Abstract
The CpG island methylator phenotype (CIMP or CIMP-high) with extensive promoter methylation seems to be a distinct epigenotype of colorectal cancer. However, no study has comprehensively examined features of colorectal cancer with less extensive promoter methylation (designated as “CIMP-low”). Using real-time polymerase chain reaction (MethyLight), we quantified DNA methylation in five CIMP-specific gene promoters [CACNA1G, CDKN2A (p16), CRABP1, MLH1, and NEUROG1] in 840 relatively unbiased, population-based colorectal cancer samples, obtained from two large prospective cohort studies. CIMP-low (defined as 1/5 to 3/5 methylated promoters) colorectal cancers were significantly more common among men (38 versus 30% in women, P = 0.01) and among KRAS-mutated tumors (44 versus 30% in KRAS/BRAF wild-type tumors, P = 0.0003; 19% in BRAF-mutated tumors, P < 0.0001). In addition, KRAS mutations were significantly more common in CIMP-low tumors (47%) than in CIMP-high tumors (with ≥4/5 methylated promoters, 12%, P < 0.0001) and CIMP-0 tumors (with 0/5 methylated promoters, 37%, P = 0.007). The associations of CIMP-low tumors with male sex and KRAS mutations still existed after tumors were stratified by microsatellite instability status. In conclusion, CIMP-low colorectal cancer is associated with male sex and KRAS mutations. The hypothesis that CIMP-low tumors are different from CIMP-high and CIMP-0 tumors needs to be tested further.
Transcriptional inactivation by cytosine methylation at promoter CpG islands of tumor suppressor genes is thought to be an important mechanism in human carcinogenesis.1 A number of tumor suppressor genes, such as CDKN2A (the p16/INK4a gene), MGMT, and MLH1, have been shown to be silenced by promoter methylation in colorectal cancers.1,2,3 In fact, a subset of colorectal cancers have been shown to exhibit promoter methylation in multiple genes, which is referred to as the CpG island methylator phenotype (CIMP).2,4,5 CIMP colorectal tumors have a distinct clinical, pathological, and molecular profile, such as associations with proximal tumor location, female sex, mucinous and poor tumor differentiation, microsatellite instability (MSI), and high BRAF and low p53 mutation rates.5,6,7,8,9,10,11,12 Promoter CpG island methylation has been shown to occur early in colorectal carcinogenesis.13,14,15,16,17
Although CIMP (which we designate as “CIMP-high” to be distinguished from “CIMP-low”) appears to be a distinct biological subtype of colorectal cancer, no study to date has comprehensively examined features of colorectal cancer with less extensive CIMP-specific promoter methylation (which we designate as “CIMP-low”). In this study using quantitative DNA methylation analysis (MethyLight) and a large number of relatively unbiased, population-based colorectal cancer samples,11 we have examined molecular features of CIMP-low tumors (defined as the presence of methylation in 1/5 to 3/5 promoters) compared with those of CIMP-0 tumors (with 0/5 methylated promoters) and CIMP-high tumors (with ≥4/5 methylated promoters). MethyLight assays can reliably distinguish high from low levels of DNA methylation, the latter of which likely have little or no biological significance.18,19
Materials and Methods
Study Group
To recruit patients into this study, we used the databases of two large prospective cohort studies; the Nurses’ Health Study (N = 121,700 women followed since 1976)20 and the Health Professional Follow-up Study (N = 51,500 men followed since 1986).21 Informed consent was obtained from all participants before inclusion in the cohorts. All cohort participants were free of cancer (except for non-melanoma skin cancer) at the time of study entry. A subset of the cohort participants developed colorectal cancers during prospective follow-up. Thus, these colorectal cancers represented population-based, relatively unbiased samples. Based on availability of tissue samples and results at the time of the study, a total of 840 colorectal cancer cases (362 from the men’s cohort and 478 from the women’s cohort) were included, among which 460 cases were examined in our previous study.11 Tissue collection and analyses were approved by the Dana-Farber Cancer Institute and Brigham and Women’s Hospital Institutional Review Boards.
Genomic DNA Extraction
After tumor areas were marked on a hematoxylin and eosin (H&E)-stained section with a pen, tumor tissue was dissected manually from additional tissue sections by a sterile needle. Normal colonic tissue for microsatellite analysis was obtained from the margins of the resection specimens. The dissected tissue was placed in buffered proteinase K solution at 56°C for 3 hours. Genomic DNA was then extracted using QIAmp DNA Mini Kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions.
Real-Time Polymerase Chain Reaction (PCR) (MethyLight) for Quantitative DNA Methylation Analysis
Sodium bisulfite treatment on genomic DNA was performed as previously described.18 For DNA methylation analysis, we typically used one to two tissue sections (10-μm thick) when large tumor sections were available. Real-time PCR to measure DNA methylation (MethyLight) was performed as previously described.22,23,24 We used ABI 7300 (Applied Biosystems, Foster City, CA) for quantitative real-time PCR. Using five sets of primers and probes, we amplified five CIMP-specific promoters [calcium channel, voltage-dependent, T type alpha-1G subunit (CACNA1G); cyclin-dependent kinase inhibitor 2A (CDKN2A) (p16/INK4A); cellular retinoic acid binding protein 1 (CRABP1); MLH1; and neurogenin 1 (NEUROG1)].11 COL2A1 (the collagen 2A1 gene) was used to normalize for the amount of input bisulfite-converted DNA.18,24 Primers and probes were previously described for the following genes: CACNA1G, CRABP1, and NEUROG1;11 CDKN2A and COL2A1;24 and MLH1.18 The percentage of methylated reference (PMR, ie, degree of methylation) at a specific locus was calculated by dividing the GENE/COL2A1 ratio of the amounts of templates in a sample by the GENE/COL2A1 ratio in M. SssI-treated human genomic DNA (presumably fully methylated) and multiplying this value by 100.25 A PMR cutoff value of 4 was based on previously validated data.18,22,23,24,25,26 Based on the distribution of PMR values at the CRABP1 locus,11 we raised PMR cutoff to 6 for CRABP1, which improved specificity of CRABP1 for the prediction of overall CIMP status (data not shown). Precision and performance characteristics of bisulfite conversion and subsequent MethyLight assays have been previously evaluated, and the assays have been validated.18
Microsatellite Instability (MSI) Analysis
For MSI analysis, whole genome amplification of genomic DNA was performed by PCR using random 15-mer primers.27 Methods to determine MSI status have been previously described.28 In addition to the recommended MSI panel consisting of D2S123, D5S346, D17S250, BAT25, and BAT26,29 we also used BAT40, D18S55, D18S56, D18S67, and D18S487 (ie, 10-marker panel).28 A “high degree of MSI” (MSI-H) was defined as the presence of instability in ≥30% of the markers in the 10-marker panel. A low degree of MSI (MSI-L) was defined as the presence of instability in <30% of the markers, and “microsatellite-stable” (MSS) tumors were defined as tumors without an unstable marker.
Sequencing of KRAS and BRAF
Methods of PCR and sequencing targeted for KRAS codons 12 and 13 and BRAF codon 600 have been previously described.28,30 All forward sequencing results were confirmed by reverse sequencing. Pyrosequencing methods for KRAS and BRAF sequencing were implemented since the study began and performed on a subset of cases. Methods of KRAS pyrosequencing have been validated as described.27 BRAF pyrosequecing was performed using the PSQ96 HS System (Biotage AB and Biosystems, Uppsala, Sweden) according to the manufacturer’s instructions. For BRAF pyrosequencing, PCR primers were 5′-CAGTAAAAATAGGTGATTTTG-3′ (forward) and biotin-5′-CAACTGTTCAAACTGATGGG-3′ (reverse), pyrosequencing primer was 5′-TGATTTTGGTCTAGCTACA-3′, and the dispensation order was TGAGTCAGTCAGTCAGTCAGTCAGTC.
Tissue Microarray (TMA) Construction and Immunohistochemistry for p53
TMAs were constructed as previously described31 using the Automated Arrayer (Beecher Instruments, Sun Prairie, WI). We analyzed whole tissue sections for cases in which there was not enough tumor tissue for TMAs or no definitive results by TMA immunohistochemistry. Methods for p53 immunohistochemistry were previously described.28 Only strong and unequivocal nuclear staining in 50% or more of tumor cells was interpreted as positive. Appropriate positive and negative controls were included in each run of immunohistochemistry. All slides were interpreted by a pathologist (S.O.) blinded from any other laboratory data.
Statistical Analysis
For statistical analysis, the χ2 test (or Fisher’s exact test for categories with an N value of less than 10) was performed on categorical data using the SAS program (version 9.1; SAS Institute, Cary, NC). All P values were two-sided, and statistical significance was set at P values of ≤0.05.
Results
Criteria for CIMP-High, CIMP-Low, and CIMP-0
We obtained 840 colorectal cancer specimens and quantified DNA methylation in the five CIMP-specific gene promoters (CACNA1G, CDKN2A, CRABP1, MLH1, and NEUROG1) by MethyLight technology. We have previously validated the selection and use of these five loci for the determination of CIMP-high in colorectal cancer.11 We also examined normal mucosa from resected segments of colon in a subset of cases and shown infrequent, at most low-levels, of DNA methylation in these loci (data not shown). Distributions of PMR values were bimodal, and only rare cases showed PMR within the range of PMR cutoff ±1 (data on the first 460 cases11 and data not shown for all 840 cases). Thus, the methylation status (positive or negative) at each locus could be unequivocally determined for a vast majority of the cases.
Table 1 shows the distributions of the number of methylated promoters (from 0 to five) in all 840 colorectal cancers composed of 362 from the men’s cohort and 478 from the women’s cohort. As in Table 1, 122 MSI-H tumors in this study showed a striking bimodal distribution with only one tumor (0.8%), exhibiting 3/5 methylated promoters. Based on this bimodal distribution, CIMP-high was defined as the presence of ≥4/5 methylated promoters. Overall, 130 (15%) of all 840 tumors were CIMP-high. The proportion of CIMP-high cases increased progressively from MSS (6.0%) to MSI-L (11%) and MSI-H (69%) tumor status (MSI-H versus MSI-L or MSS, P < 0.0001). CIMP-low tumors, which were defined as tumors with 1/5 to 3/5 methylated promoters, constituted 33% (279/840) of all tumors. CIMP-0 tumors, defined as tumors with no methylated promoter, constituted 51% (431/840) of all tumors.
Table 1.
Distribution of Colorectal Cancers According to the Number of Methylated Promoters
| Number of methylated promoters
|
CIMP-low 1 to 3 | CIMP-high≥4 | ||||||
|---|---|---|---|---|---|---|---|---|
| 0 (CIMP-0) | 1 | 2 | 3 | 4 | 5 | |||
| All cases (N = 840) | 431 (51%) | 143 | 90 | 46 | 54 | 76 | 279 (33%) | 130 (15%) |
| Men (N = 362) | 189 (52%) | 73 | 43 | 21 | 15 | 21 | 137 (38%); P = 0.01 | 36 (9.9%); P = 0.0001 |
| Women (N = 478) | 242 (51%) | 70 | 47 | 25 | 39 | 55 | 142 (30%); P = 0.01 | 94 (20%); P = 0.0001 |
| MSI-H (N = 122) | 22 (18%) | 9 | 5 | 1 | 16 | 69 | 15 (13%) | 85 (70%) |
| MSI-L (N = 72) | 36 (50%) | 11 | 9 | 8 | 5 | 3 | 28 (39%) | 8 (11%) |
| MSS (N = 621) | 356 (57%) | 120 | 72 | 36 | 33 | 4 | 228 (37%) | 37 (6.0%) |
P values are based on comparisons of CIMP-low and CIMP-high frequencies between men and women.
Characteristics of CIMP-High, CIMP-Low, and CIMP-0
We examined relations of CIMP-0, CIMP-low, and CIMP-high with sex. Whereas CIMP-high tumors were more common in women (20%) than men (9.9%, P = 0.0001), CIMP-low tumors were significantly more common in men (38%) than in women (30%, P = 0.01) among all 840 tumors (Table 1). No significant difference was observed in the frequencies of CIMP-0 tumors between men and women. Next, we stratified tumors according to MSI status. CIMP-high tumors were still significantly more common in women than men among MSI-H tumors (77% in women versus 53% in men, P = 0.007) and MSS tumors (8.1% in women versus 3.3% in men, P = 0.02). In contrast, CIMP-low tumors were more common in men than women among MSS tumors (42% in men versus 32% in women, P = 0.01) and MSI-H tumors (19% in men versus 8.9% in women, although statistical significance was not reached).
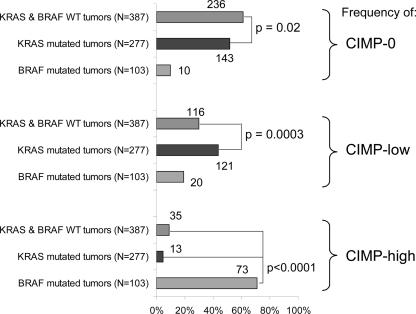
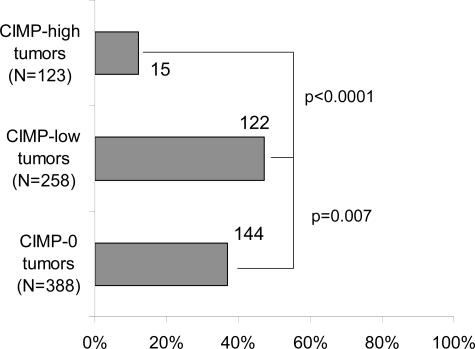
We also examined relations between CIMP status and KRAS and BRAF mutations. We subclassified tumors into KRAS-mutated tumors (with wild-type BRAF) (N = 277), BRAF-mutated tumors (with wild-type KRAS) (N = 103), and tumors with both wild-type KRAS and BRAF (N = 387). The results of tumors with mutations in both KRAS and BRAF are not shown because of the small number of such tumors (N = 5). Figure 1 shows the frequencies of CIMP-0, CIMP-low, and CIMP-high among KRAS/BRAF wild-type tumors, KRAS-mutated tumors, and BRAF-mutated tumors. In contrast to CIMP-0, which was more common in KRAS/BRAF wild-type tumors (61%) than in KRAS-mutated tumors (52%, P = 0.02), CIMP-low was more common in KRAS-mutated tumors (44%) than in KRAS/BRAF wild-type tumors (30%, P = 0.0003). BRAF-mutated tumors showed a very high frequency of CIMP-high (71%) compared with KRAS/BRAF wild-type tumors (9.0%, P < 0.0001) and KRAS-mutated tumors (4.7%, P < 0.0001). We also examined the frequencies of KRAS mutations among CIMP-high, CIMP-low, and CIMP-0 tumors. CIMP-low tumors showed a significantly higher KRAS mutation rate (47%) than CIMP-high tumors (12%, P < 0.0001) and CIMP-0 tumors (37%, P = 0.007) (Figure 2).
Figure 1.
Frequencies of CIMP-0, CIMP-low, and CIMP-high in KRAS/BRAF wild-type (WT) tumors, KRAS-mutated (BRAF-wild-type) tumors, and BRAF-mutated (KRAS-wild-type) tumors. Integer numbers beside bars indicate the actual number of cases with a specific feature (such as CIMP-0, CIMP-low, or CIMP-high). Whereas the frequency of CIMP-0 is higher in KRAS/BRAF wild-type tumors than in KRAS-mutated tumors, the frequency of CIMP-low is higher in KRAS-mutated tumors than in KRAS/BRAF wild-type tumors.
Figure 2.
Frequencies of KRAS mutations among CIMP-0, CIMP-low, and CIMP-high tumors. The KRAS mutation frequency is significantly higher in CIMP-low tumors than in CIMP-high and CIMP-0 tumors.
We examined whether the relation between KRAS mutations and CIMP-low was modified by p53 status (because p53 mutations have been inversely related to CIMP-high).31 After tumors were stratified by p53 status, the frequencies of CIMP-low were higher among KRAS-mutated (BRAF wild-type) tumors than KRAS/BRAF wild-type tumors (Table 2). Thus, KRAS mutations were associated with CIMP-low independent of p53 status. There was no significant difference in CIMP-low frequencies between p53-negative tumors (31%) and p53-positive tumors (35%), although there was a difference in CIMP-high frequencies among p53-negative tumors (22%) and p53-positive tumors (9.1%, P < 0.0001).
Table 2.
Frequency of CIMP among Colorectal Cancer with Various p53, KRAS, and BRAF Status
| p53 | KRAS | BRAF | Total | CIMP-0 | CIMP-low | CIMP-high |
|---|---|---|---|---|---|---|
| Negative | Wild-type | Wild-type | 216 | 121 (56%) | 62 (29%) | 33 (15%) |
| Mutant | Wild-type | 175 | 95 (54%) | 72 (41%) | 8 (4.6%) | |
| Wild-type | Mutant | 79 | 7 (8.9%) | 12 (15%) | 60 (76%) | |
| Mutant | Mutant | 3 | 1 | 1 | 1 | |
| Total p53 negative tumors | 473 | 224 (47%) | 147 (31%) | 102 (22%) | ||
| Positive | Wild-type | Wild-type | 212 | 142 (67%) | 65 (31%) | 5 (2.4%) |
| Mutant | Wild-type | 116 | 56 (48%) | 54 (47%) | 6 (5.2%) | |
| Wild-type | Mutant | 34 | 3 (8.8%) | 9 (26%) | 22 (65%) | |
| Mutant | Mutant | 2 | 1 | 1 | 0 | |
| Total p53 positive tumors | 364 | 202 (55%) | 129 (35%) | 33 (9.1%) | ||
Numbers and percentages in bold indicate that the association of KRAS mutations with CIMP-low appear to be independent of p53 status. Also note that the association of BRAF mutations with CIMP-high is independent of p53 status.
Nine Subtypes of Colorectal Cancer According to MSI/CIMP Status
We subclassified colorectal cancers into nine subtypes according to status of both MSI and CIMP to examine effects of CIMP status (in particular, CIMP-low) on genetic status independent of MSI status. The nine subtypes were as follows: MSI-H CIMP-high, MSI-H CIMP-low, MSI-H CIMP-0, MSI-L CIMP-high, MSI-L CIMP-low, MSI-L CIMP-0, MSS CIMP-high, MSS CIMP-low, and MSS CIMP-0.
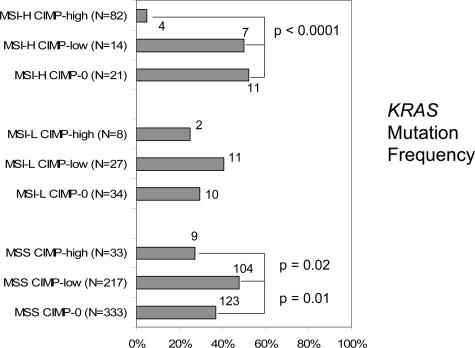
Figure 3 shows the frequencies of KRAS mutations among the nine subtypes of colorectal cancer. Among the MSS tumors, KRAS mutations were significantly more common in MSS CIMP-low tumors (48%) than in MSS CIMP-0 (37%, P = 0.01) and MSS CIMP-high tumors (27%, P = 0.02). Likewise, among the MSI-L tumors, KRAS mutations were more common in MSI-L CIMP-low tumors (41%) than in MSI-L CIMP-high (25%) and MSI-L CIMP-0 (29%), although statistical significance was not reached. Among the MSI-H tumors, there was no significant difference in KRAS mutation rates between MSI-H CIMP-low tumors (N = 14) and MSI-H CIMP-0 tumors (N = 21), partly due to the small numbers of cases.
Figure 3.
Frequencies of KRAS mutations in the nine subtypes of colorectal cancers. The KRAS mutation frequency is higher in CIMP-low tumors than in CIMP-high and CIMP-0 tumors among MSI-L/MSS tumors.
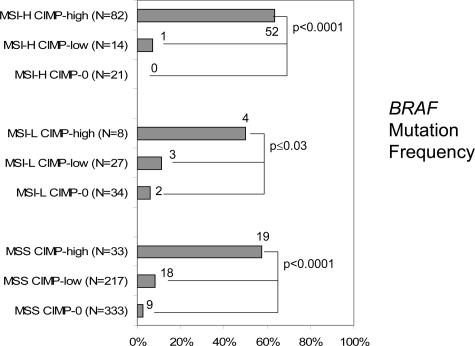
Figure 4 illustrates the frequencies of BRAF mutations in the nine subtypes of colorectal cancer. Regardless of MSI status, BRAF mutations were strikingly more frequent in CIMP-high tumors compared with CIMP-low and CIMP-0 tumors. Among MSS tumors, BRAF mutations were significantly more common in MSS CIMP-low tumors (8.6%) than in MSS CIMP-0 tumors (2.7%, P = 0.004).
Figure 4.
Frequencies of BRAF mutations in the nine subtypes of colorectal cancers. The BRAF mutation frequency is very high in CIMP-high tumors and low in CIMP-low and CIMP-0 tumors, regardless of MSI status.
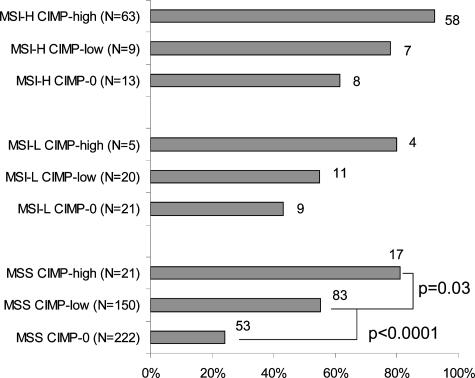
We also examined the frequencies of right-sided tumors among the nine subtypes of colorectal cancer (Figure 5). Among the MSS tumors, there was a statistically significant trend toward higher frequencies of right-sided tumors from CIMP-0 to CIMP-low and CIMP-high tumors (CIMP-high/CIMP-low versus CIMP-0, P < 0.0001; CIMP-high versus CIMP-low, P = 0.03). Similar trends were also present among MSI-H and MSI-L tumors, but differences were not statistically significant.
Figure 5.
Frequencies of right-sided tumors in the nine subtypes of colorectal cancers. The frequency of right-sided tumors seems to be higher in CIMP-high tumors than CIMP-0 tumors, and CIMP-low tumors show intermediate frequencies within each MSI type.
Discussion
We conducted this study to examine features of CIMP-low colorectal cancers compared with those of CIMP-high and CIMP-0 tumors. Our resource of a large number of relatively unbiased samples of colorectal cancer, obtained from two large prospective cohorts, has enabled us to precisely estimate the frequency of colorectal cancers with specific molecular features (such as MSI-H, CIMP-high, KRAS mutations, etc) at a population level.11 We have shown that CIMP-low tumors are associated with male sex, whereas CIMP-high tumors are associated with female sex. These intriguing opposite sex predilections of CIMP-low and CIMP-high tumors are independent of MSI status. We have also shown that CIMP-low tumors exhibit a higher KRAS mutation rate than CIMP-high and CIMP-0 tumors and that the association of CIMP-low with KRAS mutations appears to be independent of MSI status. In contrast to CIMP-low, CIMP-high is associated with female sex and BRAF mutations, and CIMP-0 is associated with wild-type KRAS/BRAF and does not have any sex predilection. These data do not support the hypothesis that CIMP-low is merely a mixture of CIMP-high and CIMP-0 tumors. If this hypothesis were true, one would expect CIMP-low tumors to show biological characteristics between CIMP-high and CIMP-0 tumors (such as a weak female predilection and a KRAS mutation rate between those of CIMP-high and CIMP-0 tumors). However, the associations of CIMP-low with a high KRAS mutation rate and male sex predilection are not compatible with this hypothesis. Thus, our data might raise an alternative hypothesis that CIMP-low tumors constitute a subtype of colorectal cancer that is different from CIMP-high and CIMP-0.
We admit that differences between CIMP-low and CIMP-0 tumors are not as clear-cut as those between CIMP-high and CIMP-low/CIMP-0. Further investigations are necessary to confirm or refute the existence of CIMP-low subtype of colorectal cancer. If CIMP-low indeed exists, further studies may also be necessary to develop a better panel of markers specific for CIMP-low to be clearly distinguished from CIMP-0. Nonetheless, some of the aforementioned associations were highly significant and less likely chance events. Our database of population-based samples from the prospective cohorts also makes the possibility of selection bias less likely.
We have introduced the term “CIMP-0” for tumors with 0/5 methylated promoters to be clearly distinguished from “CIMP-low.” Alternative terms for CIMP-0 would be “CIMP-negative” or “non-CIMP.” However, these alternative terms could be very confusing. So far there has been no study to address differences between CIMP-low and CIMP-0, and the terms “CIMP-negative” and “non-CIMP” have been used for both CIMP-low and CIMP-0 tumors. We believe that the term “CIMP-0” is unequivocal and, at this time, is the best term for tumors with no methylation in any of the five CIMP-specific promoters.
In this study, we determined CIMP status using five carefully selected gene promoters, including CACNA1G, CDKN2A (p16), CRABP1, MLH1, and NEUROG1,11 using MethyLight.22,23,24 Compared with qualitative methylation-specific PCR, MethyLight assays can reliably distinguish high from low levels of DNA methylation, the latter of which is likely have little or no biological significance.18,19 We have demonstrated a high degree of sensitivity (>90%) and/or specificity (>90%) of each of these five markers for the prediction of the overall CIMP status.11 Our criteria for CIMP positivity (tumors with ≥4/5 methylated promoters) were based on the finding of a clear bimodal distribution of the number of methylated markers11 and trends of KRAS and BRAF mutation frequencies according to the number of methylated promoters. In this study, we have found that all but one MSI-H tumors (N = 121) showed either ≥4/5 methylated promoters (CIMP-high) or ≤2/5 methylated promoters (CIMP-low/CIMP-0). In contrast, the definitions of CIMP used in most previous studies (≥50% methylated loci) were less rigid6,8 and may misdiagnose a substantial number of CIMP-low tumors as CIMP-high.
In fact, there are conflicting data in the literature regarding KRAS mutations in CIMP-high tumors. The original study on CIMP showed a higher KRAS mutation rate in CIMP-positive colorectal cancers than in CIMP-negative tumors.5 Independent of MSI status, higher frequencies of KRAS mutations in CIMP-high tumors compared with CIMP-low/negative tumors were demonstrated in some studies,7,8 whereas lower frequencies of KRAS mutations in CIMP-high tumors have been shown by our studies (Ref. 11 and this study). In this study, our results of high KRAS mutation rates in CIMP-low tumors can explain these conflicting data in the literature. Previous studies7,8 using the criteria for CIMP-high (≥2/4 or ≥2/5 methylated loci, respectively) were not strict, so a considerable number of CIMP-low tumors (frequently KRAS-mutated) might have been mixed into CIMP-high tumors (with low KRAS mutation rates).
Although substantial data on CIMP-high colorectal cancers have accumulated, there are little data on CIMP-low versus CIMP-0 colorectal cancers in the literature. Kambara et al9 described the frequencies of KRAS and BRAF mutations in CIMP-high (≥3/4 methylated markers), CIMP-low (1/4 to 2/4 methylated markers), and CIMP-negative (0/4 methylated markers) tumors. The authors showed a high KRAS mutation rate in CIMP-low tumors (43% = 19/44) compared with CIMP-negative tumors (29% = 10/34), although statistical significance was not reached. In contrast to our study, Kambara et al9 used methylation-specific PCR assays on MINT1, MINT2, MINT12, and MINT31 in 104 colorectal cancers and did not examine the effects of CIMP status independent of MSI status. An inverse association of KRAS mutation and MSI-H has been previously shown,32 and the effect of CIMP status on the KRAS mutation rate independent of MSI status should be evaluated.
The prognostic significance of CIMP-low has not been studied. Although biological significance of CIMP-low is still speculative at this point, analysis of patient survival or other outcomes will shed lights into biological significance to CIMP-low compared with CIMP-high and CIMP-0. Our prospective cohort studies, the Nurses’ Health Study (N = 121,700 followed since 1976)20 and the Health Professional Follow-up Study (N = 51,500 followed since 1986)21 are currently ongoing. Thus, relational data on patient survival and CIMP-high and CIMP-low will be available in the future.
In conclusion, CIMP-low colorectal cancer is associated with male sex and KRAS mutations independent of MSI status. Our data might raise the hypothesis that CIMP-low colorectal cancer is a different subtype from CIMP-high and CIMP-0 tumors. Further studies are necessary to confirm or refute this hypothesis.
Note:
Three markers (CDKN2A, CRABP1, and NEUROG1) have recently been shown to be associated with KRAS mutations after exclusion of BRAF-mutated tumors, suggesting that perhaps a separate KRAS-associated CIMP subgrouping exists with an overlapping set of methylation markers.33 These results are in agreement with our data. Compared to CACNA1G and MLH1, specificities of the three markers are slightly lower11 (ie, more frequently positive in CIMP-low tumors); hence, there are positive associations between the three markers and KRAS mutations.
Acknowledgments
We thank the Nurses’ Health Study and Health Professionals Follow-up Study cohort participants who have generously agreed to provide us with biological specimens and information through responses to questionnaires. We thank Graham Colditz, Walter Willett, and many other staff members who implemented and have maintained the cohort studies. We deeply thank Peter Laird, Daniel Weisenberger, and Mihaela Campan for their assistance in the development of the MethyLight assays.
Footnotes
This work was supported by National Institutes of Health grants P01 CA87969-03 and P01 CA55075-13.
No conflict of interest is declared.
References
- Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14 Spec No 1:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- Rashid A, Issa JP. CpG island methylation in gastroenterologic neoplasia: a maturing field. Gastroenterology. 2004;127:1578–1588. doi: 10.1053/j.gastro.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijnsoever M, Grieu F, Elsaleh H, Joseph D, Iacopetta B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002;51:797–802. doi: 10.1136/gut.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O’Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- Samowitz W, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, Kambara T, MacPhee DG, Young J, Leggett BA, Jass JR, Tanaka N, Matsubara N. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol. 2004;22:4584–4594. doi: 10.1200/JCO.2004.02.154. [DOI] [PubMed] [Google Scholar]
- Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner G, Weisenberger DJ, Campan M, Laird PW, Loda M, Fuchs CS. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Odze RD, Kawasaki T, Brahmandam M, Kirkner GJ, Laird PW, Loda M, Fuchs CS. Correlation of pathologic features with CpG island methylator phenotype (CIMP) by quantitative DNA methylation analysis in colorectal carcinoma. Am J Surg Pathol. 2006;30:1175–1183. doi: 10.1097/01.pas.0000213266.84725.d0. [DOI] [PubMed] [Google Scholar]
- Chan AO, Issa JP, Morris JS, Hamilton SR, Rashid A. Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol. 2002;160:529–536. doi: 10.1016/S0002-9440(10)64872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823–1830. doi: 10.1016/S0002-9440(10)61128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid A, Shen L, Morris JS, Issa J-PJ, Hamilton SR. CpG Island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129–1135. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jass JR. Serrated adenoma of the colorectum and the DNA-methylator phenotype. Nat Clin Pract Oncol. 2005;2:398–405. doi: 10.1038/ncponc0248. [DOI] [PubMed] [Google Scholar]
- Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573–580. doi: 10.1136/gut.2003.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, Fuchs C. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman WB, Rivenbark AG. Quantitative DNA methylation analysis: the promise of high-throughput epigenomic diagnostic testing in human neoplastic disease. J Mol Diagn. 2006;8:152–156. doi: 10.2353/jmoldx.2006.060026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colditz GA, Hankinson SE. The Nurses’ Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- Wei EK, Giovannucci E, Fuchs CS, Willett WC, Mantzoros CS. Low plasma adiponectin levels and risk of colorectal cancer in men: a prospective study. J Natl Cancer Inst. 2005;97:1688–1694. doi: 10.1093/jnci/dji376. [DOI] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Siegmund KD, Muller HM, Fiegl H, Marth C, Muller-Holzner E, Jones PA, Laird PW. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester TR, Skinner KA, Laird PW. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, Laird PW, Skinner KA. Fields of aberrant CpG island hypermethylation in Barrett’s esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–5026. [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Brahmandam M, Cantor M, Namgyal C, Kawasaki T, Kirkner G, Meyerhardt JA, Loda M, Fuchs CS. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- Ogino S, Meyerhardt JA, Cantor M, Brahmandam M, Clark JW, Namgyal C, Kawasaki T, Kinsella K, Michelini AL, Enzinger PC, Kulke MH, Ryan DP, Loda M, Fuchs CS. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005;11:6650–6656. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]
- Ogino S, Brahmandam M, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. Combined analysis of COX-2 and p53 expressions reveals synergistic inverse correlations with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Neoplasia. 2006;8:458–464. doi: 10.1593/neo.06247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, Robertson MA, Nichols MF, Gruenthal KM, Lynch BJ, Leppert MF, Slattery ML. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am J Pathol. 2001;158:1517–1524. doi: 10.1016/S0002-9440(10)64102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]