Abstract
Laboratory testing for dengue virus is used to confirm the diagnosis of dengue virus infection and to differentiate dengue from other febrile tropical illnesses. There are few data on the clinical use of reverse transcription-polymerase chain reaction (RT-PCR) for diagnosis of dengue virus infection. We prospectively evaluated 121 consecutive patients with possible dengue who had samples submitted for RT-PCR, IgM serology, and virus culture. Results were compared with the final discharge diagnosis. Semi-nested RT-PCR was performed using genus- and serotype-specific NS3 consensus primers. Results of 112 patients were available for the final analysis. The RT-PCR was positive in 40 of 62 patients with dengue. Patients who were RT-PCR-positive alone showed a mean of 4.4 days to RT-PCR positivity compared with 5.9 days in patients who were RT-PCR-negative and IgM serology-positive (P = 0.03, Mann-Whitney U-test). The sensitivity, specificity, negative predictive value, and positive predictive value were 70, 100, 84, and 100%, respectively, for samples analyzed within 5 days of illness onset. The RT-PCR also provided epidemiological data regarding the prevailing dengue virus serotypes: 25 with Den-2, eight with Den-3, and seven with Den-1 infection. We propose an algorithm of dengue testing that uses RT-PCR within 5 days of illness onset, whereas IgM capture enzyme-linked immunosorbent assay is preferred for those presenting later.
Dengue fever (DF) and dengue hemorrhagic fever (DHF) are caused by the dengue virus (family Flaviviridae, genus Flavivirus). Worldwide, it is considered to be the most important arthropod-borne disease with an estimated 50 to 100 million cases of DF annually and over 2.5 billion people at risk of the infection.1,2 DF and DHF are caused by four antigenically similar viruses, DEN-1 to DEN-4, but infection with one virus does not provide cross-immunity to infection with other serotypes. Symptoms of dengue fever are often nonspecific, frequently mimicking other illnesses such as influenza, malaria, chikungunya, leptospirosis, measles, typhus, or typhoid.3 Consequently, laboratory testing for DF and DHF is needed to confirm the diagnosis and to differentiate dengue from other febrile illnesses.
DF can be diagnosed by virus isolation, but the procedure is time consuming and generally requires a week of incubation. Currently, the most common method used by laboratories for the diagnosis of DF is detection of dengue antibodies using a capture enzyme-linked immunosorbent assay (ELISA) for IgM antibody or the detection of a rise in IgG antibody titer in paired sera. However, convalescent-paired sera for IgG antibodies are of little practical use in early management of patients. Furthermore, IgM antibodies are often absent in the acute phase of the illness and may be totally absent in secondary dengue virus infection. In addition, IgM antibodies may cross-react with other arboviral infections, eg, yellow fever or Japanese encephalitis. Therefore, there is a need to develop rapid laboratory diagnostic tools that will allow for early and accurate diagnosis and treatment.
The development of the polymerase chain reaction has facilitated the introduction of a number of rapid diagnostic assays for detection of viruses, including several for dengue viruses.4,5,6,7 However, most of these studies have been retrospective in design and were conducted on known culture-positive human samples or used for detection of dengue viral RNA from cell cultures and mosquitoes. Nevertheless, they confirm that nucleic-acid-based assays are promising tools that may shorten the time required for results and improve early diagnosis of DF. In addition, they may permit virus serotyping for epidemiological studies and can also be used to construct phylogenetic trees to monitor the evolution of geographic strains of dengue virus.8 However, these tests are rarely available outside a research center and are not routinely used in the clinical management of patients.
In an attempt to validate the diagnostic sensitivity, specificity, and appropriate clinical application of the RT-PCR for detection of dengue viruses in patients, we prospectively evaluated a semi-nested multiplex RT-PCR that is group- and serotype-specific for dengue viruses types 1 to 4. Simultaneous dengue virus culture and IgM capture ELISA serology were also performed.
Materials and Methods
Between 10 October 2002 and 31 January 2003, we enrolled consecutive patients with a febrile illness suspicious for DF or DHF at the National University Hospital, Singapore. This is a 921-bed tertiary referral hospital that is also the teaching hospital for the National University of Singapore Medical School. Patients who had samples submitted for both dengue virus RT-PCR and IgM serology were considered eligible for the study. Patients were followed prospectively until discharge from the hospital, and clinical and laboratory data were entered into a standardized Dengue Data Form. Patient demographics, clinical symptoms and signs, laboratory findings, and diagnoses were entered. Statistical calculations were performed using SPSS version 10.0 (SPSS, Chicago, IL).
Dengue Serology
Dengue serology was determined using a dengue IgM capture ELISA assay (Focus Technologies, Cypress, CA) according to the manufacturer’s recommendations. The test has a stated sensitivity of 85 to 95% and a specificity of 98%.
RT-PCR
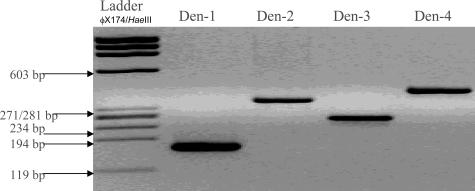
Genomic RNA was extracted from plasma samples using the Qiagen viral RNA mini kit (Qiagen GmbH, Heiden, Germany). Complementary DNA (cDNA) synthesis was performed by reverse transcription using 12.5 μl of the RNA template in a 25-μl final reaction volume with 200 units of Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA). Amplification was performed using a 5-μl sample cDNA in a 25-μl final reaction volume with 1 unit of Hotstar TaqDNA polymerase (Qiagen GmbH), 0.5 mmol/L MgCl2, 0.4 mmol/L each of dNTPs, and 0.4 μmol/L of each primer. A highly conserved area within the NS3 region of the dengue genome was amplified using consensus primers DV-1 (5′-GGRACKTCAGGWTCTCC-3′) and DV-3 (5′-AARTGIGCYTCRTCCAT-3′). The samples were amplified as follows: initial denaturation at 94°C for 10 minutes, followed by 35 cycles of denaturation (94°C for 1 minute), annealing (50°C for 30 seconds), and DNA molecule extension (72°C for 1 minute). Semi-nested PCR was then performed using serotype-specific primers DSP-1 (5′-AGTTTCTTTTCCTAAACACCTCG-3′), DSP-2 (5′-CCGGTGTGCTCRGCYCTGAT-3′), DSP-3 (5′-TTAGAGTYCTTAAGCGTCTCTTG-3′), and DSP-4 (5′-CCTGGTTGATGACAAAAGTCTTG-3′) and DV-1 (5′-GGRACKTCAGGWTCTCC-3′) (Table 1). Amplification was performed using the GeneAmp PCR Systems 9700 Thermocycler (Applied Biosystems). The detection limits of the semi-nested RT-PCR using the serotype-specific primers are 1 plaque-forming unit for dengue serotypes 1, 2, and 4 and 0.01 TCID50 for serotype 3. The dengue virus serotype-specific primers have been previously shown not to amplify DNA from the related flaviviruses Japanese encephalitis, West Nile (Kunjin), and yellow fever viruses.9 The samples were amplified as follows: initial denaturation at 94°C for 10 minutes, followed by 35 cycles of denaturation (94°C for 1 minute), annealing (50°C for 30 seconds), and DNA molecule extension (72°C for 1 minute). Amplified product was analyzed by electrophoresis in a 3% agarose gel (Figure 1).
Table 1.
Oligonucleotide Primers Used to Amplify and Type Dengue Viruses
| Primer | Sequence | Genome position | Size in bp of amplified DNA product |
|---|---|---|---|
| DV1 | 5′-GGRACKTCAGGWTCTCC-3′ | 4918 to 4934 | |
| DV3 | 5′-AARTGIGCYTCRTCCAT-3′ | 5384 to 5368 | 470 |
| DSP1 | 5′-AGTTTCTTTTCCTAA ACACCTCG-3′ | 5067 to 5045 | 169 |
| DSP2 | 5′-CCGGTGTGCTCRGCYCTGAT-3′ | 5279 to 5260 | 362 |
| DSP3 | 5′-TTAGAGTYCTTAAGCGTCTCTTG-3′ | 5174 to 5152 | 265 |
| DSP4 | 5′-CCTGGTTGATGACAAAAGTCTTG-3′ | 5342 to 5320 | 169 |
I, inosine; K, G/T; R, A/G; W, A/T; Y, C/T.
Figure 1.
Ethidium bromide-stained agarose gel of amplified DNA products. φX174/HaeIII DNA Ladder was used for size determination of bands.
Virus Culture
Virus culture was done using C6/36 Aedes albopictus cell line (CRL-1660; American Type Culture Collection Manassas, VA). In brief, an aliquot of patient plasma that was used for RT-PCR was inoculated onto a 90% confluent monolayer of cells. Liebovitz medium (Sigma-Aldrich, St. Louis, MO) supplemented with 5% fetal calf serum was added after adsorbing the serum to the cells at 32°C for 1 hour. The culture was incubated at 32°C and observed daily for cytopathic effect. At 7 days, the culture supernatant was harvested if cytopathic effect was observed in 75% or more of the monolayer. If no cytopathic effect was seen, the supernatant was passaged onto a fresh monolayer of C6/36 cells, and this was repeated twice. After harvesting the supernatant, the cell monolayer was scraped off with a rubber policeman and washed with phosphate-buffered saline and spotted onto a Teflon-coated slide. Flavivirus-, dengue complex-, and dengue serotype-specific antibodies (HB-112, HB-114, HB-46, HB-47, HB-48, and HB-49; American Type Culture Collection) were then used to confirm the isolation of dengue virus via indirect immunofluorescence.
Results
A total of 121 patients were considered eligible for the study. Nine patients were excluded from the final analysis; four did not have simultaneous dengue virus serology, and five patients did not have RT-PCR submitted. The results of 112 patients were available for analysis. Overall, 62 patients (55%) were assessed by investigators to have dengue fever based on established clinical criteria.2 The mean age of patients was 35.4 years; there were 41 males (66%) and 21 females (34%).
RT-PCR
The RT-PCR was positive in 40 of 62 dengue patients. The overall sensitivity and specificity were 64.5 and 100%, respectively. The median time to RT-PCR positivity was 4.5 days (mean, 4.5 days; range, 2 to 7 days). Analysis of blood samples examined within 5 days after the onset of fever showed an improved RT-PCR sensitivity of 70% with specificity of 100%. The positive predictive value and negative predictive value were 100 and 84%, respectively. In addition, the RT-PCR was positive in 15 patients (24%) who were assessed to have clinical dengue but were IgM serology-negative. Analysis of a subgroup of patients who were RT-PCR-positive alone showed a mean of 4.4 days to RT-PCR positivity compared with 5.9 days in patients who were RT-PCR-negative and IgM serology-positive (P = 0.03, Mann-Whitney U-test). The prevailing dengue serotypes were as follows 25 with Den-2, eight with Den-3, and seven with Den-1 infection.
Serology
IgM serology was positive on initial testing in 43 of 62 (69%) dengue patients (mean number of days of illness: 5.5 days, range 3 to 11 days). Another 13 patients had positive IgM serology on repeat convalescent testing of samples. The overall sensitivity and specificity of the IgM serology was 90.3 and 80%, respectively. The positive predictive value and negative predictive value were 85 and 87%, respectively. False positive IgM serology results were found in 10 patients: three patients with typhus, one with HIV infection, one with acute hepatitis B, one with malaria, one with systemic lupus erythematosus flare, and three with bacterial sepsis.
Virus Culture
Virus culture was performed on 105 plasma samples. Seven samples were not examined for culture because of insufficient plasma. Only 11 plasma samples were positive by virus culture. All these samples were also positive by RT-PCR, and there was 100% concordance between the dengue serotypes identified by virus culture and RT-PCR.
Discussion
RT-PCR has been developed for the diagnosis of several infectious diseases, and in recent years, it has been revolutionizing the laboratory diagnosis of viral infections. This method is rapid, sensitive, specific, and can be used for genome detection in human clinical samples, biopsies, autopsy tissues, or mosquitoes.10,11 In this study, we prospectively evaluated the use of a semi-nested dengue virus RT-PCR assay with IgM serology and virus culture to determine the appropriate clinical application and diagnostic value of RT-PCR for the diagnosis of DF/DHF. In Singapore, both DF and DHF are legally notifiable diseases based on clinical criteria without waiting for laboratory confirmation, and hence, DF and DHF is primarily a clinical diagnosis.12
We found that the RT-PCR assay was a useful diagnostic tool for samples collected in the first 5 days of illness with a sensitivity of 70% (mean of 4.4 days to RT-PCR positivity compared with 5.9 days in patients who were RT-PCR-negative, P = 0.03, Mann-Whitney U-test). The RT-PCR also detected dengue virus in 15 patients in the acute phase of illness who were assessed to have clinical dengue but were dengue virus IgM ELISA-negative. Overall, our results agree with the observations made by Gubler et al,13 Vaughn et al,14 and Lanciotti et al4 that virus isolation or RT-PCR is only successful in the early phase of illness as dengue viremia is short-lived.
Uncomplicated DF is generally a 5- to 7-day illness, and IgM antibodies in primary dengue virus infection appear within 2 days of defervescence and peak at 2 weeks but may be absent in secondary dengue infection.15 We observed a similar result in our study; the IgM capture ELISA was only positive in 43 of 62 (69%) dengue patients during initial testing (mean number of days of illness at positive testing 5.5 days). Repeat convalescent testing yielded another 13 positive samples for an overall sensitivity of 90.3%. In addition, we found that serological diagnosis of dengue was less specific than RT-PCR, 80% (with 10 false positive results) versus 100%, respectively. Dengue IgM antibodies may remain detectable for up to 3 months after infection, and interpretation of positive serology can be difficult.16 It is thus worth emphasizing the importance of correlating serology results with patient clinical data and recognizing the study limitations involved in the use of dengue IgM antibody testing alone.
The relatively low sensitivity of RT-PCR in our study may be due to the fact that the majority of samples were submitted after day 5 of illness onset. The mean calculated number of days at RT-PCR submission was 6.3 days. This is a reflection of the fact that patients were hospitalized late in their illness, mean of 4.5 days, which is consistent with the findings of other studies of hospitalized dengue patients in Singapore.17,18 Our findings suggest that an improved sensitivity may be obtained if RT-PCR is used early during illness, ideally within 5 days of the illness, or in the outpatient setting when patients are first assessed (the use of RT-PCR in outpatients is currently being evaluated in the EDEN study—Early Dengue infection—in Singapore and has shown excellent sensitivity; B. Sil, personal communication). In addition, our RT-PCR protocol did not control for the presence of PCR inhibitors that might account for some loss of sensitivity.
Although there was 100% concordance for dengue virus serotyping between RT-PCR and virus culture, the RT-PCR was much faster, 8 hours compared with >7 days, and we found virus culture to have a lower sensitivity in our study. This may be due to the fact that most other studies comparing virus culture and RT-PCR were conducted retrospectively using patient samples that were known to be dengue virus culture-positive. We wanted to limit the discomfort of repeated venepuncture in our study, so virus culture was performed using the remaining plasma that had been collected for RT-PCR. The plasma was stored at −70°C before transport to the virology laboratory where it was thawed for virus culture. The delay in set-up for virus culture, freeze-thawing of samples and the use of plasma instead of serum for culture may account for the lower rates of virus isolation.
We found that a useful characteristic of the RT-PCR assay is its ability to accurately identify the dengue serotype responsible for ongoing disease transmission. Changes in the dominant circulating dengue serotype have been associated with epidemics of dengue infection and are thus of epidemiological relevance.19 In addition, a prompt response to vector control is an important component of interrupting dengue transmission. However, this is dependent on rapid case notification. Therefore, RT-PCR may reduce the time between case recognition and the initiation of emergency control operations needed to identify vector-breeding sites and to begin source control and fogging.
Laboratory testing for dengue using a combination of RT-PCR and IgM serology allows for rapid diagnosis and is able to differentiate dengue virus infection from other febrile tropical illnesses. We propose that for hospitalized patients a useful algorithm for dengue confirmation is to reserve RT-PCR testing for those patients who are admitted within 5 days of illness onset and to use the IgM capture ELISA in those patients admitted after day 5 of illness. This has now become an accepted protocol for dengue testing at our hospital and is an evidence-based approach to test selection. We believe that this testing algorithm should also be applicable in nonendemic countries such as the United States, where the disease is likely to be increasingly encountered in returning travelers.20
Footnotes
None of the authors have any potential conflict of interest.
Current address of K.S.: Rush University Medical Center, Chicago, IL.
Current address of E.E.O.: Defense Medical and Environmental Research Institute, DSO National Laboratories, Singapore.
References
- Clarke T. Dengue virus: break-bone fever. Nature. 2002;416:672–674. doi: 10.1038/416672a. [DOI] [PubMed] [Google Scholar]
- Rigau-Perez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, Vorndam AV. Dengue and dengue hemorrhagic fever. Lancet. 1998;352:971–977. doi: 10.1016/s0140-6736(97)12483-7. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Dengue and dengue hemorrhagic fevers of southeast Asia. Yale J Biol Med. 1965;37:434–454. [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Calisher CH, Gubler DJ, Chang G, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu PY, Chang SF, Kuo YC, Yueh YY, Chien LJ, Sue CL, Lin TH, Huang JH. Development of group and serotype specific one step SYBR green I-based real time reverse transcriptase PCR assay for dengue virus. J Clin Microbiol. 2003;41:2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Takasaki T, Yamada K, Nerome R, Tajima S, Kurane I. Development and evaluation of fluorogenic TaqMan reverse transcriptase PCR assays for detection of dengue virus types 1 to 4. J Clin Microbiol. 2004;42:5935–5937. doi: 10.1128/JCM.42.12.5935-5937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudiro TM, Ishiko H, Green S, Vaughn DW, Nisalak A, Kalayanarooj S, Rothman AL, Raengsakulrach B, Janus J, Kurane I, Ennis FA. Rapid diagnosis of dengue viremia by reverse transcriptase polymerase chain reaction using 3′-noncoding region universal primers. Am J Trop Med Hyg. 1997;56:424–429. doi: 10.4269/ajtmh.1997.56.424. [DOI] [PubMed] [Google Scholar]
- Chow VTK, Seah CLK, Chan YC. Comparative analysis of NS3 sequences of temporally separated dengue 3 virus strains isolated from Southeast Asia. Intervirology. 1994;37:252–258. doi: 10.1159/000150386. [DOI] [PubMed] [Google Scholar]
- Seah CLK, Chow VTK, Chan YC. Semi-nested PCR using NS3 primers for the detection and typing of dengue viruses in clinical serum specimens. Clin Diagn Virol. 1995;4:13–20. doi: 10.1016/0928-0197(94)00063-z. [DOI] [PubMed] [Google Scholar]
- Deubel V, Laille M, Hugnot JP, Chungue E, Guesdon JL, Drouet MT, Bassot S, Chevrier D. Identification of dengue sequences by genomic amplification: rapid diagnosis of dengue virus serotypes in peripheral blood. J Virol Methods. 1990;30:41–54. doi: 10.1016/0166-0934(90)90042-e. [DOI] [PubMed] [Google Scholar]
- Deubel V, Pierre V. Molecular techniques for rapid and more sensitive detection and diagnosis of flavivirus. Spencer RC, Wright EP, Newsom SWB, editors. Andover, UK: Intercept; Rapid Methods and Automation in Microbiology and Immunology. 1994:227–237. [Google Scholar]
- Goh KT. Epidemiology of dengue haemorrhagic fever in Singapore. Asian J Infect Dis. 1978;2:25–29. [Google Scholar]
- Gubler DJ, Suharyono W, Tan R, Abidin M, Sie A. Viremia in patients with naturally acquired dengue infection. Bull World Health Organ. 1981;59:623–630. [PMC free article] [PubMed] [Google Scholar]
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Rothman AL, Ennis FA, Nisalak A. Dengue in the early febrile phase: viremia and antibody response. J Infect Dis. 1997;176:322–330. doi: 10.1086/514048. [DOI] [PubMed] [Google Scholar]
- Kuno G, Gomez I, Gubler DJ. An ELISA procedure for the diagnosis of dengue infections. J Virol Methods. 1991;33:101–113. doi: 10.1016/0166-0934(91)90011-n. [DOI] [PubMed] [Google Scholar]
- Gubler DJ. Laboratory Diagnosis of dengue virus infections. Gubler DJ, Kuno G, editors. Oxford: CABI; Dengue and Dengue Haemorrhagic fever. 1997:313–333. [Google Scholar]
- Tai DYH, Chee YC, Chan KW. The natural history of dengue illness based on a study of hospitalized patients in Singapore. Singapore Med J. 1999;40:238–242. [PubMed] [Google Scholar]
- Heng BH, Goh KT, Ong YW, Teo D, Ooi PL. Clinical epidemiology of dengue fever and dengue hemorrhagic fever in Singapore. Goh KT, editor. Singapore: Institute of Environmental Epidemiology, Ministry of the Environment, Singapore; Dengue in Singapore. 1998:80–86. [Google Scholar]
- Gubler DJ. History of dengue and dengue hemorrhagic fever. Gubler DJ, Kuno G, editors. Oxford: CABI; Dengue and Dengue Haemorrhagic fever. 1997:1–22. [Google Scholar]
- Grobusch MP, Niedrig M, Gobels K, Klipstein-Grobusch K, Teichmann D. Evaluation of the use of RT-PCR for the early diagnosis of dengue fever. Clin Microbiol Infect Dis. 2006;12:395–397. doi: 10.1111/j.1469-0691.2006.01353.x. [DOI] [PubMed] [Google Scholar]