Abstract
During neurogenesis of the mammalian neocortex, neural progenitor cells divide to generate daughter cells that either become neurons or remain as progenitor cells. The mouse numb (m-numb) gene encodes a membrane-associated protein that is asymmetrically localized to the apical cell membrane of dividing cortical progenitor cells and may be segregated to only the apical daughter cell that has been suggested to remain as a progenitor cell. To examine m-numb function during neural development, we generated a loss-of-function mutant allele of m-numb. Mice homozygous for this mutation exhibit severe defects in cranial neural tube closure and precocious neuron production in the forebrain and die around embryonic day 11.5 (E11.5). These findings suggest that m-numb is an essential gene that plays a role in promoting progenitor cell fate during cortical neurogenesis.
The mammalian neocortex contains a wide variety of neurons that are organized into six distinct layers roughly parallel to the cortical surface. During a restricted period of neurogenesis, which in mice is between embryonic day 11 and 17 (E11 and E17), these different neurons are generated by progenitor cells that occupy the ventricular zone (inner layer) of the developing cerebral wall (telencephalic neuroepithelium). Cortical neurons are generated at precise times of development in an “inside–out” fashion. Neurons that occupy deeper layers of the mature neocortex are generated first, followed by those in more superficial layers (1–3). Little is known about the molecular mechanism that allows distinct classes of neurons to be generated at precise times and in correct numbers from a morphologically indistinguishable population of progenitor cells during development.
It has been suggested that asymmetric division by cortical progenitor cells, in which a neuron and a daughter progenitor cell are generated, is at least partly responsible for cortical neurogenesis. Early evidence came from lineage-tracing experiments, in which clonally related neurons were shown to occupy multiple cortical layers that are generated at different times of development (4–13). More direct evidence came from time-lapse imaging of 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI)-labeled ventricular zone cells in living ferret brain slices, which further suggests that the orientation of cell cleavage planes predicts whether a cell divides symmetrically or asymmetrically (14). It has been postulated that cortical progenitor cells with vertical cleavage planes (perpendicular to the ventricular surface) divide symmetrically to produce two neural progenitor cells, whereas those with horizontal cleavage planes (parallel to the ventricular surface) divide asymmetrically to produce two different daughter cells, a basal daughter cell that becomes a neuron and an apical daughter that remains as a progenitor cell (14, 15). What remains largely unknown are the precise roles that asymmetric division plays and its molecular mechanism.
During Drosophila neurogenesis, asymmetric division by neural precursor cells is responsible for generating sensory organs in the peripheral nervous system and neuronal diversity in the central nervous system. In dividing neural precursor cells, the numb (d-numb) gene product, a membrane-associated protein, becomes localized to only one pole of the cell membrane and, as a result, is segregated primarily to one daughter cell. This segregation of d-Numb during cell division is necessary for the daughter cells to adopt distinct fates (16–18). We and others have isolated a highly conserved mouse homologue of d-numb, m-numb, which is expressed by cortical progenitor cells (19, 20). When a cortical progenitor cell enters mitosis at the ventricular surface, the m-Numb protein becomes asymmetrically localized to the apical cell membrane and, after a horizontal division, is likely to be segregated to the apical daughter cell (19) that has been suggested to remain as a progenitor cell (14). We have postulated that this asymmetric segregation of the m-Numb protein allows the apical daughter cell to adopt the progenitor fate (19, 21). When expressed in Drosophila, the m-Numb protein, like its fly counterpart, localizes asymmetrically in dividing neural precursor cells, thereby restoring the ability of their daughter cells to adopt distinct fates in numb loss-of-function mutant embryos (19, 20, 22).
To analyze in vivo functions of m-numb during cortical neurogenesis, we generated a loss-of-function allele of m-numb. We report here the characterization of this mutant allele. We show that mice homozygous for this mutation exhibit severe defects in cranial neural tube closure and precocious neuron production in the forebrain, and die around E11.5. These findings demonstrate that m-numb is an essential gene for embryonic development and are consistent with the notion that m-numb plays a role in promoting progenitor cell fate during cortical neurogenesis.
Materials and Methods
Construction of Targeting Vector.
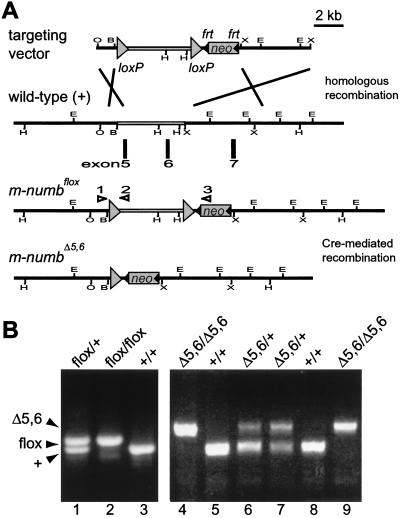
Over 50 kb of the m-numb locus, represented in six clones, was isolated from a 129/Sv genomic library (a gift from D. Littman, New York University Medical School) and characterized using a combination of restriction enzyme digestion and Southern blotting (data not shown). Exons 5 and 6 were mapped to a 6-kb EcoRI–XbaI fragment at the 5′ end of one genomic clone, with the EcoRI site (next to an endogenous BamHI site) that came from the vector. To construct the targeting vector, two linkers containing the loxP site (23) were synthesized and used to flox the 6-kb fragment. The floxed 6-kb fragment was then cloned into a vector containing a 1.5-kb XhoI–BamHI m-numb genomic fragment, which is immediately 5′ to the 6-kb EcoRI–XbaI fragment in the mouse genome. The resultant 7.5-kb floxed m-numb fragment was then cloned into a vector containing a flrted pgk-neo cassette (24), a 6.2-kb XbaI fragment, which is immediately 3′ to the floxed m-numb fragment in the mouse genome, and a pgk-thymidine kinase (tk) cassette.
Generation of Mutant Animals.
Recombinant embryonic stem (ES) clones and m-numbflox heterozygous mice were generated as previously described (25). m-numbΔ5,6 heterozygous mice were generated by mating m-numbflox mice with β-actin-Cre transgenic mice (a gift from M. Lewandoski and G. Martin, Univ. of California, San Francisco). Genotyping of ES cells and mutant mice were carried out using Southern blotting and PCR (primer 1, TCAGCAGTTTCTGAGTTCAGTTCCC; primer 2, TAAAAACGCAGTCGAGAAAC; primer 3, ACGAGTTCTTCTGAGGGGATCGGC). As shown in Fig. 1, primers 1 and 2 generate a 0.4-kb fragment that represents either wild-type or floxed m-numb allele (the latter is 45 bp longer), whereas primers 1 and 3 produce a 0.6-kb fragment indicative of the m-numbΔ5,6 allele.
Figure 1.
Targeted disruption of the m-numb gene. (A) Schematic drawings of the targeting vector, the wild-type, the floxed, and the mutant m-numb alleles. Exons 5 and 6 encode amino acids 68–139 of the m-Numb protein. All EcoRI (E) and HindIII (H) sites are shown, whereas only the relevant BamHI (B), XhoI (O), and XbaI (X) sites are shown. Open triangles represent PCR primers as described in Materials and Methods. (B) PCR analysis of the wild-type (+), floxed (flox), and mutant (Δ5,6) m-numb alleles.
Immunoblot, in Situ Hybridization, and Immunostaining.
Immunoblot, in situ hybridization, and immunostaining were performed on paraffin or O.C.T. sections using methods as previously described (19, 21). Primary antibodies were: anti-Neurofilament (150 kDa; Chemicon), rabbit polyclonal; anti-MAP2 (Boehringer Mannheim), mouse monoclonal; and anti-Nestin (Developmental Studies Hybridoma Bank), mouse monoclonal. For BrdUrd-labeling experiments, BrdUrd (Sigma) was injected i.p. into pregnant mice (100 μg/g of body weight) 2 h before sacrifice. Embryos were fixed in 4% paraformaldehyde for 3 h and embedded in OCT Sections (8 μm) were treated with 2 M HCl for 30 min at room temperature and rinsed several times with PBS before adding mouse anti-BrdUrd (Chemicon). Probes for in situ hybridization were: Pax6, 260-bp EcoRI–NheI cDNA fragment; BF-1, 1.4-kb NotI–KpnI cDNA fragment; Otx2, 260-bp HaeIII cDNA fragment; and Fgf17, 540-bp NcoI–SpeI fragment.
Results
Generation of the m-numbΔ5,6 Mutant Allele.
We first characterized the m-numb locus, which spans over 50 kb of the genomic DNA (data not shown). Because the phosphotyrosine binding (PTB) domain is essential for d-numb function in Drosophila (26, 27), we decided to delete only exons 5 and 6, which reside within a 6-kb genomic fragment and encode the middle portion of the PTB domain. Through homologous recombination in ES cells, we inserted two loxP sites to flank the 6-kb genomic DNA, along with the selection marker neo, which is flanked by the recombinase Flp binding site frt (23, 24) (Fig. 1A). One heterozygous ES cell line was used for blastocyst injection that resulted in germline chimeras. Mice either heterozygous or homozygous for the floxed m-numb allele (m-numbflox) are viable and appear normal, indistinguishable from their wild-type littermates.
To generate mice carrying a mutant allele of m-numb, we mated m-numbflox mice with transgenic mice expressing the Cre recombinase under the control of the β-actin promoter. As expected, the presence of Cre resulted in the deletion of the 6-kb genomic fragment containing exons 5 and 6, as demonstrated by PCR (Fig. 1B) and genomic Southern blot (data not shown). Because β-actin is expressed in germ cells, the resultant m-numb allele, m-numbΔ5,6, was transmitted to the next generation, generating mice heterozygous for this mutation. The heterozygous mice (m-numbΔ5,6/+) are viable and appear normal, indistinguishable from their wild-type littermates.
m-numbΔ5,6 Mutation Causes Embryonic Lethality.
To generate homozygous m-numb mutant mice (m-numbΔ5,6/Δ5,6) and examine the consequence of m-numb loss in cortical neurogenesis, we intercrossed m-numbΔ5,6/+ mice. Of the 136 progeny we initially genotyped 3–4 wk after birth, 37 were wild type and 99 were heterozygotes, but no homozygous mutants were recovered, suggesting that the m-numb mutation causes embryonic lethality. To determine the timing of death, we genotyped embryos from heterozygous intercross matings. Although we were able to recover homozygous mutant embryos as late as E11.5 (6/30), by E13.5, no recognizable homozygous mutant embryos could be recovered (n = 32), suggesting that m-numb mutant embryos die around E11.5.
m-numbΔ5,6 Is a Null Allele of m-numb.
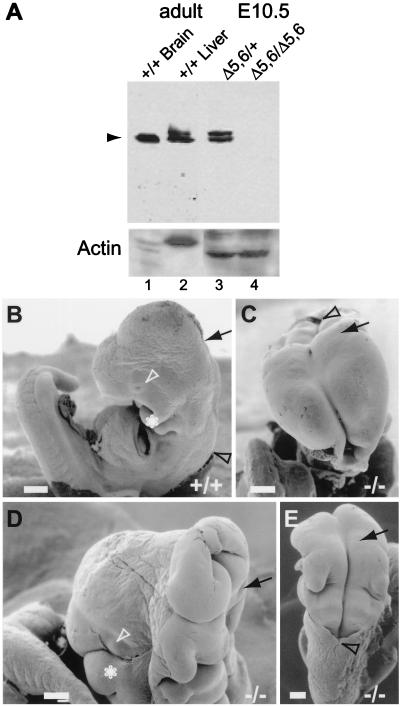
To examine whether deleting exons 5 and 6 produces a null allele of m-numb, we prepared protein extracts from wild-type adult tissues and E10.5 heterozygous and homozygous embryos. We used an antibody against the C terminus of m-Numb (19) to analyze the protein extracts by immunoblot. This antibody should recognize any mutant proteins that might arise from potential alternative splicing between exons 4 and 7 in the absence of the exons 5 and 6, which encode only 72 amino acids (≈12% of the m-Numb protein) near the N terminus. As shown in Fig. 2A, no m-Numb immunoreactivity could be detected in extracts from homozygous embryos. The complete loss of m-Numb immunoreactivity in mutant embryos lacking exons 5 and 6 could be because of unstable mutant mRNA or protein. It is also possible that the presence of a flrted neo cassette affects the splicing between exon 4 and 7. Regardless, m-numbΔ5,6 is likely a null allele of m-numb.
Figure 2.
Neural tube closure defects in m-numb homozygous mutant embryos. (A) Immunoblot analysis of m-Numb protein expression in wild-type (+/+), heterozygous (Δ5,6/+), and homozygous mutant (Δ5,6/Δ5,6) mice using an antibody against the C-terminal m-Numb (19). Anti-Actin antibody was used as a control. (B–E) Scanning electron micrograph of wild-type (B, E10.5, lateral view) and m-numb homozygous mutant (C, E10.5, frontal view; D, E11.5, lateral view; E, E10.5, dorsal view) embryos. (Scale bar is 200 μm.) Arrow marks the midbrain–hindbrain junction; black arrowhead marks the hindbrain–spinal cord junction; white arrowhead marks the eye; and ∗ marks the first branchial arch.
m-numbΔ5,6 Homozygous Embryos Exhibit Defects in Cranial Neural Tube Closure.
The m-numbΔ5,6 homozygous embryos that survive to E11.5 display several gross abnormalities. Most noticeably, mutant embryos are one-half to two-thirds the size of their wild-type or heterozygous littermates (Fig. 3 A and D). Furthermore, all mutant embryos exhibit defects in cranial tube closure (Fig. 2 B–E). In wild-type embryos, cranial neural tube (forebrain, midbrain, and hindbrain) closure is completed by E9. In mutant embryos, however, the cranial neural tube remains open even at E11.5. Occasionally, the mutant forebrain is closed (Figs. 2D and 4R). However, the telencephalic vesicle in these embryos is not as prominent as that of the wild type. These gross morphological defects are unlikely a result of developmental arrest before E9. Although about one-third of the mutant embryos we recovered at E9.5 were still undergoing axial rotation, the majority of the mutant embryos had completed the process. The external landmarks (such as the optic vesicle, the otic vesicle, branchial arches, and limb buds) and the number of somites in these mutant embryos were also indicative of E9.5 embryos. Caudal neural tube (spinal cord) closure, on the other hand, appears to be normal. Some of the mutant embryos appear necrotic at E9.5, although most survive until E10.5. What causes m-numb mutant embryos to die around E11.5 remains to be determined. We frequently observe mutant embryos with extensive bleeding (Fig. 3A, and data not shown), suggesting that death is likely the result of vascular defects.
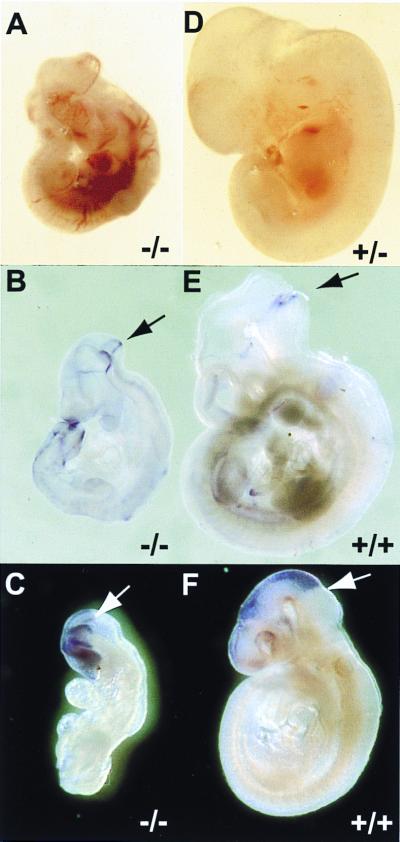
Figure 3.
Neural tube patterning in wild-type and m-numb mutant embryos. (A and D) E11.5 m-numbΔ5,6 heterozygous (+/−) and homozygous (−/−) embryos. Notice the extensive bleeding in the homozygous mutant embryo. (B and E) Fgf17 expression at the midbrain–hindbrain junction (arrow). (C and F) Otx2 expression in the midbrain and forebrain. Arrow marks the midbrain–hindbrain junction.
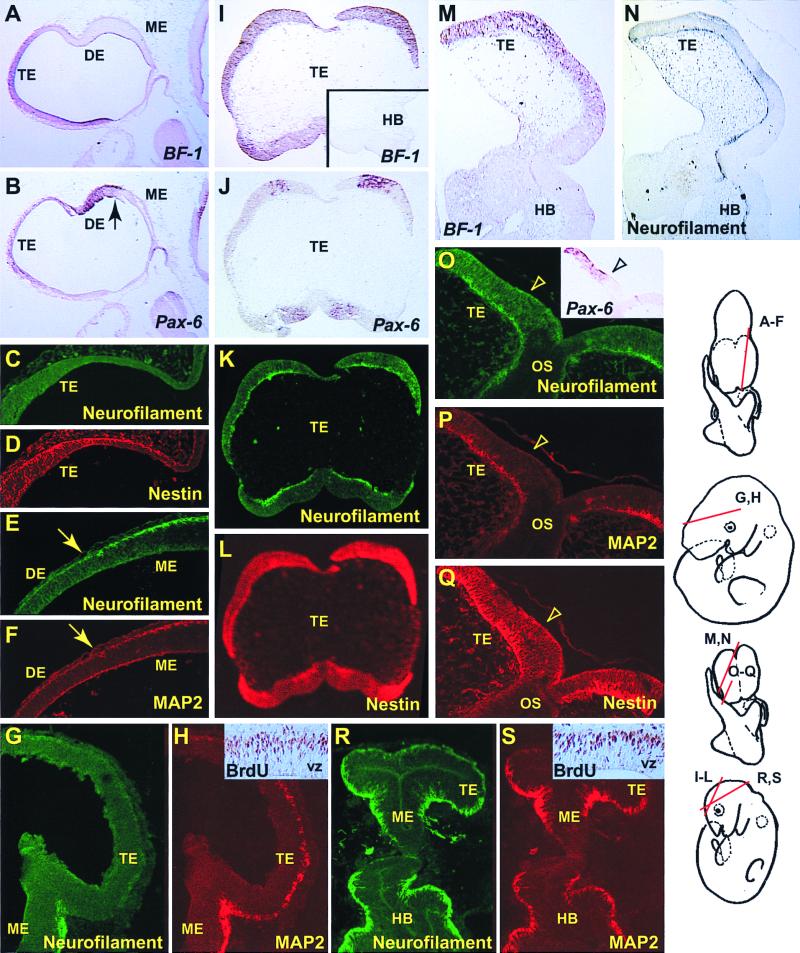
Figure 4.
Precocious production of neurons in the m-numb mutant forebrain at E10.5. (A–H) Heterozygous control (m-numbΔ5,6/+). (I–S) Homozygous mutant (m-numbΔ5,6/Δ5,6). Because of the defects in cranial neural tube closure, molecular markers (BF-1 and Pax6) are used to identify the mutant telencephalon. I–L, M and N, and O–Q are adjacent sections, respectively. C and D, E and F, G and H, K and L, O and P, and R and S are double-staining of the same section, respectively. Notice that sections from the wild-type forebrain (telencephalon and diencephalon) have very few cells that show immunostaining for Neurofilament (C, E, and G) or MAP2 (F and H), both of which are neuronal markers, whereas those from the mutant forebrain have Neurofilament (K, N, O, and R) and MAP2 (P and S)-positive cells throughout the forebrain. Both in the wild-type (D) and the mutant (L and Q) forebrain, most of the cells in the neuroepithelium are positive for Nestin, a neural progenitor cell marker. Insets in H (wild type) and S (mutant) show BrdUrd incorporation in the forebrain after a 2-h pulse. The arrow in B, E, and F marks the junction between mesencephalon and diencephalon, whereas the arrowhead in O, P, and Q marks the border of Pax-6 expression domain (O, Inset). TE, telencephalon; DE, diencephalon; ME, mesencephalon; HB, hindbrain; OS, optic stalk; VZ, ventricular zone.
Patterning of the Neural Tube Is Unaffected in m-numbΔ5,6 Mutant Embryos.
Despite the defects in neural tube closure, subdivision of the rostral neural tube into various brain regions appears to have taken place in m-numbΔ5,6 mutant embryos, as indicated by the expression of various molecular markers. Fgf17 is expressed at the junction between midbrain and hindbrain in wild-type E9.5 embryos (28) (Fig. 3E). This Fgf17 expression domain is also present in mutant embryos (Fig. 3B). Otx2 is expressed in most of the forebrain and midbrain neuroepithelium, with a sharp boundary at the midbrain-hindbrain junction (29) (Fig. 3F). As shown in Fig. 3C, in m-numb mutant embryos at E9.5, Otx2 is indeed expressed in the presumptive forebrain and midbrain region.
To verify the presence of the lateral and dorsal telencephalon, which is the region of the forebrain that gives rise to the cerebral cortex (neocortex and hippocampus), we examined the expression of BF-1 and Pax-6. In wild-type embryos, BF-1 expression within the neural tube is restricted to the telencephalon, along the entire dorsal–ventral axis (30) (Fig. 4A). Although Pax-6 is more widely expressed in the developing neural tube, its expression within the telencephalon is restricted to the dorsal and lateral neuroepithelium; it is absent from the basal (ventral) telencephalon that gives rise to the striatum (31) (Fig. 4B). Pax-6 expression is also absent from the midbrain. In m-numbΔ5,6 homozygous mutant embryos at E10.5, BF-1 is expressed in the presumptive forebrain region (n = 3), further confirming the presence of the telencephalon (Fig. 4 I and M). Moreover, Pax-6 expression can also be detected in the forebrain in adjacent sections (Fig. 4 J and O), suggesting the presence of dorsal–ventral patterning in the mutant forebrain. We made similar observations using other lateral and dorsal telencephalon markers, such as Emx-1 and Emx-2 (32) (data not shown). Therefore, it appears that the m-numbΔ5,6 mutation does not affect the subdivision of the neural tube into different brain regions, nor its dorsal–ventral patterning.
Precocious Neuron Production in m-numbΔ5,6Mutant Forebrain.
We examined neuron production in m-numb mutant embryos at E10.5, which is at the very beginning of cortical neurogenesis. In wild-type E10.5 embryos, very few neurons can be detected in the forebrain using two neuronal markers, anti-Neurofilament (150 kDa) and anti-MAP2 (Fig. 4 C, E, and G and Fig. 4F, respectively), consistent with what has been previously reported (33). In a few forebrain sections, scattered MAP2-positive cells can be identified (Fig. 4H), suggesting that postmitotic neurons are being generated in the forebrain at this stage. In E10.5 mutant embryos, however, Neurofilament and MAP2-positive cells can be clearly identified throughout the forebrain (n = 4) (Fig. 4 K, N, O, and R and Fig. 4 P and S, respectively). Furthermore, staining of sections adjacent to those with BF-1 and Pax-6 expression also reveals an abnormally large number of neurons in the presumptive neocortex of the m-numbΔ5,6 mutant (Fig. 4 K and O). Consistent with this possibility, in one mutant embryo with closed forebrain and, therefore, identifiable telencephalic vesicles, neurons are precociously produced in the neocortex (Fig. 4 R and S). We also attempted to examine neuron production in E11.5 mutation embryos (n = 3), but were unable to obtain staining using a wide variety of neuronal and progenitor cell markers, suggesting that E11.5 mutant embryos are already undergoing necrosis.
Despite being generated precociously, neurons in the mutant neocortex lay outside of the ventricular zone in the external mantle layer where neurons normally reside, suggesting that m-numb mutation has little effect on the placement of early neurons. Most cells in the ventricular zone of the E10.5 mutant neuroepithelium appear to be neural progenitor cells, as indicated by the expression of BF-1 (Fig. 4 I and M) and Pax-6 (Fig. 4 J and O) and staining with an antibody against the Nestin protein (Fig. 4 L and Q), a neural progenitor cell marker. Indeed, BrdUrd-positive nuclei appear after a short pulse of labeling in the outer half of the ventricular zone of both mutant and wild-type embryos; no gross difference in the density or location of these proliferating cells has been observed (n = 2; Fig. 4 H and S, Insets). Previous studies indicate that only a small fraction (<20%) of the cell divisions in the neuroepithelium is asymmetric at the beginning of cortical neurogenesis (14, 15). Therefore, it is not surprising that the majority of the neural progenitor cells that divide symmetrically to produce more progenitor cells appear not to be affected by the m-numbΔ5,6 mutation.
Discussion
In Drosophila, asymmetric segregation of the Numb protein is crucial for daughter cells to adopt distinct fates after asymmetric divisions. We have shown that the m-Numb protein is asymmetrically localized to the apical cell membrane of dividing cortical progenitor cells (19). After a horizontal division, the majority of the m-Numb protein is likely to be segregated to the apical daughter cell that has been suggested to remain as a progenitor cell (14).
Our observation of precocious neuron production in the mutant forebrain is consistent with the notion that m-numb plays a role in promoting the progenitor cell fate (19, 21). The precocious neuron production could reflect cell fate alteration resulting from the absence of m-Numb in progenitor cells that undergo asymmetric division. However, we could not determine definitively in these mutant embryos whether neurons were produced at the expense of progenitor cells or simply because of acceleration of differentiation because the early defects in neural tube closure made it difficult to quantitatively compare neuron production between wild-type and mutant forebrains. These questions can be better addressed in the future when transgenic mice that express Cre recombinase only in cortical progenitor cells become available. Mating such transgenic mice with mice carrying the m-numbflox allele will allow us to bypass the early embryonic lethality and target the m-numbΔ5,6 mutation specifically to cortical progenitor cells.
Interestingly, the neurogenesis defect we observed in the loss-of-function mutant forebrain is opposite to and, therefore, consistent with findings from gain-of-function experiments in which Numb was overexpressed in neural progenitor cells (22, 34). For example, the murine embryonic carcinoma cell line P19 undergoes neural differentiation when aggregated cells are treated with retinoic acid. It has been reported that overexpression of PRRL-containing human Numb proteins, which are the Numb isoforms expressed by undifferentiated progenitor cells, results in an increase in the number of proliferating P19 cells (22, 35). It is worth pointing out that Numb overexpression has no effect on proliferation in P19 cells that have not been induced for neural differentiation. Similar results have also been obtained using neural crest stem cells (22). An avian homologue of m-numb, c-numb, has also been isolated (34). The c-Numb protein was reported to localize to the basal membrane in chick neuroepithelial cells, unlike m-Numb, and was suggested to promote neuronal fate in asymmetric divisions. However, it is of interest to note that the reported overexpression of c-Numb in chick neuroepithelial cells resulted in a significant reduction of neuron production and an increase in the number of proliferating progenitor cells. Therefore, our loss-of-function study in mice and the reported gain-of-function study in chick and mammalian cell lines both point to a numb function in promoting the progenitor fate.
Although more experiments are necessary to further examine Numb localization in different species, one possibility that might account for the reported difference between mice and chick is antiserum specificity. The antiserum used in the chick study is unlikely to be c-Numb-specific. The peptide selected to raise the c-Numb antibody is highly conserved in mouse Numblike (15 of 20 and, more importantly, 14 of the C-terminal 15 amino acids are identical). In fact, the reported immunostaining pattern of c-Numb in the developing neural tube (34) resembles that of Numblike, not Numb, in corresponding mouse tissues (21). Moreover, an antibody raised against the C-terminal 14 amino acids of m-Numb (20), all of which are conserved in the c-Numb peptide, has been reported to crossreact with mouse Numblike (35). Localization of FLAG-tagged exogenous c-Numb was examined, and 3 of the 11 dividing cells examined reportedly showed basal localization. However, the dividing cell as shown in the paper appears to be at prophase with significant amount of cytoplasm between the nucleus and the FLAG-tagged c-Numb immunoreactivity (34). Normally, there is very little cytoplasm between the nucleus and the cell membrane in interphase and prophase cortical progenitor cells in the ventricular zone, raising concerns as to whether the immunoreactivity might belong to a neighboring cell.
In Drosophila, numb function is not limited to the nervous system. For example, asymmetric segregation of d-Numb protein is required for muscle progenitor cells to divide asymmetrically to generate distinct muscle founders (36, 37). Asymmetric d-Numb segregation is also necessary during Malpighian tubule development for the tip mother cell to generate two distinct daughter cells, a tip cell and a sibling cell (38). As in Drosophila, m-numb is expressed in many tissues during mouse embryogenesis and likely plays important roles in cell fate specification outside of the nervous system, as indicated by the extensive bleeding and abnormally small size of the mutant embryo.
It should be noted that, in addition to cell fate specification, m-numb may also function in later events of neural development. Recent findings suggest that Notch signaling is required in differentiating neurons (39–41). In fact, translocation of the Notch intracellular domain into the nucleus, long considered a hallmark of Notch signaling, is observed in differentiating cortical neurons but not progenitor cells. It has been further suggested that m-Numb can regulate this function of Notch. For example, m-Numb can rescue the effect of Notch1 intracellular domain on neurite outgrowth in cultured cortical neurons (39), consistent with the notion that m-Numb also plays a role in neuronal differentiation (21). The knockout construct reported in this paper should allow us to examine the potential roles of m-numb in many developmental processes in future conditional knockout experiments.
Acknowledgments
We thank L. Ackerman for technical assistance with scanning electron microscopy. We also thank E. Lai, P. Gruss, A. Simeone, and D. Ornitz for various cDNA clones, and M. Lewandoski and G. Martin for β-actin-Cre transgenic mice. This work was supported by a National Institute of Mental Health grant to the Silvio Conte Center for Neuroscience Research at UCSF and a set-up fund from Yale University (to W.Z.). W.Z. was supported by a postdoctoral fellowship from the American Cancer Society. R.A.P. was supported by National Institutes of Health/National Institute of Child Health and Human Development. M.S. is a postdoctoral fellow, and L.Y.J. and Y.N.J. are investigators of the Howard Hughes Medical Institute.
Abbreviations
- m-numb
mouse numb
- d-numb
Drosophila numb
- En
embryonic day n
- ES cells
embryonic stem cells
References
- 1.Rakic P. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 2.Caviness V S, Jr, Takahashi T, Nowakowski R S. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 3.McConnell S K. Neuron. 1995;15:761–768. doi: 10.1016/0896-6273(95)90168-x. [DOI] [PubMed] [Google Scholar]
- 4.Luskin M B, Pearlman A L, Sanes J R. Neuron. 1988;1:635–647. doi: 10.1016/0896-6273(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 5.Luskin M B, Parnavelas J G, Barfield J A. J Neurosci. 1993;13:1730–1750. doi: 10.1523/JNEUROSCI.13-04-01730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price J, Thurlow L. Development (Cambridge, UK) 1988;104:473–482. doi: 10.1242/dev.104.3.473. [DOI] [PubMed] [Google Scholar]
- 7.Walsh C, Cepko C L. Science. 1988;241:1342–1345. doi: 10.1126/science.3137660. [DOI] [PubMed] [Google Scholar]
- 8.Walsh C, Cepko C L. Science. 1992;255:434–440. doi: 10.1126/science.1734520. [DOI] [PubMed] [Google Scholar]
- 9.Parnavelas J G, Barfield J A, Franke E, Luskin M B. Cereb Cortex. 1991;1:463–468. doi: 10.1093/cercor/1.6.463. [DOI] [PubMed] [Google Scholar]
- 10.Grove E A, Williams B P, Li D-Q, Hajihosseini M, Friedrich A, Price J. Development (Cambridge, UK) 1993;117:553–561. doi: 10.1242/dev.117.2.553. [DOI] [PubMed] [Google Scholar]
- 11.Minoe M C, Danevic C, Boardman P, Harris B, Parnavelas J G. J Neurosci. 1994;14:107–123. doi: 10.1523/JNEUROSCI.14-01-00107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornack D R, Rakic P. Neuron. 1995;15:311–321. doi: 10.1016/0896-6273(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 13.Reid C B, Liang I, Walsh C. Neuron. 1995;15:299–310. doi: 10.1016/0896-6273(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 14.Chenn A, McConnell S K. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 15.Smart I H M. J Anat. 1973;116:67–91. [PMC free article] [PubMed] [Google Scholar]
- 16.Uemura T, Shepherd S, Ackerman L, Jan L Y, Jan Y N. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 17.Rhyu M S, Jan L Y, Jan Y N. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 18.Spana E P, Kopczynski C, Goodman C S, Doe C Q. Development (Cambridge, UK) 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- 19.Zhong W, Feder J N, Jiang M-M, Jan L Y, Jan Y N. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 20.Verdi J M, Schmandt R, Bashirullah A, Jacob S, Salvino R, Craig C G, Program A E, Lipshitz H D, McGlade C J. Curr Biol. 1996;6:113–145. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhong W, Jiang M-M, Weinmaster G, Jan L Y, Jan Y N. Development (Cambridge, UK) 1997;124:1887–1897. doi: 10.1242/dev.124.10.1887. [DOI] [PubMed] [Google Scholar]
- 22.Verdi J M, Bashirullah A, Goldhawk D E, Kubu C J, Jamali M, Meakin S O, Lipshitz H D. Proc Natl Acad Sci USA. 1999;96:10472–10476. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu M, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 24.Meyers E N, Lewandoski M, Martin G R. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 25.Qiu M, Bulfone A, Martinez S, Meneses J J, Shimamura K, Pedersen R A, Rubenstein J L R. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- 26.Frise E, Knoblich J A, Younger-Shepherd S, Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 1996;93:11925–11923. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaikie P, Immanuel D, Wu J, Li N, Yajnik V, Margolis B. J Biol Chem. 1994;269:32031–32034. [PubMed] [Google Scholar]
- 28.Maruoka Y, Ohbayashi M, Hoshikawa M, Itoh N, Hogan B L M, Furuta Y. Mech Dev. 1998;74:175–177. doi: 10.1016/s0925-4773(98)00061-6. [DOI] [PubMed] [Google Scholar]
- 29.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nature (London) 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- 30.Tao W, Lai E. Neuron. 1992;8:967–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- 31.Walther C, Gruss P. Development (Cambridge, UK) 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 32.Simeone A, Gulisano M, Acampora D, Stornaiuolo A, Rambaldi M, Boncinelli E. EMBO J. 1992;11:2541–2550. doi: 10.1002/j.1460-2075.1992.tb05319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishibashi M, Ang S-L, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 34.Wakamatsu Y, Maynard T M, Jones S U, Weston J A. Neuron. 1999;23:71–81. doi: 10.1016/s0896-6273(00)80754-0. [DOI] [PubMed] [Google Scholar]
- 35.Dho S E, French M B, Woods S A, McGlade C J. J Biol Chem. 1999;274:33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz G M, Bate M. Development (Cambridge, UK) 1997;124:4857–4866. doi: 10.1242/dev.124.23.4857. [DOI] [PubMed] [Google Scholar]
- 37.Carmena A, Murugasu-Oue B, Mennon D, Jimenez F, Chia W. Genes Dev. 1998;12:304–315. doi: 10.1101/gad.12.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan S, Cato A M, Skaer H. Dev Biol. 2000;217:153–165. doi: 10.1006/dbio.1999.9499. [DOI] [PubMed] [Google Scholar]
- 39.Sestan N, Artavanis-Tsakonas S, Rakic P. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 40.Berezovska O, McLean P, Knowles R, Frosh M, Lu F M, Lux S E, Hyman B T. Neuroscience. 1999;93:433–439. doi: 10.1016/s0306-4522(99)00157-8. [DOI] [PubMed] [Google Scholar]
- 41.Redmond L, Oh S R, Hicks C, Weinmaster G, Ghosh A. Nat Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]