Abstract
Many uveal melanoma patients die of metastasis despite ocular treatment. Transcriptomic profiling of enucleated tumors can identify patients at high metastatic risk. Because most uveal melanomas do not require enucleation, a biopsy would be required for this analysis. Here, we establish the feasibility of transcriptomic analysis of uveal melanomas from fine needle aspirates. Transcriptomic profiles were analyzed from postenucleation “mock” needle biopsies and matching tumors from eight enucleated eyes and from fine needle aspirates in 17 uveal melanomas before radiotherapy. Predictive accuracy was assessed using a weighted voting classifier optimized for probe set selection using a minimal redundancy/maximum relevance algorithm. Transcriptomic profiles from mock biopsies were highly similar to those from their matching tumor samples (P < 0.0001). Transcriptomic profiles from fine needle aspirates clustered into two classes with discriminating probe sets that overlapped significantly with those for our published classification (P < 0.00001). No loss of predictive accuracy was identified among eight needle aspirates obtained from a distant location. Thus, it is feasible to obtain RNA of adequate quality and quantity to perform transcriptomic analysis on uveal melanoma samples obtained by fine needle biopsy. This method can be applied to specimens obtained from distant geographic locations and can stratify uveal melanoma patients based on metastatic risk.
Uveal melanoma is the most common primary cancer of the eye and has a strong predilection for hematogenous metastasis to the liver.1 Our laboratory and others have described a highly robust transcriptomic classification of uveal melanomas based on RNA analysis of primary tumors.2,3 The class 1 signature was associated with an excellent prognosis, whereas the class 2 signature portended a high risk of metastatic death.2 We showed that the class 2 signature was strongly associated with other predictors of poor prognosis, such as epithelioid cytology, looping extracellular matrix patterns, and monosomy 32,4; in addition, class 2 tumors exhibited a shift from a neural crest/melanocyte phenotype to an epithelial-like phenotype.5 This molecular classification represents a potentially valuable prognostic tool to identify high-risk patients and to treat micrometastatic disease before overt clinical presentation. Transcriptomic profiling has been reported only on larger pieces of uveal melanoma tissue obtained at enucleation. Enucleation is performed in only approximately 10% of uveal melanoma patients. For the remaining ∼90% of patients who are treated with globe-sparing modalities, such as radiotherapy,1 transcriptomic profiling would be more useful if it could be performed on fine needle aspirates before eye-sparing treatments.
There were a number of possible obstacles to the successful transfer of transcription-based classifications to a biopsy platform. Because intraocular biopsy requires a very small needle size (25-gauge),6 it was unclear whether the material obtained would be sufficient for microarray-based transcriptomic profiling. In addition, the effects of the small needle size on sampling errors were unknown. Furthermore, the methodological differences in the preparation of RNA from solid tumor tissue versus fine needle aspirates could affect the measurement of RNA transcripts. The purpose of this study was to explore the feasibility of transcription-based classification of uveal melanomas using fine needle aspirates.
Materials and Methods
Preparation of RNA Samples
All studies were approved by the Human Studies Committee at Washington University, and informed consent was obtained from each subject. Fine needle biopsies were performed using a 25-gauge needle on uveal melanomas before radiotherapy as previously described.6 Fine needle aspirates were divided into samples for cytologic diagnosis and RNA analysis. The samples for RNA analysis were expelled into an empty RNase-free tube in the operating room. The empty syringe was filled with 200 μl of extraction buffer from the PicoPure RNA isolation kit (Arcturus, Mountain View, CA), which was then transferred to the same tube to collect any additional tumor cells lodged in the needle hub. The contents of the tube were incubated at 42°C for 30 minutes. Immediately following enucleation, and before opening the eye, mock biopsies were obtained through the sclera in a fashion identical to the actual biopsies. The eye was then opened, and a large piece of matching tumor tissue was obtained, snap frozen, and prepared for RNA analysis as previously described.2 RNA was isolated using the PicoPure kit (including the optional DNase step), which yielded about 100 ng to 1.5 μg of total RNA per aspirate using the NanoDrop 1000 system (Wilmington, DE). RNA samples were stored at −80°C until sent to the Siteman Cancer Center Gene Chip Facility for amplification using the Affymetrix Genechip 3′ Amplification 2-Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA). Amplified cDNA samples were hybridized overnight to Hu133Av2 chips in the Genechip Hybridization Oven Model 640. Chips were washed using the GeneChip Fluidics station 450, and gene expression measured on the Affymetrix Genechip Scanner. For specimens sent to St. Louis from San Francisco, tubes were placed on dry ice and mailed by overnight courier, after which they were incubated at 42°C for 30 minutes and handled as described for the other biopsy samples. No RNA degradation was observed for these samples.
Analysis of Microarray Expression Profiles
All new transcriptomic profiles obtained on Hu133Av2 chips and our previous profiles obtained on Hu133A chips2 were normalized by Robust Multiarray Averaging using RMAexpress (http://rmaexpress.bmbolstad.com). The Tschentscher dataset obtained on HG-U95Av2 chips3 was kindly provided by the authors and was not normalized with RMA. For all analyses except minimal redundancy/maximal relevance (mRMR), expression data were log2 transformed. Probe sets were filtered for a median significance P value <0.05 and gene expression variance >1 across all specimens (to eliminate genes that did not vary substantially between tumors). Hierarchical clustering was performed with Dchip (http://biosun1.harvard.edu/complab/dchip/) using 1-correlation for the distance metric, centroid linkage method, and gene ordering by cluster tightness. Probe sets were ranked for ability to discriminate tumor classes by signal-to-noise algorithm using GeneCluster2 software (http://www.broad.mit.edu/cancer/software/genecluster2/gc2.html), which was also used to generate a predictive test using a weighted voting algorithm. Class predictions were analyzed by leave-one-out cross validation. Confidence scores were calculated by GeneCluster software as described.7 Rank order of discriminating probe sets to be entered stepwise into the predictive test for cross validation were determined with mRMR software (http://miracle.lbl.gov/proj/mRMR/), using mutual information difference as the feature selection scheme and ±0.5 standard deviations as a threshold for discretizing expression values. Significance of discriminating probe set overlap was determined using hypergeometric probability using the PROBHYBR function of SAS 9.0 statistical software (SAS Institute, Cary, NC) as previously described.8
Results
Analysis of RNA from Ocular Biopsy Specimens
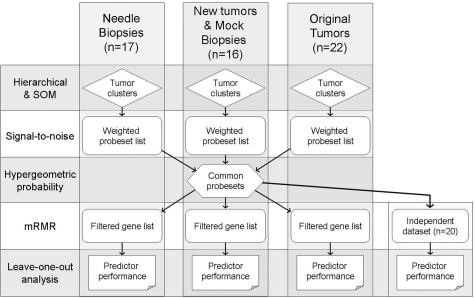
The study goal was to evaluate the feasibility of molecular prognostic testing in uveal melanomas using transcriptomic profiles from fine needle biopsy specimens. We used a series of statistical techniques to analyze four independent transcriptomic profile datasets derived from 1) eight uveal melanomas obtained at eye removal (denoted by the prefix MM) and eight mock needle biopsy samples from the same tumors (denoted MB), 2) 17 fine needle biopsies of uveal melanomas before radiotherapy (denoted NB), 3) 22 uveal melanomas from our original published dataset (denoted OrigMM),2 and 4) 20 uveal melanomas published by another laboratory (denoted Tschentscher).3 The study design is summarized in Figure 1. Clinical and pathological features are summarized in Table 1 and detailed in Supplementary Table S1 (see supplemental material at http://jmd.amjpathol.org). To conserve the small amounts of RNA obtained from the needle aspirates, RNA concentrations were not measured directly, but rather, the RNA quality and hybridization characteristics were assessed by the background score and scaling factor supplied by the Affymetrix software. The background score, an indicator of nonspecific binding to the array, was only slightly higher for the NB group (mean, 2.3 units ± 0.1 SE) compared with the MB group (mean, 1.7 units ± 0.04 SE) and the MM group (mean, 1.6 units ± 0.07 SE). The scaling factor, a multiplication function that is inversely proportional to the overall chip hybridization signal intensity, was lower for the NB group (mean, 9.6 units ± 2.2 SE) compared with the MB group (mean, 17.5 units ± 4.9 SE) and the MM group (mean, 13.1 units ± 0.8 SE). Therefore, the NB chips exhibited good hybridization characteristics with acceptable background, indicating that the fine needle aspirates yielded RNA of adequate quantity and quality for direct transcriptomic profiling. Of note, eight specimens obtained from the San Francisco location and shipped to St. Louis showed no decrease in RNA quality or hybridization characteristics compared with the St. Louis specimens.
Figure 1.
Overview of study design.
Table 1.
Summary of Clinical-Pathologic Features
| Feature | Datasets
|
||
|---|---|---|---|
| OrigMM | MM-MB | NB | |
| Mean age (range) | 61 (25–82) | 59 (37–74) | 60 (25–88) |
| Mean largest tumor diameter, millimeters (range) | 18 (10–26) | 17 (10–22) | 13 (5–18) |
| Mean tumor thickness, millimeters (range) | 10 (4–16) | 10 (5–14) | 9 (3–18) |
| Tumor location | Anterior, 9 (41%) | Anterior, 5 (63%) | Anterior, 8 (47%) |
| Posterior, 13 (59%) | Posterior, 3 (37%) | Posterior, 9 (53%) | |
| Cytology | Spindle, 8 (36%) | Spindle, 2 (25%) | Spindle, 6 (35%) |
| Mixed, 8 (36%) | Mixed, 2 (25%) | Mixed, 4 (24%) | |
| Epithelioid, 6 (27%) | Epithelioid, 4 (50%) | Epithelioid, 4 (24%) | |
| Not available, 3 (17%) | |||
| Molecular class | Class 1, 14 (64%) | Class 1, 3 (37%) | Class 1, 9 (53%) |
| Class 2, 8 (36%) | Class 2, 5 (63%) | Class 2, 8 (47%) | |
Assessment of Sampling Error Using Postenucleation Mock Biopsy Specimens
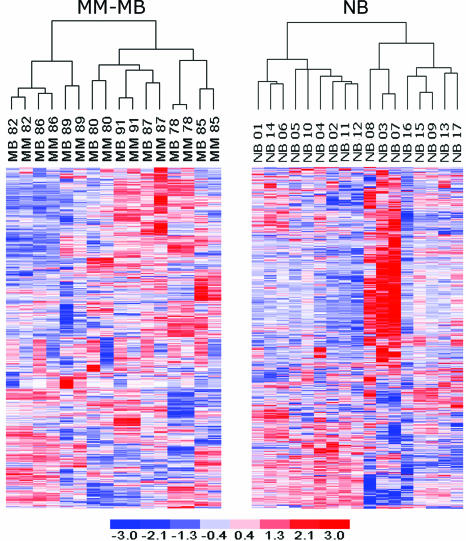
The possibility that tumor cells obtained in a biopsy specimen may not be representative of the entire tumor is a concern with the small amount of material obtained from intraocular biopsies.9 We evaluated the similarity of transcriptomic profiles between the 16 MM and MB samples using unsupervised hierarchical clustering. Because the MM samples represented about 25% of the total tumor volume, they were assumed to represent adequately the overall transcriptomic profile of the tumor. Probe sets were filtered for a median significance P value <0.05 and gene expression variance >1 across all specimens (to eliminate genes that did not vary substantially between tumors), which resulted in 806 probe sets. For each MB specimen, the matching MM specimen demonstrated the shortest linkage distance (ie, the most similar transcriptomic profile) (Figure 2), indicating that each MB was more similar to its paired MM than to any of the other MB or MM specimens (P < 0.0001). This similarity was remarkable, considering the relative homogeneity in gene expression between allogeneic uveal melanomas and the methodological differences in RNA preparation for the MM versus the MB samples. These results show that transcriptomic profiles obtained from fine needle aspirates closely approximate larger tumor samples from which they originate and that sampling error is unlikely to represent an obstacle to biopsy-based transcriptomic profiling in uveal melanoma.
Figure 2.
Heat maps showing unsupervised hierarchical clustering of uveal malignant melanomas (MM), matching mock biopsy specimens (MB), and needle biopsy specimens (NB) using 806 probe sets filtered for a median significance P value <0.05 and gene expression variance >1.
Supervised Analysis of Matched Mock Biopsy/Solid Tumor Samples
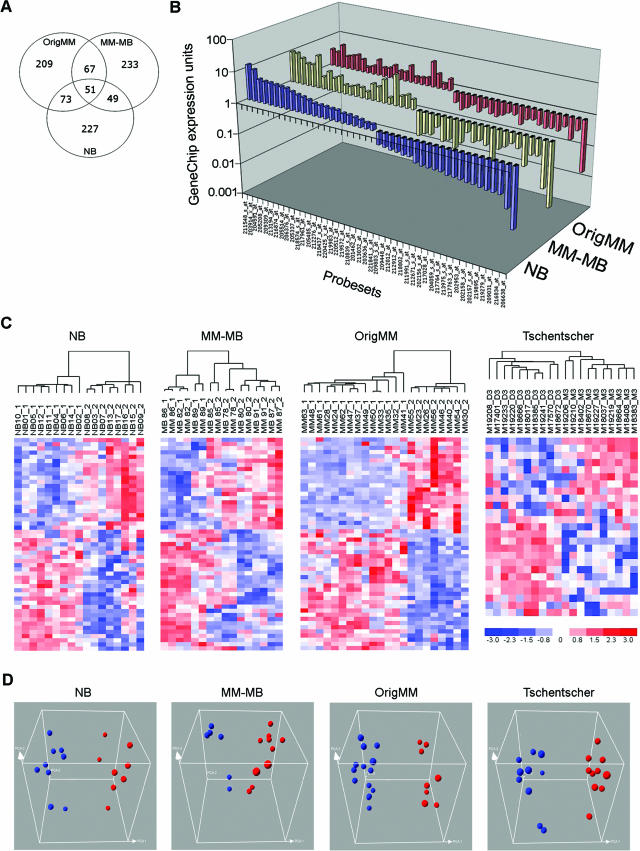
Hierarchical clustering of the MM and MB samples also revealed a higher order of tumor aggregation into two groups of six and 10 specimens each (Figure 2). Self-organizing maps, another unsupervised analytical technique, grouped the specimens into the same clusters (data not shown). To determine whether the dichotomous clustering of MM-MB specimens may correspond to the prognostically validated class 1 and class 2 designations previously described in the OrigMM dataset,2 we compared the discriminating probe sets for the two datasets. For this analysis, we filtered the probe sets for a median significance P value <0.05 and included the top 5000 most variable probe sets. Discriminating probe sets were generated for both datasets using the Marker Selection algorithm of the GeneCluster2 software.7 The distance function was prescribed by the signal-to-noise metric and the class estimate by median gene expression. The 400 top-ranked probe sets from each dataset were compared with each other for overlap. A highly significant overlap of 118 concordant probe sets was observed between the MM-MB and origMM lists (P < 0.00001) (Figure 3A). These findings suggest that the transcriptomic profiles obtained from the MM-MB dataset correspond to the prognostically significant tumor classes identified with the origMM dataset, and they demonstrate the feasibility of molecular prognostic testing in uveal melanoma based on fine needle aspirates.
Figure 3.
A: Venn diagram showing concordant probe sets between the NB, MM-MB, and origMM datasets. B: Comparison of GenChip expression units for the 45-probe set list in the indicated datasets. C: Hierarchical clustering and (D) principal component analysis of the indicated datasets using the 45-probe-set list (blue spheres, class 1 tumors; red spheres, class 2 tumors). The two classes indicated in the Tschentscher dataset refer to monosomy versus disomy for chro-mosome 3.
Analysis of Needle Biopsy Samples
Needle biopsy (NB) specimens were obtained at two locations (nine from St. Louis and eight from San Francisco) and processed at the Washington University site. The 17 samples were normalized together using RMA and then filtered for median significance P value <0.05 and gene expression variance >1 across all specimens. Hierarchical clustering identified two tumor groups of eight and nine specimens (Figure 2). The same dichotomous clustering was observed with self-organizing maps (data not shown). These results were consistent with those obtained with the origMM, MM, and MB datasets, and they suggested that the NB specimens segregated into prognostically significant classes based on transcriptomic profile. To support this interpretation, we compared the discriminating gene list with those from the MM-MB and origMM datasets. As described above for the MM-MB dataset, we filtered the NB dataset for probe sets with median significance P value <0.05, included the top 5000 most variable probe sets, and performed Marker Selection using the signal-to-noise metric and the class estimate by median gene expression. The 400 top-ranked probe sets were then compared with the MM-MB and origMM datasets. There was a highly significant overlap of 100 concordant probe sets between the NB and MM-MB datasets and an even more significant overlap of 124 probe sets between the NB and origMM datasets (for both comparisons, P < 0.00001) (Figure 3A). Importantly, epithelioid cytology was strongly associated with the class 2 molecular profile in the OrigMM and MM-MB datasets, where enucleation specimens were available for histopathological assessment (P < 0.0001), but this association was less evident when cytology was assessed from biopsy specimens alone in the NB dataset (P = 0.24). In fact, three cases where the biopsy sample was insufficient for accurate cytologic diagnosis (NB3, NB4, and NB16) nevertheless provided sufficient material for accurate molecular profiling. This ability to use extremely small amounts of tumor material highlights a potential advantage of molecular profiling.
Development of a Predictive Model
To identify a feature set (group of probe sets) that was sufficiently robust to classify individual tumors accurately from any of our datasets (OrigMM, MM-MB, and NB), we took the intersection of the three discriminating probe set lists, which resulted in 51 probe sets, which was further reduced to 45 probe sets after eliminating six that had a fold change <1.5 in one or more datasets (Figure 3, A and B, and Supplementary Table S2—see supplemental material at http://jmd.amjpathol.org). All tumors from all three datasets were grouped correctly by hierarchical clustering and principal component analysis using only the information contained in the 45-probe set list (Figure 3, C and D).
To validate further this feature set, we analyzed an independent dataset published by Tschentscher et al, in which an association was shown between transcriptomic profile and monosomy 3, a strong predictor of metastasis.3 Using the Affymetrix “Best Match” file, we were able to identify matches for 25 of our 45 probe sets from this dataset, which was generated using the HG-U95Av2 chip. The expression data, which were presented as fold change, were normalized to mean = 0 and unit variance. Unsupervised hierarchical clustering and principal component analysis using these 25 probe sets correctly grouped all tumors according to chromosome 3 status (Figure 3, C and D). Thus, a small feature set can accurately classify a broad range of uveal melanomas from whole-tissue and biopsy specimens.
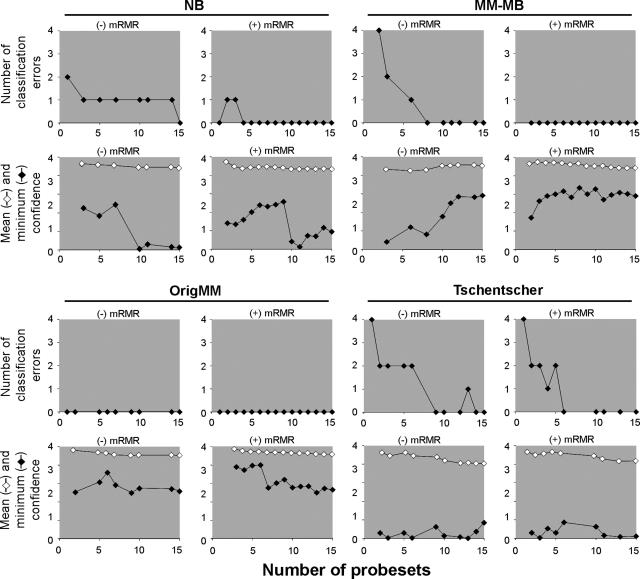
We then tested the performance of this feature set in a predictive model by entering the probe sets stepwise in a random fashion into a weighted voting algorithm and evaluating class assignment by leave-one-out cross-validation using GeneCluster software. The predictor correctly classified all tumors with 15 probe sets for the NB dataset, eight for the MM-MB dataset, one for the OrigMM dataset, and nine for the Tschentscher dataset (Figure 4). The number of probe sets required for class assignment was reduced even further by ranking them for minimal redundancy and maximal relevance using the mRMR algorithm10 and entering them stepwise into the predictor. Using this technique, all of the tumors were classified correctly with as few as one probe set for the NB dataset, two for the MM-MB dataset, one for the OrigMM dataset, and six for the Tschentscher dataset (Figure 4).
Figure 4.
Predictive model for classifying tumor samples for the indicated datasets. The predictive model was evaluated for class assignment by transcriptome signature in all datasets except the Tschentscher dataset, which was evaluated for monosomy 3. Probe sets were entered randomly (−mRMR) or by minimum redundancy and maximum relevance (+mRMR) into a weighted voting algorithm. Classification errors calculated by leave-one-out cross-validation are plotted on the upper graph. The mean and minimum confidence scores are plotted on the lower graph.
Discussion
This study supports the feasibility of prognostic testing in uveal melanoma by transcriptomic profiling of fine needle biopsy specimens. These results show that RNA of sufficient quantity and quality can be obtained from fine needle ocular aspirates to generate microarray-based transcriptomic profiles that closely resemble those obtained from large tissue sections from the same tumor. Furthermore, we show that accurate molecular profiles can be obtained from extremely small biopsy samples that cannot be assessed confidently by cytologic examination alone. These studies also demonstrate that RNA from fine needle aspirates can be shipped safely to distant locations before processing, which would be required for large-scale clinical testing and multicenter trials. Although the short time frame of our study did not allow us to assess directly whether transcriptomic profiles obtained from fine needle aspirates could predict metastasis, several lines of evidence suggest that this is the case. First, the list of discriminating probe sets that assigned NB samples to the correct class was highly similar to that for the OrigMM dataset in which class label associated strongly with metastatic death.2 This technique of comparing probe sets rather than raw expression data allowed us to compare datasets generated at different times using different array platforms. Second, the discriminating probe sets from the NB samples could be used to classify tumors by chromosome 3 status (Tschenstcher dataset) and metastatic death (OrigMM dataset). Ultimately, the prognostic value of biopsy-derived transcriptomic profiles will require a prospective study, and the feasibility of conducting such a study is established by our results.
This study also demonstrates effective strategies for reducing three sources of error in transcriptome-based machine learning and predictive testing: overfitting, redundancy, and instability of predictive feature sets.10,11 The impact of overfitting (the degree to which a predictive model is representative of the tumors used to train the model but not of all tumors on which the predictor may be applied), can be reduced by applying the predictor to multiple independent datasets.11 Hence, we have filtered and cross-validated our feature set and predictive model through four independent datasets (NB, MM-MB, OrigMM, and Tschentscher). Classification accuracy can also be affected adversely by feature set redundancy (the tendency for multiple genes in a feature set to provide overlapping predictive information), which can be addressed using algorithms such as mRMR to eliminate redundant features and to retain those with maximal predictive power.10 In each of our four datasets, mRMR allowed us to reduce substantially the number of probe sets in the predictive model without sacrificing accuracy. Feature set instability (the tendency for feature sets to vary between independent datasets) has been a major obstacle to transcriptome-based clinical testing and has led to the suggestion that feature selection should be approached as a process of finding groups of potential feature sets, rather than a single “best” feature set.12 In fact, a third of the top 15 probe sets ranked by mRMR appeared in only one dataset, and only one appeared in all three datasets, making it unlikely that a single marker will be found that contains the full predictive information of a refined feature set.
In summary, we have identified a list of 45 probe sets from the intersection of class discriminators in three different datasets to provide a pool of features that can be used to classify future datasets, thereby laying the groundwork for a clinical test that can be validated prospectively in a multicenter study to guide management of individual uveal melanoma patients. Following prospective validation, this prognostic assay could improve substantially the management of uveal melanoma patients by identifying and treating high-risk patients before overt presentation of metastatic disease.
Supplementary Material
Acknowledgments
We thank Yang Sun for assistance with collection of clinical data, and Hanchuan Peng for assistance with mRMR algorithm. We have no financial interest in the materials or findings discussed in this article.
Footnotes
Supported by grants from the Tumori Foundation, Macula Society, Barnes-Jewish Hospital Foundation, and Research to Prevent Blindness, Inc. (to J.W.H.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Harbour JW. Clinical overview of uveal melanoma: introduction to tumors of the eye. Albert DM, Polans A, editors. New York: Marcel Dekker,; Ocular Oncology. 2003:1–18. [Google Scholar]
- Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschentscher F, Husing J, Holter T, Kruse E, Dresen IG, Jockel KH, Anastassiou G, Schilling H, Bornfeld N, Horsthemke B, Lohmann DR, Zeschnigk M. Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Res. 2003;63:2578–2584. [PubMed] [Google Scholar]
- Onken MD, Lin AY, Worley LA, Folberg R, Harbour JW. Association between microarray gene expression signature and extravascular matrix patterns in primary uveal melanomas. Am J Ophthalmol. 2005;140:748–749. doi: 10.1016/j.ajo.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Onken MD, Ehlers JP, Worley LA, Makita J, Yokota Y, Harbour JW. Functional gene expression analysis uncovers phenotypic switch in aggressive uveal melanomas. Cancer Res. 2006;66:4602–4609. doi: 10.1158/0008-5472.CAN-05-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner-Jones BE, Foster WJ, Harbour JW, Smith ME, Davila RM. Fine needle aspiration biopsy with adjunct immunohistochemistry in intraocular tumor management. Acta Cytol. 2005;49:297–308. doi: 10.1159/000326153. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Wackerly DD, Mendenhall W, Scheaffer RL. Belmont, CA: Duxbury Press,; Mathematical Statistics with Applications. 1996:107 p. [Google Scholar]
- Char DH, Kroll SM, Stoloff A, Kaleta-Michaels S, Crawford JB, Miller TR, Howes EL, Jr, Ljung BM. Cytomorphometry of uveal melanoma. Comparison of fine needle aspiration biopsy samples with histologic sections. Anal Quant Cytol Histol. 1991;13:293–299. [PubMed] [Google Scholar]
- Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- Pusztai L, Hess KR. Clinical trial design for microarray predictive marker discovery and assessment. Ann Oncol. 2004;15:1731–1737. doi: 10.1093/annonc/mdh466. [DOI] [PubMed] [Google Scholar]
- Sima C, Braga-Neto U, Dougherty ER. Superior feature-set ranking for small samples using bolstered error estimation. Bioinformatics. 2005;21:1046–1054. doi: 10.1093/bioinformatics/bti081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.