated for 12 h and centrifuged at 10,000 g for 1 h. The sediment was dried, and 5 g of the resultant acetone powder was diluted in 250 mL buffer A (0.05 M Tris-HCl, pH 7.1) and mixed for 2 h. The insoluble fraction was separated out by centrifugation at 10,000 g for 30 min, and the supernatant was applied to a DEAE-MacroPrep column (Bio-Rad) equilibrated with buffer A. After loading, the column was washed with the same buffer. Protein was eluted with a 0–0.5 M NaCl gradient in buffer A. After adding NaCl to a final concentration of 5 M, DSN-containing fractions were loaded onto a phenyl-agarose column (Amersham Pharmacia Biotech) equilibrated with buffer A containing 5 M NaCl. After washing the column with the same high-salt buffer, DSN was eluted with a 5–3 M NaCl gradient in buffer A. DSN-containing fractions were pooled, diluted twofold with 0.035 M Tris-HCl (pH 8.1), and applied to a hydroxyapatite column (Bio-Rad) equilibrated with 0.045 M Tris-HCl (pH 7.5). The fractions were directly eluted with 0.025 M sodium phosphate buffer, pH 7.5, combined, and dialyzed overnight against 0.01 M Tris-HCl (pH 7.1) containing 0.001 M MgCl2. Dialyzed fractions were loaded onto a heparin-Sepharose column (Amersham Pharmacia Biotech), equilibrated with the 0.01 M Tris-HCl (pH 7.1) buffer containing 0.001 M MgCl2, and eluted with a 0–0.3 M NaCl gradient after washing. DSN-containing fractions were concentrated to 1 mL on a Biomax-5K membrane (Millipore), transferred to buffer A, and subjected to gel filtration on a Sephadex G-75 (Sigma) column. Purified DSN was concentrated on a Biomax-5K membrane, diluted with one volume of glycerol, heated at 70°C for 10 min under mineral oil, and incubated at 4°C overnight. About 0.15 mg DSN protein (2700 Kunitz units) was purified from 5 g acetone powder. DSN was stored at −20°C for long-term use.
DSN Activity Analysis
DNAse activity was measured using the Kunitz assay (Kunitz 1950) modified by Liao (1974), in 1 × DSN buffer (7 mM MgCl2, 50 mM Tris-HCl, pH 8.0) containing 400 μM calf thymus DNA. To determine the influence of Mg2+ on crab DSN, the assay was performed with varying concentrations of MgCl2 (0.1–10 mM final concentration). The dependence of DSN activity on pH was analyzed using the Kunitz assay with 50 mM sodium formate (pH 3.0–3.5), 50 mM sodium acetate (pH 3.5–6.0), 50 mM Mes-NaOH (pH 6.0–7.0), 50 mM Tris-HCl (pH 7.0–9.5), and 50 mM glycine-NaOH (pH 9.0–10.0) in the presence of 7 mM MgCl2. The dependence of DSN activity on temperature was analyzed at different reaction temperatures, ranging between 20° and 90°C (at 5°C intervals).
DSN activity on λ dsDNA and phage M13 ssDNA was compared by agarose gel electrophoresis. The reaction was performed in a total volume of 10 μL comprising 1 × DSN buffer, 0.06 Kunitz units DSN, 150 ng λ DNA, and 50 ng M13 DNA. To prevent ds structure formation in phage M13 DNA, the reaction mixture was incubated at 70°C for 1, 5, or 5 min. The digestion products were visualized on a 0.9% agarose gel, following ethidium bromide staining.
For analysis of DSN activity on synthetic oligonucleotide substrates, oligonucleotides labeled with a fluorescent donor at the 5′ end and a fluorescent quencher at the 3′ end were used as ssDNA. To generate ds substrates, labeled oligonucleotides were mixed with equimolar amounts of complementary nonlabeled oligonucleotides. The cleavage reaction was performed in a total volume of 20 μL comprising 1 × DSN buffer, 0.6 Kunitz units DSN, and 0.3 μM oligonucleotide substrate. DNase activity was evaluated by estimating the change in fluorescence intensity of the reaction mixture during incubation with DSN. Fluorescence intensity was measured on a spectrofluorometer Cary Eclypse (Varian).
RNase activity assay was measured essentially as described by Ho et al. (1998). The reaction mixture (20 μL) contained 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, and 0.6 Kunitz units of DSN. After warming the mixture at 55°C for 5 min, freshly prepared RNA (1.2% mass/vol, Baker's yeast, Sigma) was added and the incubation continued for 1 h. Following incubation, 0.1 volumes of ammonium acetate (7.5 M) and 3 volumes of 96% ethanol were added. The entire solution was mixed and centrifuged. The supernatant was diluted fivefold with water and absorbance at 260 nm was determined.
DSNP Assay
PCR
PCR reactions were performed with the Advantage 2 PCR Kit (Clontech). Each PCR reaction (25 μL) contained 1 × Advantage 2 Polymerize mix (Clontech), 1 × reaction buffer, 200 μM dNTPs, 0.3 μM each gene-specific primer, and 10 ng human total DNA. For model experiments, the following PCR conditions and gene-specific primers were employed:
p53 (C309T polymorphism): 30 PCR cycles (95°C for 7 sec; 65°C for 20 sec; 72°C for 20 sec) were performed with primers 5′-aaggggagcctcaccacg-3′ and 5′-ccacggatctgaagggt gaa-3′.
prothrombin (20210 G-to-A polymorphism): 30 cycles (95°C for 7 sec; 63°C for 20 sec; 72°C for 20 sec), primers 5′-atggttcccaataaaagtgac-3′ and 5′-aatagcactgggagcattga-3.
MTHFR (C677T polymorphism): 30 PCR cycles (95°C for 7 sec; 63°C for 20 sec; 72°C for 20 sec), primers 5′-cttgaaggagaaggtgtctg-3′ and 5′-aagaaaagctgcgtgatgatg-3′.
Fragments (69, 534, and 952 bp) comprising the 7028 C-T site of the COX1 gene were amplified using three pairs of primers at the following positions: Dir1 (6540–6559) and Rev1 (7492–7473); Dir2 (6792–6811) and Rev2 (7345–7326); Dir3 (6991–7010) and Rev3 (7060–7041) (primer positions are specified according to GenBank accession no. NC 001807). In each case, 26 PCR cycles were performed at 95°C for 7 sec, 63°C for 20 sec, and 72°C for 30 sec.
Plasmid Target Preparation
Plasmid target DNA samples for the DSNP assay were prepared using the pT-Adv vector (Clontech) with insertions of 69-bp COX1 PCR fragments. Plasmid DNA was prepared using the QiaPrep Spin mini-kit (QIAGEN), according to the manufacturer's instructions.
SNP Typing
A 5-μL aliquot of PCR products containing about 75 ng DNA (or 500 ng plasmid DNA in 50 mM Tris-HCl, pH 8.0) was mixed with 1.5 μL 10 × KF(exo-) buffer (Fermentas MBI), signal probes (to a final concentration of 0.3 μM), 0.5 Kunitz unit DSN, 2 U KF(exo-) (Fermentas MBI), and milliQ water (to a final volume of 15 μL), and incubated for 1 h at 35°C. Fluorescence intensity was measured on a spectrofluorometer Cary Eclypse (Varian). Plates were photographed using an Olympus SZX12 fluorescent stereomicroscope. The following signal probes were used in model experiments:
p53 (C309T polymorphism): p53WT (wild-type-specific), 5′-Fl-tgggcagtgc-DABCYL-3′ and p53M (mutant-specific) 5′- TAMRA-tggacagtgc-DABCYL-3′.
prothrombin (20210 G-to-A polymorphism): F2WT, 5′-Fl-gctcgctgag-DABCYL-3′ and F2M, 5′-TAMRA-gcttgctgag-DABCYL-3′.
MTHFR (C677T polymorphism): MTHFRWT, 5′-Fl-tcggctcccg-DABCYL-3′ and MTHFRM, 5′-TAMRA-tcgactcccg-DABCYL-3′.
COX1 gene: 7028 T variant-specific probe (Mit2T), 5′-Fl-gtgagctaca-DABCYL-3′.
WEB SITE REFERENCES
http://www.ncbi.nlm.nih.gov/BLAST; NCBI BLAST home page.
http://smart.embl-heidelberg.de; SMART—Simple Modular Architecture Research Tool.
Acknowledgments
We are grateful to Prof. N.K. Yankovsky (Vavilov Institute of General Genetics, Moscow, Russia), and Dr. A.V. Polyakov (Research Centre for Medical Genetics, Moscow, Russia) for helpful discussions and providing materials. We also thank Dr. G.N. Rudenskaya for considerable assistance in DSN purification. This work was supported by the Evrogen JSC and Russian Foundation for Support Domestic Science (grant to S.L.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL luk@ibch.ru; FAX 7 (095) 330-7056.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.547002.
REFERENCES
- Abarzua P, LoSardo JE, Gubler ML, Neri A. Microinjection of monoclonal antibody PAb421 into human SW480 colorectal carcinoma cells restores the transcription activation function to mutant p53. Cancer Res. 1995;55:3490–3494. [PubMed] [Google Scholar]
- Brookes AJ. The essence of SNPs. Gene. 1999;234:177–186. doi: 10.1016/s0378-1119(99)00219-x. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chou MY, Liao TH. Shrimp hepatopancreatic deoxyribonuclease—Purification and characterization as well as comparison with bovine pancreatic deoxyribonuclease. Biochim Biophys Acta. 1990;1036:95–100. doi: 10.1016/0304-4165(90)90019-s. [DOI] [PubMed] [Google Scholar]
- Friedhoff P, Meiss G, Kolmes B, Pieper U, Gimadutdinow O, Urbanke C, Pingoud A. Kinetic analysis of the cleavage of natural and synthetic substrates by the Serratia nuclease. Eur J Biochem. 1996;241:572–580. doi: 10.1111/j.1432-1033.1996.00572.x. [DOI] [PubMed] [Google Scholar]
- Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP, et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- Gite SU, Shankar V. Single-strand-specific nucleases. Crit Rev Microbiol. 1995;21:101–122. doi: 10.3109/10408419509113536. [DOI] [PubMed] [Google Scholar]
- Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robbins CM, et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- Harlow, E. and Lane, D. 1988. Immunoblotting. Antibodies: Laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Ho HC, Liao TH. Protein structure and gene cloning of Syncephalastrum racemosum nuclease. Biochem J. 1999;339:261–267. [PMC free article] [PubMed] [Google Scholar]
- Ho HC, Shiau PF, Liu FC, Chung JG, Chen LY. Purification, characterization and complete amino acid sequence of nuclease C1 from Cunninghamella echinulata var. echinulata Eur J Biochem. 1998;256:112–118. doi: 10.1046/j.1432-1327.1998.2560112.x. [DOI] [PubMed] [Google Scholar]
- Housman D, Ledley FD. Why pharmacogenomics? Why now? Nat Biotechnol. 1998;16:492–493. doi: 10.1038/nbt0698-492. [DOI] [PubMed] [Google Scholar]
- Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- Kruglyak L. The use of a genetic map of biallelic markers in linkage studies. Nat Genet. 1997;17:21–24. doi: 10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Kunitz M. Crystalline desoxyribonuclease. I. Isolation and general properties, spectrophotometric method for the measurement of desoxyribonuclease activity. II. Digestion of thymus nucleic acid (desoxyribonucleic acid). The kinetics of the reaction. J Gen Physiol. 1950;33:349–363. doi: 10.1085/jgp.33.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok PY. High-throughput genotyping assay approaches. Pharmacogenomics. 2000;1:95–100. doi: 10.1517/14622416.1.1.95. [DOI] [PubMed] [Google Scholar]
- Landegren U, Nilsson M, Kwok PY. Reading bits of genetic information: Methods for single-nucleotide polymorphism analysis. Genome Res. 1998;8:769–776. doi: 10.1101/gr.8.8.769. [DOI] [PubMed] [Google Scholar]
- Liao T-H. Bovine pancreatic deoxyribonuclease D. J Biol Chem. 1974;249:2354–2356. [PubMed] [Google Scholar]
- Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Hilfiker R. The use of single-nucleotide polymorphism maps in pharmacogenomics. Nat Biotechnol. 2000;18:505–508. doi: 10.1038/75360. [DOI] [PubMed] [Google Scholar]
- Meiss G, Franke I, Gimadutdinow O, Urbanke C, Pingoud A. Biochemical characterization of Anabaena sp. strain PCC 7120 non-specific nuclease NucA and its inhibitor NuiA. Eur J Biochem. 1998;251:924–934. doi: 10.1046/j.1432-1327.1998.2510924.x. [DOI] [PubMed] [Google Scholar]
- Menzorova NI, Rasskazov VA. Substrate specificity of Ca2+, Mg2+-dependent DNAase from sea urchin (Strongylocentrotus intermedius) embryos. Biokhimiia. 1981;46:872–880. [PubMed] [Google Scholar]
- Menzorova NI, Zinatulin RF, Favorov VV, Rasskazov VA. Isolation and characterization of Ca,Mg-dependent endonuclease from hepatopancreas of Paralithodes Camchatika. Biokhimia. 1993;58:681–691. [Google Scholar]
- Nielsena R. Estimation of population parameters and recombination rates from single nucleotide polymorphisms. Genetics. 2000;154:931–942. doi: 10.1093/genetics/154.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poort SR, Rosendaal FR, Reitsma PH, Bertina RM. A common genetic variation in the 3′ untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood. 1996;88:3698–3703. [PubMed] [Google Scholar]
- Reddy LG, Shankar V. Immobilized nucleases. Crit Rev Biotechnol. 1993;13:255–273. doi: 10.3109/07388559309041320. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Roses AD. Pharmacogenetics. Hum Mol Genet. 2001;10:2261–2267. doi: 10.1093/hmg/10.20.2261. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: A Web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singwi S, Joshi S. Potential nuclease-based strategies for HIV gene therapy. Front Biosci. 2000;5:D556–D579. doi: 10.2741/singwi. [DOI] [PubMed] [Google Scholar]
- Suck D. DNA recognition by structure-selective nucleases. Biopolymers. 1997;44:405–421. doi: 10.1002/(SICI)1097-0282(1997)44:4<405::AID-BIP5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Syvanen AC. Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat Rev Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- Wang WY, Liaw SH, Liao TH. Cloning and characterization of a novel nuclease from shrimp hepatopancreas, and prediction of its active site. Biochem J. 2000;346:799–804. [PMC free article] [PubMed] [Google Scholar]
- Williams RJ. Restriction endonucleases and their uses. Methods Mol Biol. 2001;160:409–429. doi: 10.1385/1-59259-233-3:409. [DOI] [PubMed] [Google Scholar]