Abstract
Mutation V600E of BRAF, a kinase-encoding gene from the RAS/RAF/MAPK pathway, in colorectal carcinoma (CRC) suggests a sporadic origin of the disease, providing an exclusion criterion for hereditary nonpolyposis colorectal cancer. Here we describe detection of this mutation by real-time chemistry TaqMan MGB probes, confirmed by direct DNA sequencing as the gold standard. DNA was extracted from paraffin-embedded tissue from 112 tumors obtained from the EPICOLON study. Seventy-two tumors were CRC with defective DNA mismatch repair (MMR; microsatellite instability and/or loss of protein expression by immunohistochemical analysis), and 40 were proficient MMR controls. BRAF mutation was detected in 20/72 (27.8%) CRC with defective MMR and in 3/40 (7.5%) proficient MMR controls (P = 0.011). BRAF mutation was detected in 19/51 (37.3%) tumors with loss of MLH1 expression and in none of the tumors with loss of MSH2 expression (0/13). BRAF mutation was not found in cases with germline mutation of MLH1 (4/112) or MSH2 (3/112) genes. The sensitivity and specificity of our real-time chemistry were both 100% for detecting the V600E mutation. Because real-time chemistry methodology has advantages in cost, time, and labor, we consider it a valuable alternative to automatic direct sequencing, particularly for serial measurements.
Hereditary nonpolyposis colorectal cancer (HNPCC) is the most common hereditary colon cancer syndrome, accounting for 0.9 to 2.5% of all colorectal cancer (CRC).1 HNPCC is due to a germline mutation of DNA mismatch repair (MMR) genes, mainly MLH1 or MSH2, and cancers occurring in this condition show high-level DNA microsatellite instability (MSI-H). According to the international criteria for HNPCC diagnosis, cancer patients with Amsterdam clinical criteria or Bethesda criteria showing MSI-H should receive genetic counseling and be offered testing for germline mutations of MMR genes.2 Furthermore, 5 to 10% of unselected CRC show MSI-H phenotype that is primarily due to hypermethylation of the MLH1 gene promoter, not germline mutation.3 Thus, it would be desirable to exclude this group from MMR gene testing.
Recently, an oncogenic V600E hotspot mutation in BRAF, a kinase-encoding gene from the RAS/RAF/MAPK pathway, has been found in diverse human cancers,4 especially in CRC with MMR deficiency.5 Interestingly, BRAF mutations have not been detected in those cases with a germline mutation in MLH1, MSH2, or MSH6; thus, detection of a positive BRAF-V600E mutation in CRC suggests a sporadic origin of the disease.6 These findings have potential impact for the genetic testing for HNPCC diagnostics and suggest a potential use of BRAF as an exclusion criterion for HNPCC.7,8
So far, detection of this mutation has been achieved by restriction fragment length polymorphism analysis,9 allele-specific polymerase chain reaction (PCR),10 single-strand conformation analysis,11 and direct automatic sequencing.8 The last method is widely accepted as the standard, but this technique is very time consuming. For this reason, and to perform routine detection of V600E mutation, we assessed the performance of alternative real-time chemistry TaqMan MGB probes in a 7500 sequence detection system designed for allelic discrimination of single nucleotide polymorphisms. The aim of this research was to validate and determine the cost effectiveness of real-time chemistry methodology.
Materials and Methods
Colorectal Cancer Tissue Samples and DNA Extraction
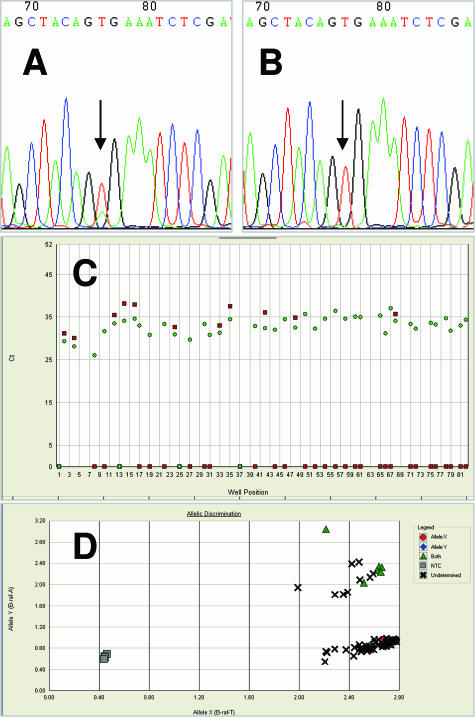
CRC specimens were obtained from researchers participating in the EPICOLON study12,13 at Hospital del Mar (M.A., X.B.). The study was approved by the institutional ethics committee, and informed written consent for molecular studies was obtained from all patients. Specimens were previously sequenced for the detection of BRAF V600E mutation (Figure , 1Aand 1B). DNA was extracted from paraffin-embedded tissue after deparaffination by the standard procedure involving digestion with proteinase K and phenol-chloroform extraction. Then, extracted DNA was checked and quantified in a Biophotometer (Eppendorf AG, Hamburg, Germany) and stored at −20°C until later use.
Figure 1.
Mutation analysis of the BRAF gene. An example of sequencing electropherogram with BRAF codon V600E heterozygous (A) and wild type (B). C: Amplification plot that displays threshold cycle (Ct) as a function of well position; Ct 0 means no amplification with specific probe, circle is allele T, and square is allele A. D: Allelic discrimination plot that determines the genotype of each tumor. NTC is no template control, Allele X means homozygous allele T, Allele Y means homozygous allele A, and Both means heterozygous alleles T and A.
Primer-Probe Design
We designed two different sets of primers-probes with Primer Express Software version 2 (Applied Biosystems, Foster City, CA). We generated two different amplicons, one of 176 bp and another of 125 bp. The primers were designed within introns to avoid amplification of a pseudogene located in chromosome X. The chosen primers and probes were subjected to Basic Local Alignment Search Tool (BLAST) N, BLAST X, and BLAST P database searches to find any sequence similarities. The chosen reporter fluorophores for TaqMan MGB probes were VIC (Applied Biosystems) for detecting the wild-type allele and 6-carboxyfluorescein (FAM) for the mutant allele. The two sets of primers-probes were as follows: set 1, BRAF-51F (forward) 5′-CTACTGTTTTCCTTTACTTACTACACCTCAGA-3′, BRAF-176R (reverse) 5′-ATCCAGACAACTGTTCAAACTGATG-3′, mutant probe 5′-FAM-CTACAGaGAAATCTC-3′, and wild-type probe 5′-VIC-AGCTACAGtGAAATC-3′; and set 2, BRAF-16F (forward) 5′-GCTTGCTCTGATAGGAAAATGAGATC-3′, BRAF-176R (reverse) 5′-ATCCAGACAACTGTTCAAACTGATG, mutant probe 5′-FAM-TAGCTACAGaGAAATC-3′, and wild-type probe 5′-VIC-CTAGCTACAGtGAAATC-3′. Every possible combination of primers and sets of probes were tested with controls of DNA from the HT29 cell line, which harbors the V600E BRAF heterozygotic mutation,4 commercial human DNA (Roche Diagnostics GmbH, Mannheim, Germany), and from six previously sequenced DNAs from CRC tissues. We considered the best option the one that was able to discriminate the wild type from heterozygous genotype and that provided the shorter amplicon; thus we chose the primers from set 1 and the probes from set 2.
Real-Time PCR
Once we had chosen our primers and TaqMan MGB probes, real-time PCR was performed with DNA extracted from CRC tissues. Real-time PCR was performed in a final reaction volume of 20 μl containing 10 μl of 2× TaqMan Universal PCR Master Mix (Applied Biosystems), 900 nmol/L of each primer, 250 nmol/L of each probe, and 5 μl of DNA solution. PCR was performed in MicroAmp optical 96-well plates with optical adhesive covers (Applied Biosystems). Amplification and detection were performed with an ABI prism 7500 sequence detection system (Applied Biosystems). The amplification conditions were 2 minutes at 50°C for AmpErase uracil-N-glycosylase activity and 10 minutes at 95°C for AmpliTaq Gold activation, followed by 50 cycles of 15 seconds at 92°C for denaturation and 1.5 minutes at 60°C for annealing and extension. After amplification, end point detection of fluorescence was performed at 60°C. The fluorescence data were analyzed with the allelic discrimination software of the ABI Prism 7500 instrument, and amplification plots produced in the PCR reaction were also checked. A time, cost, and labor analysis was also performed to establish the most efficient technique per sample comparing real-time PCR in an ABI Prism 7500 versus automatic sequencing in ABI Prism 310. For real-time PCR versus automatic sequencing, time was established at 4 minutes (6 hours/96 samples) versus 10 hours, cost at 2 € ($2.60) versus 8 € ($10.40), and labor at 1.25 minutes (2 hours/96 samples) versus 16 minutes, respectively.
Results and Discussion
We tested DNA from 112 tumors obtained from the EPICOLON study, a clinical epidemiology survey aimed at establishing the incidence of HNPCC in Spain.12,13 Seventy-two tumors were CRC with defective MMR (MSI and/or loss of protein expression in immunohistochemical analysis), and 40 were proficient MMR controls. Mutation analysis of MLH1/MSH2 was performed in all cases with defective MMR.
BRAF mutation was detected in 20/72 (27.8%) CRC with defective MMR and in 3/40 (7.5%) proficient MMR controls (P = 0.011) (Figure , 1Cand 1D). In tumors with loss of MLH1 expression, BRAF mutation was detected in 19/51 (37.3%) tumors and in none of the tumors with loss of MSH2 expression (0/13). BRAF mutation was not found in those cases with germline mutation of the MLH1 (4/112) or MSH2 (3/112) genes. These findings are along the same lines as those of other authors7,8.
Considering as gold standard the direct sequencing methodology, the sensitivity and specificity of the real-time PCR procedure were both 100%. Therefore, this real-time PCR method with TaqMan MGB probes is a robust and convenient way to determine the presence of V600E mutation. The use of TaqMan MGB probes gives clear discrimination of genotypes independent of the amount of input DNA.
In conclusion, real-time PCR is a highly sensitive and specific methodology for detecting V600E mutation. Because real-time chemistry methodology has advantages in cost, time, and labor, we consider that it may be an alternative to automatic direct sequencing, particularly for serial measurements. The combination of TaqMan MGB probes with universal assay conditions allows an efficient workflow when the number of different single-nucleotide polymorphisms that are detected in the clinical laboratory increases.
Acknowledgments
We thank Dolores Durá, Juan Pulido, and Estefania Rojas for technical assistance.
Footnotes
This work was supported by grant from the Sociedad Española de Oncología Médica (SEOM) and Instituto de Salud Carlos III (CO3/02).
C.A. and S.B. are recipients of a research contract of Spanish Ministry of Health, Instituto de Salud Carlos III (CM04/00035 and 01/3080).
References
- Kerber RA, Neklason DW, Samowitz WS, Burt RW. Frequency of familial colon cancer and hereditary nonpolyposis colorectal cancer (Lynch syndrome) in a large population database. Fam Cancer. 2005;4:239–244. doi: 10.1007/s10689-005-0657-x. [DOI] [PubMed] [Google Scholar]
- Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: rAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- Deng G, Bell I, Crawley S, Gum J, Terdiman JP, Allen BA, Truta B, Sleisenger MH, Kim YS. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- Domingo E, Laiho P, Ollikainen M, Pinto M, Wang L, French AJ, Westra J, Frebourg T, Espin E, Armengol M, Hamelin R, Yamamoto H, Hofstra RM, Seruca R, Lindblom A, Peltomaki P, Thibodeau SN, Aaltonen LA, Schwartz S., Jr BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E, Niessen RC, Oliveira C, Alhopuro P, Moutinho C, Espin E, Armengol M, Sijmons RH, Kleibeuker JH, Seruca R, Aaltonen LA, Imai K, Yamamoto H, Schwartz S, Jr, Hofstra RM. BRAF-V600E is not involved in the colorectal tumorigenesis of HNPCC in patients with functional MLH1 and MSH2 genes. Oncogene. 2005;24:3995–3998. doi: 10.1038/sj.onc.1208569. [DOI] [PubMed] [Google Scholar]
- McGivern A, Wynter CV, Whitehall VL, Kambara T, Spring KJ, Walsh MD, Barker MA, Arnold S, Simms LA, Leggett BA, Young J, Jass JR. Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Fam Cancer. 2004;3:101–107. doi: 10.1023/B:FAME.0000039861.30651.c8. [DOI] [PubMed] [Google Scholar]
- Young J, Barker MA, Simms LA, Walsh MD, Biden KG, Buchanan D, Buttenshaw R, Whitehall VL, Arnold S, Jackson L, Kambara T, Spring KJ, Jenkins MA, Walker GJ, Hopper JL, Leggett BA, Jass JR. Evidence for BRAF mutation and variable levels of microsatellite instability in a syndrome of familial colorectal cancer. Clin Gastroenterol Hepatol. 2005;3:254–263. doi: 10.1016/s1542-3565(04)00673-1. [DOI] [PubMed] [Google Scholar]
- Ikehara N, Semba S, Sakashita M, Aoyama N, Kasuga M, Yokozaki H. BRAF mutation associated with dysregulation of apoptosis in human colorectal neoplasms. Int J Cancer. 2005;115:943–950. doi: 10.1002/ijc.20957. [DOI] [PubMed] [Google Scholar]
- Pinol V, Castells A, Andreu M, Castellvi-Bel S, Alenda C, Llor X, Xicola RM, Rodriguez-Moranta F, Paya A, Jover R, Bessa X. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- Pinol V, Andreu M, Castells A, Paya A, Bessa X, Rodrigo J. Frequency of hereditary non-polyposis colorectal cancer and other colorectal cancer familial forms in Spain: a multicentre, prospective, nationwide study. Eur J Gastroenterol Hepatol. 2004;16:39–45. doi: 10.1097/00042737-200401000-00007. [DOI] [PubMed] [Google Scholar]