Abstract
Variation in mtDNA has been used extensively to draw inferences in phylogenetics and population biology. In the majority of eukaryotes investigated, transmission of mtDNA is uniparental and clonal, with genotypic diversity arising from mutation alone. In other eukaryotes, the transmission of mtDNA is biparental or primarily uniparental with the possibility of “leakage” from the minority parent. In these cases, heteroplasmy carries the potential for recombination between mtDNAs of different descent. In fungi, such mtDNA recombination has long been documented but only in laboratory experiments and only under conditions in which heteroplasmy is ensured. Despite this experimental evidence, mtDNA recombination has not been to our knowledge documented in a natural population. Because evidence from natural populations is prerequisite to understanding the evolutionary impact of mtDNA recombination, we investigated the possibility of mtDNA recombination in an organism with the demonstrated potential for heteroplasmy in laboratory matings. Using nucleotide sequence data, we report here that the genotypic structure of mtDNA in a natural population of the basidiomycete fungus Armillaria gallica is inconsistent with purely clonal mtDNA evolution and is fully consistent with mtDNA recombination.
Keywords: Armillaria gallica, basidiomycetes, heteroplasmy, population
In populations of eukaryotic organisms, transmission of the nuclear genome between generations is often sexual, with genotypic diversity arising from mutation and recombination. In contrast, transmission of mtDNA in the majority of eukaryotes is clonal (1), with genotypic diversity arising from mutation alone. However, in considering mitochondrial genome evolution (1), there are two reasons why strict clonality need not be the rule. First, the molecular machinery required for mtDNA recombination is widespread. Homologous DNA recombination activity has been detected in human mitochondria (2), and intermolecular mtDNA recombination has been documented in plants (3) and animals (4); this recombination, however, is between mtDNA sequences sharing the same history of descent. In contrast, recombination between mtDNAs of different descent is documented amply in fungi (5–17) but only in laboratory experiments and only under conditions in which heteroplasmy is ensured. Second, although mtDNA is most often transmitted to the zygote from only one parent, there are many cases in which mtDNA is transmitted from both parents, albeit in varying proportions (1). Despite the widespread occurrence of heteroplasmy and the presence of molecular machinery necessary for recombination, we know of no documented example of genetic exchange and recombination among mtDNAs in a population of any organism in nature. We therefore investigated the possibility of mtDNA recombination in a natural population of Armillaria gallica, a basidiomycete fungus in which heteroplasmy and mtDNA recombination have been demonstrated in laboratory matings (18).
The life history of A. gallica provides two advantages in addressing the question of mtDNA recombination. One advantage is that the stability of any genetic marker can be assessed by collecting isolates, within an individual, that represent separation by a large number of mitotic divisions. Genetic individuals of this fungus are both large and old (19). Each individual arises in a unique mating event and then grows vegetatively by the extension of specialized aggregates of hyphae called “rhizomorphs” throughout the humus layer of forest soils. One such individual covers at least 15 hectares and is estimated to be several centuries old (19); more recent data indicate that the size and age of this individual are not unique in this species (18).
The other advantage is that the population structure and mating mechanism of A. gallica provide opportunities for mtDNA genetic exchange. The genotypic structure of the population from which A. gallica individuals are drawn is consistent with random mating (18). This type of population structure offers the best chance of detecting genetic exchange in mtDNA because the majority of matings bring together mtDNAs of different descent. In laboratory matings of Armillaria and other basidiomycete fungi, fusion of undifferentiated, haploid, vegetative hyphae is followed by reciprocal exchange and migration of fertilizing nuclei throughout the existing mycelium of the opposing mate. While nuclei migrate, mitochondria remain stationary, resulting in a uniformly dikaryotic or diploid colony that is mosaic for mtDNA type (17, 20–23). Heteroplasmy and mtDNA recombination have been observed in the region where the hyphae of different mates fuse (12, 17, 18). In the field, however, only one mtDNA is observed in any given genetic individual of Armillaria (18, 21, 24). All but one mtDNA type is lost due to stochastic processes associated with the turnover of mycelium or mtDNA molecules or to differential fitness effects at the level of mycelia or mtDNA molecules. The mtDNA that is fixed within the newly formed individual may therefore be either parental or recombinant. In natural populations, both clonality and recombination are expected to occur in the mitochondrial genome.
Previous population genetic data based on restriction fragment length polymorphisms are consistent with mtDNA recombination in a natural population of A. gallica (18), and similar data exist for other fungal populations (5, 25–27). Interpreting these data as evidence of mtDNA recombination is, however, provisional because of the possibility of multiple, independent origins of any allelic state defined by restriction fragment length polymorphisms. This study therefore assayed the variation in nucleotide sequences present in all mtDNA haplotypes within the population sample. Because of the low level of nucleotide diversity observed, each nucleotide at a variable site was attributed to a common origin. Genotypic data based on nucleotide sequences of A. gallica were inconsistent with a strictly clonal pattern of mtDNA evolution and were fully consistent with mtDNA recombination.
MATERIALS AND METHODS
The population sample of 121 genetic individuals collected from four locations in eastern North America is the same as that used earlier to investigate the A. gallica population structure (18). That study shows that genotype frequencies at seven nuclear loci are consistent with Hardy–Weinberg expectations and that allele frequencies are not significantly different between regions. Therefore, this sample was treated as representing a single population.
DNA isolation, purification, and cloning were as described (18). DNA sequences of cloned fragments and PCR products were obtained by cycle sequencing (Bethesda Research Laboratory). Oligonucleotide hybridizations were carried out on PCR products subjected to agarose gel electrophoresis and transferred to nylon membranes (Genescreen Plus, Dupont/NEN). Prehybridizations were performed in 6× SSC, 0.5% SDS, and 10× Denhardt’s solution, and hybridizations were performed in 6× SSC and 0.5% SDS. Oligonucleotides were end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (28). Hybridizations were carried out at 2°C below the estimated Tm (29). Washes were performed in a stepwise fashion in 6× SSC and 0.5% SDS at temperatures starting at the hybridization temperature and increasing by 1 or 2°C with the final wash temperature as high as 5°C over the estimated Tm depending on the oligonucleotide used.
For assay of single-strand conformational polymorphisms and single-nucleotide length variation, PCR products were synthesized by using primers end-labeled with [γ-33P]ATP and T4 polynucleotide kinase (28). Products were subjected to electrophoresis on 390 × 300 × 0.4 mm of 6% nondenaturing polyacrylamide gels (40 acrylamide:0.8 bisacrylamide) at a constant 200 V for 22–24 h and on 390 × 300 × 0.4 mm of 6% denaturing polyacrylamide gels (i.e., sequencing gels) at a constant power of 55 W for 3–4 h, respectively. Gels were dried and then were visualized by autoradiography.
The multilocus association test was done with the popgen 0.1 program provided by A. Burt (Imperial College, U.K.). Phylogenetic analyses were done by using the test version 4.0dxx of paup with permission from David L. Swofford (Smithsonian Institution, Washington, D.C.).
RESULTS
A restriction map was used to select cloned mtDNA fragments that represent well separated regions of the A. gallica mitochondrial genome (18). These clones were sequenced, and the DNA sequences (11.4 kb total) were used to design primers for amplification of the corresponding regions from 10 different genetic individuals. This subset of individuals was selected, based on restriction fragment length polymorphism data, to represent the range of mtDNA variation present in the population sample.
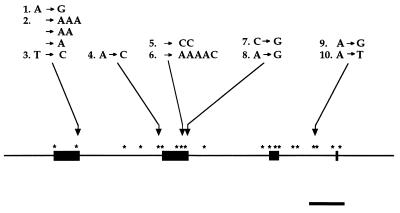
Fig. 1 summarizes the variation found by sequencing 6.8 kb of mtDNA from each of 10 genetic individuals in the subsample. In these sequences, only 10 variable positions were identified (Table 1). At each position, the nucleotide variation detected likely represented a neutral change. blastn and blastx comparisons to entries in GenBank (30) revealed the following. The B466 amplicon (sites 1, 2, and 3) shows similarity to NADH dehydrogenase chain 1; sites 1 and 2 are upstream of the initiation codon, and site 3 is a synonymous substitution in the third position of an Arg codon within the reading frame. The R247 amplicon (site 4) contains a portion of a pseudogene with sequence similarity to DNA-directed RNA polymerases; the variable site is outside the region of similarity. The R213 amplicon (sites 5 and 6), B376 amplicon (sites 7 and 8), and B638 amplicon (sites 9 and 10) show no significant sequence similarity to any entry in GenBank.
Figure 1.
Linearized map of the circular mtDNA genome of A. gallica strain 442–4. Thick lines represent hybridization probes used by Saville et al.(18). Asterisks represent sequenced regions in strain 442–4 totaling 11.4 kb. The numbers represent the 10 variable sites in the 6.8 kb of the sequence from each of 10 individuals; changes are relative to the same strand of DNA in strain 442–4. The locations of sites within amplicons are listed in Table 1.
Table 1.
Amplified regions of A. gallica mtDNA
| Amplicon | Primers | Size, bp | Site | Position |
|---|---|---|---|---|
| B466 | GCAAGGAATAACAGCCC | 472 | 1 | 376 |
| (AF035388) | AAGCCTTCTCAAGCAAGCCTGG | 2 | 373 | |
| 3 | 295 | |||
| R247 | CTCCCCTACTTCTTCAAAGG | 471 | 4 | 63 |
| (AF035389) | CGTTAATTACCTCTCCTTCAGC | |||
| R213 | GGTTTAACGAAGGTACCACTGGC | 344 | 5 | 192–193 |
| (AF035390) | CCAAGGTAGTTAACGGGCCTAGC | 6 | 242–246 | |
| B376 | AAGTGTTGGATTGACTCGCACC | 700 | 7 | 357 |
| (AF035391) | GCCTACGTACTGGATGTAGGTAGTGC | 8 | 97 | |
| B638 | GGATCCTCATGGTTAAGCATTAC | 244 | 9 | 148 |
| (AF035392) | CTAATATTGAGACTAATGTATCCGCTG | 10 | 146 |
The GenBank accession numbers appear in parentheses under the amplicon name. The orientation of sites relative to one another is shown in Fig. 1.
The entire sample of 121 genetic individuals was assayed for the variable sites shown in Fig. 1, as well as for the presence of further variation, by hybridization with allele-specific oligonucleotides or by direct DNA sequencing. The haplotype assignments are listed in Table 2. No further variation was detected in the entire sample relative to the subsample. For all sites with simple substitutions, only two of the four possible nucleotides were present. For site 2 with multiple alleles, only varying numbers of adenine residues were present. This low level of variation in nucleotide sequences implies that parallel change was highly unlikely, especially for the nine sites with only two alleles each.
Table 2.
mtDNA haplotypes by abundance
| Sites
|
Individuals, n | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| A | ––– | T | A | –– | ––––– | C | A | A | A | 48*†‡ |
| A | A–– | T | A | –– | ––––– | C | A | A | A | 24*‡ |
| A | ––– | T | A | –– | ––––– | C | G | A | A | 11*‡ |
| A | ––– | T | A | –– | ––––– | G | A | G | A | 7* |
| A | ––– | T | A | –– | ––––– | G | A | A | A | 7*‡ |
| A | ––– | T | C | –– | ––––– | G | A | G | A | 3* |
| A | ––– | T | A | § | ––––– | G | A | A | A | 2 |
| A | AAA | C | A | –– | ––––– | C | A | A | A | 2†‡ |
| A | ––– | T | A | CC | ––––– | C | A | A | A | 2†‡ |
| A | A–– | T | A | –– | AAAAC | G | A | A | A | 1* |
| A | ––– | T | A | –– | ––––– | C | A | G | A | 1* |
| G | ––– | T | A | –– | AAAAC | C | A | A | A | 1*‡ |
| A | –––¶ | T | A | –– | ––––– | C | A | A | A | 1 |
| A | ––– | T | C | –– | ––––– | C | A | A | A | 1* |
| G | ––– | T | A | –– | ––––– | C | A | A | A | 1* |
| A | A–– | T | A | –– | ––––– | G | A | A | A | 1* |
| A | ––– | T | A | –– | ––––– | G | G | G | A | 1* |
| –‖ | –––‖ | T | A | –– | ––––– | G | A | A | A | 1 |
| A | ––– | T | A | –– | ––––– | C | A | A | T | 1 |
| A | A–– | T | C | –– | ––––– | G | A | A | A | 1*‡ |
| A | A–– | T | A | –– | ––––– | G | A | G | A | 1* |
| A | A–– | T | A | –– | ––––– | C | G | A | A | 1* |
| A | AA– | T | A | –– | ––––– | G | A | A | A | 1 |
| A | ––– | T | A | –– | ––––– | A | A | 1 | ||
Numbered sites are the same as in Fig. 1. The two most frequent alleles at each site are designated 1 and 2, respectively, below and in Table 2. Sites 2, 5, and 6 represent insertion/deletions in which the absence of a base is indicated by a dash. Blanks for Sites 7 and 8 in the last haplotype indicate missing data; genotypic data from this line were included in the pairwise comparisons of loci (Table 3) but were omitted from the multilocus association test (see text).
Designates the 16 haplotypes used in the phylogenetic analysis (see text).
Individuals from which multiple isolates were assayed in each of the 10 variable sites belonged to these haplotype classes.
The 10 individuals fully sequenced over 6.8 kb are characterized by these eight haplotypes.
Large insertion of undetermined sequence.
 |
 |
 |
∥
Note that this deletion encompasses sites 1 and 2.
The long life and large size of individuals within this population provided an opportunity to investigate the stability of genetic markers. We checked the stability of the 10 mtDNA sites shown in Fig. 1 and Table 2 within three genetic individuals covering large territories (18). From these individuals, 33, 17, and 33 isolates, respectively, were assayed by hybridization with all of the allele-specific oligonucleotides, by analysis of single-strand conformation polymorphisms, by determination of sequence length with single-base resolution, and by sequencing two separate PCR products for a total of 700 bases in all isolates. For each individual, this sample of isolates represented >1 Km of linear growth or >107 cell generations by earlier estimates (18). No variation was detected in any of the five amplified regions within an individual, suggesting a low rate of nucleotide change.
In five of the 10 variable sites, the frequencies of the two most common alleles were >0.04; the distributions of these alleles among genotypes were sufficient to address the question of recombination in Table 3. The other sites harbored one frequent allele and one or more rare alleles that occurred at a frequency of <0.04 and were not useful in addressing the question of recombination in the present sample because of the small expected numbers of certain genotypes. For similar reasons, the rare alleles at site 2 also are excluded from the comparisons in Table 3. Overall, there was little evidence of disequilibrium between loci. Observed genotype frequencies in seven of the 10 pairwise tests among the five sites were not significantly different from random expectation (Table 3). These P values, however, must be interpreted with caution because the multiple comparisons are nonindependent; each locus is used in more than one pairwise comparison, and a recombination in a given genetic interval may affect simultaneously more than one comparison. With the Bonferroni correction for multiple nonindependent tests (divide the standard P value for significance of 0.05 by 10), however, only two pairwise comparisons (sites 4 and 7 and sites 7 and 9) showed genotypic frequencies that were significantly different from random expectation.
Table 3.
Genotype counts for pairs of five sites
| Pairs of sites | Observed (expected)
genotypes
|
Fisher’s exact probability | |||
|---|---|---|---|---|---|
| 11 | 12 | 21 | 22 | ||
| 2 and 4 | 83 (83.3) | 4 (3.8) | 28 (27.8) | 1 (1.3) | 0.402 |
| 2 and 7 | 66 (68.1) | 20 (18.0) | 25 (23.0) | 4 (6.1) | 0.125 |
| 2 and 8 | 74 (76.3) | 12 (9.7) | 28 (25.7) | 1 (3.3) | 0.091 |
| 2 and 9 | 75 (77.2) | 12 (9.7) | 28 (25.8) | 1 (3.3) | 0.093 |
| 4 and 7 | 93 (89.3) | 22 (25.7) | 1 (4.7) | 5 (1.3) | 0.002* |
| 4 and 8 | 102 (102.5) | 13 (12.5) | 5 (4.5) | 0 (0.5) | 0.570 |
| 4 and 9 | 106 (103.5) | 10 (12.5) | 2 (4.5) | 3 (0.5) | 0.008* |
| 7 and 8 | 82 (83.0) | 12 (10.2) | 25 (23.2) | 1 (2.8) | 0.142 |
| 7 and 9 | 93 (83.8) | 1 (10.8) | 14 (23.2) | 12 (2.8) | 0.000* |
| 8 and 9 | 95 (95.4) | 12 (11.6) | 12 (11.6) | 1 (1.4) | 0.368 |
Sites are numbered as in Fig. 1 and Table 1. The numbers in parentheses are those expected under the null hypothesis of no association among the alleles at different loci. For genotypic classes, “1” refers to the most common allele and “2” refers to the second most frequent allele at a site; rare alleles were excluded from these comparisons (i.e., there is no lumping of allelic classes).
Significant at P < 0.05, with no correction made for multiple, nonindependent tests.
A multilocus disequilibrium test (31, 32) was carried out among all 10 sites in 120 individuals. The index of association (IA = 0.150) falls within the range of IA values found in 10,000 randomly permuted data sets (range, −0.262 to 0.500) but is marginally significant (P = 0.043). One source of this association is the minority allele of site 4, which occurs in all three representatives of the sixth haplotype shown in Table 2. When the sixth haplotype was counted only once for a total of 118 genotypes, the IA (0.103) was not significantly different from random expectation (P = 0.113). All other analyses counting each genotype only once, or excluding some of the sites, also gave IA values not significantly different from random expectation (data not shown).
In the absence of recombination, and with the low level of nucleotide sequence variation observed here, it should be possible to find an unrooted tree of mitochondrial haplotypes of high internal consistency, but this was not the case. When the sites were considered as “characters” and the haplotypes were considered as “taxa,” all unrooted trees had high levels of homoplasy. The reason for this is evident as shown in Table 3; for nine of the 10 comparisons of the sites, all four possible genotypes of the two most frequent alleles are present in the sample. In addition, all four possible genotypes are present in the comparison of sites 1 and 6 and sites 6 and 7 (Table 2).
For the phylogenetic analysis, we considered the 16 haplotypes defined by the two most common alleles at sites 1, 2, 4, 6, 7, 8, and 9 (Table 2). Sites 3, 5, and 10 were excluded because they were not phylogenetically informative; each of these sites had only two alleles, with the minority allele present in only one haplotype. For the same reason, haplotypes distinguished only by rare alleles at sites 1 and 2 also were excluded. Among the 16 haplotypes, all variation was appropriate for testing whether or not recombination had occurred. In this analysis, an exact search method (branch and bound) revealed 462 most parsimonious trees, each of 15 steps in total length and each with a consistency index of 0.47. The strict consensus of these trees had no phylogenetic resolution, except for a branch defined by the A-G transition at site 1, which separated two haplotypes from the remaining 14. Each tree of 15 steps assumed no recombination. In contrast, if recombination was allowed, the number of necessary mutational steps to explain the 16 haplotypes was seven. Furthermore, the test of Archie (33), used as described by Burt et al. (34), showed that the most parsimonious tree for the 16 haplotypes was no better than random. The observed minimum tree length of 15 steps was not significantly different from the minimum lengths of trees found in 1,000 randomly permuted data sets (mean = 14.3 steps, P = 0.85); this result is exactly what is expected with genetic exchange and general recombination.
The phylogenetic analysis above included site 2, which had multiple alleles consisting of varying numbers of adenine residues. Because the mechanism of mutation in site 2 may be different from that in sites with simple base substitutions, we repeated the phylogenetic analysis as above but without site 2. Among the 12 haplotypes remaining after site 2 was excluded, there were 104 most parsimonious trees, each of 11 steps. As in the analysis above, all trees were of low consistency (consistency index = 0.545) and their strict consensus had no resolution except for the same internal branch identified above. With recombination, only six steps were required. Therefore, even without site 2, the best trees still had extensive homoplasy, which is expected with recombination and not expected with strict clonality.
DISCUSSION
There are two hypotheses for the observed distribution of mitochondrial genotypes in this A. gallica population sample. The first is that mtDNA evolves strictly clonally. This hypothesis requires that, starting from any ancestral genotype, at least one of the derived allelic states must arise more than once to produce all four genotypes observed in each of 11 pairwise comparisons of loci (Tables 2 and 3); this requirement for parallel mutation clearly is evident in the phylogenetic analysis in which many most parsimonious trees were identified, each with extensive homoplasy distributed over all sites in the analysis. With the low overall level of nucleotide sequence variation, however, it is difficult to imagine how any of the variable sites could have changed more than once in the time since the most recent common ancestor of all sequences assayed here. Because the variable sites used to address the question of recombination were well distributed in the mtDNA with no two sites known to reside within the same gene, parallel change within all of these sites is not plausible; we know of no mutational bias strong enough to produce the observed genotypes in the absence of recombination. Furthermore, for all loci with simple substitutions, only two of the four possible nucleotides were present. If these sites used here were highly mutable, then we might expect to see more than two bases present at each variable site in the population. Lastly, no variation was detected in isolates separated by a large number of mitotic cell divisions in each of three individuals. Under these conditions, we can infer that each allelic state, defined by the nucleotide(s) present or absent at a site in the mtDNA (Table 2), arose only once in the population and therefore reflects identity by descent at that site. For these reasons, the observed distribution of haplotypes is difficult to reconcile with strict clonality.
The second hypothesis is that mtDNA evolves with recombination in populations of A. gallica. Under this hypothesis, starting from any ancestral genotype, each derived allelic state need arise only once by mutation; recombination then produces new combinations of existing alleles. This interpretation is fully consistent with the low overall level of nucleotide variation detected and with the closeness of observed numbers of genotypes to those expected under the assumption of random association (Table 3). Furthermore, with recombination, eight fewer mutational steps are required to explain the existing haplotypes than are required in the absence of recombination and no parallel mutation is required. Finally, there is direct observation of mtDNA recombination in A. gallica and other basidiomycetes in laboratory crosses (12, 17, 18). Considering all of the evidence, we conclude that recombination is the more parsimonious explanation for observed distribution of haplotypes than mutation alone.
The conclusion that recombination has occurred, however, does not imply that the sites are in perfect equilibrium. A surprisingly small number of recombination events can give the appearance of random mating (33); estimates of recombination rate are therefore not possible here. The evidence for disequilibrium in our data is scant and could be explained by differences in the rates of clonal transmission of intact mtDNA genotypes, genetic hitchhiking of one or more sites on nearby genes under selection, or the element of chance associated with sampling. None of these possibilities, however, conflicts with our overall interpretation that mtDNA recombination has occurred.
We speculate that genetic exchange and recombination among the mtDNAs of natural populations has not been detected previously for two reasons. First, wherever mtDNA transmission is biased strongly from one parent, the recombination frequencies are expected to be low, possibly below the limits of detection given the sample sizes used in most investigations. Second, in species in which mtDNA recombination has been established in laboratory experiments, no corresponding investigations of natural populations have been carried out. In contrast, for A. gallica, mtDNA transmission is biparental, mtDNA recombination occurs at detectable frequencies in the laboratory, and mtDNA recombination now has been investigated in a natural population. We predict that the level of mtDNA recombination detected in this population of A. gallica will be found in any population for which similar conditions exist. Furthermore, even with the low level of mitochondrial transmission from the minority parent detected in species such as mice (35) and Drosophila (36), some level of mtDNA recombination is expected.
mtDNA recombination in natural populations has two important evolutionary implications. First, in those organisms in which mtDNA recombination proves more frequent than mutation, phylogenetic inference among mtDNA haplotypes is confounded because different portions of the molecule have different histories of descent. In animals, a low level of mitochondrial transmission from the minority parent occurs more readily when that parent is a different but closely related species; therefore, the potential for heteroplasmy and mtDNA exchange may be highest at hybrid zones, those geographic locations where closely related species or long-separated subpopulations come into contact. Indeed, recombination might explain some of the discrepancies in human mtDNA phylogeny that have been attributed to introgression (37, 38). This is particularly relevant in light of the detection of homologous recombination activity in human mitochondria (2).
In addition to complicating phylogenetic inference, mtDNA recombination has implications affecting fitness and long term survival. Asexual populations of finite size are subject to a process known as Muller’s ratchet in which deleterious mutations inexorably accumulate as the genotypes with the smallest numbers of mutations are lost to drift. Under some conditions, this process ultimately leads to extinction, or “mutational meltdown” (39). Does Muller’s ratchet apply to mitochondrial genomes? In animals in which mtDNA recombination is absent, or very rare, there is molecular evidence for the accumulation of deleterious mutations (40). In those organisms with more frequent genetic exchange in mitochondria, the advance of Muller’s ratchet may be stopped as genotypes with fewer deleterious mutations are regenerated by recombination.
Acknowledgments
We thank M. G. Milgroom for discussion, A. Burt for the mutlilocus association test software, and D. Swofford for the test version 4.0dxx of paup. This study was supported by a research grant from the Natural Sciences and Engineering Research Council to J.B.A.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF03588–AF03592).
References
- 1.Birky C W. Proc Natl Acad Sci USA. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thyagarajan B, Padua R A, Campbell C. J Biol Chem. 1996;271:27536–27543. doi: 10.1074/jbc.271.44.27536. [DOI] [PubMed] [Google Scholar]
- 3.Lonsdale D M, Brears T, Hodge T P, Melville S E, Rottmann W H. Philos Trans R Soc London, Ser B. 1988;319:149–164. [Google Scholar]
- 4.Lunt D H, Hyman B C. Nature (London) 1997;387:247. doi: 10.1038/387247a0. (lett.). [DOI] [PubMed] [Google Scholar]
- 5.Barroso G, Blesa S, Labarere-J Appl Environ Microbiol. 1994;61:1187–1193. doi: 10.1128/aem.61.4.1187-1193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belcour L, Begel O. Mol Gen Genet. 1977;153:11–21. doi: 10.1007/BF01035991. [DOI] [PubMed] [Google Scholar]
- 7.Brunner A, DeCobos A T, Griffiths D E. Mol Gen Genet. 1977;152:183–191. doi: 10.1007/BF00268816. [DOI] [PubMed] [Google Scholar]
- 8.Chung K-R, Leuchtmann A, Schardl C L. Genetics. 1996;142:259–265. doi: 10.1093/genetics/142.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins R A, Saville B J. Nature (London) 1990;345:177–179. doi: 10.1038/345177a0. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda M, Harada Y, Imahori S, Fukumasa-Nakai Y, Hyashi Y. Curr Genet. 1995;27:550–554. doi: 10.1007/BF00314446. [DOI] [PubMed] [Google Scholar]
- 11.Mannella C A, Lambowitz A. Biochem Biophys Res Commun. 1978;80:673–679. doi: 10.1016/0006-291x(78)91621-2. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto T, Fukumasa-Nakai Y. Curr Genet. 1996;30:549–552. doi: 10.1007/s002940050168. [DOI] [PubMed] [Google Scholar]
- 13.Rowlands R T, Turner G. Mol Gen Genet. 1975;141:69–79. doi: 10.1007/BF00332379. [DOI] [PubMed] [Google Scholar]
- 14.Seitz-Mayr G, Wolf K, Kaudewitz F. Mol Gen Genet. 1978;164:309–320. doi: 10.1007/BF00333162. [DOI] [PubMed] [Google Scholar]
- 15.Wilkie D, Thomas D Y. Genetics. 1973;73:367–377. doi: 10.1093/genetics/73.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf K, Dujon B, Slonimski P P. Mol Gen Genet. 1973;125:53–90. doi: 10.1007/BF00292983. [DOI] [PubMed] [Google Scholar]
- 17.Baptista-Ferreira J L C, Economou A, Casselton L A. Curr Genet. 1983;7:405–407. doi: 10.1007/BF00445883. [DOI] [PubMed] [Google Scholar]
- 18.Saville B J, Yoell H, Anderson J B. Mol Ecol. 1996;4:485–497. doi: 10.1111/j.1365-294x.1996.tb00341.x. [DOI] [PubMed] [Google Scholar]
- 19.Smith M L, Bruhn J N, Anderson J B. Nature (London) 1992;356:428–431. [Google Scholar]
- 20.Specht C A, Novotny C P, Ullrich R C. Curr Genet. 1992;22:129–134. doi: 10.1007/BF00351472. [DOI] [PubMed] [Google Scholar]
- 21.Smith M L, Duchesne L C, Bruhn J N, Anderson J B. Genetics. 1990;126:575–582. doi: 10.1093/genetics/126.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May G, Taylor J W. Genetics. 1988;118:213–220. doi: 10.1093/genetics/118.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hintz W E A, Anderson J B, Horgen P A. Genetics. 1988;119:35–42. doi: 10.1093/genetics/119.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith M L, Bruhn J N, Anderson J B. Phytopathology. 1994;84:822–829. [Google Scholar]
- 25.Bates M R, Buck K W, Brasier C M. Mycol Res. 1993;97:1093–1100. [Google Scholar]
- 26.Smith M L, Anderson J B. Curr Genet. 1994;25:545–553. doi: 10.1007/BF00351676. [DOI] [PubMed] [Google Scholar]
- 27.Taylor J W, Smolich B D, May G. Evolution. 1986;40:716–739. doi: 10.1111/j.1558-5646.1986.tb00533.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Coning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Itakura K, Rossi J J, Wallace R B. Annu Rev Biochem. 1984;53:323–356. doi: 10.1146/annurev.bi.53.070184.001543. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Brown A H D, Feldman M W, Nevo E. Genetics. 1980;96:523–536. doi: 10.1093/genetics/96.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Archie J W. Syst Zool. 1989;38:239–252. [Google Scholar]
- 34.Burt A, Carter D A, Koenig G L, White T J, Taylor J W. Proc Natl Acad Sci USA. 1996;93:770–773. doi: 10.1073/pnas.93.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gyllensten U, Wharton D, Josefsson A, Wilson A C. Nature (London) 1991;352:255–257. doi: 10.1038/352255a0. [DOI] [PubMed] [Google Scholar]
- 36.Kondo R, Satta Y, Matsuura E T, Ishiwa H, Takahata N, Chigusa S I. Genetics. 1990;126:657–663. doi: 10.1093/genetics/126.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Templeton A. Am Anthropol. 1993;95:51–72. [Google Scholar]
- 38.Treisman M. J Theor Biol. 1995;173:23–29. doi: 10.1006/jtbi.1995.0039. [DOI] [PubMed] [Google Scholar]
- 39.Lynch M, Burger R, Butcher D, Gabriel W. J Hered. 1993;84:339–344. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- 40.Lynch M. Mol Biol Evol. 1996;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]