Abstract
Epstein-Barr virus (EBV) is associated with a wide range of benign and malignant diseases, including infectious mononucleosis, lymphoma, posttransplant lymphoproliferative disorder, and nasopharyngeal carcinoma. Measurement of EBV viral load in plasma is increasingly used for rapid assessment of disease status. We evaluated the performance characteristics of an EBV polymerase chain reaction assay that uses commercial reagents and instruments from Roche Diagnostics (Indianapolis, IN). DNA was extracted from plasma using a MagNaPure instrument, and viral load was measured by real-time polymerase chain reaction on a LightCycler. Analyte-specific reagents included primers and hybridization probes targeting the EBV LMP2 gene and a spiked control sequence. Accuracy and reproducibility were established using DNA from three cell lines. The assay was sensitive to approximately 750 copies of EBV DNA per milliliter of plasma and was linear across at least four orders of magnitude. The assay detected EBV DNA in three of five samples from nasopharyngeal carcinoma patients, seven of nine infectious mononucleosis samples, and 34/34 samples from immunosuppressed patients with clinically significant EBV-related disease, whereas EBV DNA was undetectable in plasma from 21 individuals without EBV-related disease. In conclusion, this LightCycler EBV assay is rapid, sensitive, and linear for quantifying EBV viral load. The assay appears to be useful for measuring clinically significant EBV levels in immunodeficient patients.
Viral load measurement is increasingly used in clinical laboratories to assist in diagnosing and monitoring virus-associated diseases. Primary Epstein-Barr virus (EBV) infection is characterized by high plasma viral load that declines to undetectable levels as the immune system recognizes and controls the infection.1,2,3 Periodic reactivation is accompanied by transient viremia and shedding of virions in saliva.4 Some patients later develop EBV-related neoplasms that are characterized by high circulating levels of EBV DNA.5 Therefore, EBV viral load assays not only detect active infection but also serve as a tumor marker for certain forms of malignancy.
Tumors that are almost universally EBV-associated include posttransplant lymphoproliferative disorder and nasopharyngeal carcinoma, whereas cancers such as acquired immune deficiency syndrome (AIDS)-related lymphoma and Hodgkin lymphoma harbor EBV in only about half of the cases.5 When a cancer is EBV-related, the viral DNA appears to be present in virtually all of the malignant cells, thus serving as a marker for the tumor clone.6 However, the EBV genome is not restricted to malignant cells as evidenced by high levels of EBV DNA in whole blood and in fractions thereof.7 Circulating EBV DNA levels are often elevated at the time of initial diagnosis and, in some cases, even before the cancer is clinically evident.5,8,9,10,11 On effective treatment, EBV load declines, suggesting that EBV DNA as a measure of tumor burden is useful for monitoring efficacy of therapy and early relapse.5,8,12,13
The advent of real-time polymerase chain reaction (PCR) has greatly facilitated the quantitation of viral DNA in human blood and tissue samples. Plasma is a convenient sample type because EBV DNA levels are usually very low or undetectable in plasma of healthy individuals, whereas levels are elevated in conjunction with active EBV infection and many EBV-related malignancies.5,14,15 The EBV found in plasma or serum usually exists in the form of naked DNA rather than as encapsidated virions, except in infectious mononucleosis where virions are also commonly present.1,16 The cell-free DNA associated with cancers is presumably derived from apoptosis or necrosis of infected malignant cells as suggested by strain identity between plasma and tumor compartments.17,18,19,20
Well-designed real-time PCR assays are sensitive, specific, reproducible, and linear across a wide dynamic range. In addition, because amplicons are sequestered inside a closed vessel, the risk of amplicon contamination is minimal. Accuracy, precision, and linearity of real-time assays are theoretically better than with end-point product quantitation methods. Technologist time is also lower than with gel-based detection, even more so when robotic systems are used to facilitate extraction. A variety of real-time probe design strategies are feasible, including TaqMan, molecular beacons, and fluorescence resonance energy transfer.21,22,23,24 The combination of two primers and one or more internal probes, as well as the potential for melt-temperature analysis of the probe binding region in certain assay designs, helps assure target specificity. Finally, real-time PCR assays are more rapid than gel-based PCR assays, thus allowing prompt interpretation of test results.
In the current study, we implemented a commercial real-time PCR assay for EBV DNA, and we evaluated its performance characteristics. The assay relies on an automated extractor, a rapid thermocycler, and analyte-specific reagents. A prior study by Ruiz et al evaluated a kit version of this assay that is marketed by Roche Molecular Diagnostics in Europe,23 and another study by Le et al demonstrated the clinical utility of the Roche EBV PCR reagents in nasopharyngeal carcinoma patients.24 Our study is the first to use EBV genomic DNA prepared from cell lines to evaluate assay sensitivity, accuracy, reproducibility, and linearity. DNA from other herpesviruses was used to test specificity. Plasma samples from patients with various EBV-related diseases and from controls without EBV viremia were used to assess clinical applicability.
Materials and Methods
EBV Viral Load Measurement
A commercial EBV viral load assay was validated using three instruments from Roche Molecular Diagnostics (Indianapolis, IN), namely a Roche MagNaPure extractor, a Roche LightCycler real-time thermocycler, and a Roche LightCycler Carousel Centrifuge. Analyte specific reagents targeting the EBV latent membrane protein 2 (LMP2) gene and a spiked control sequence were also from Roche Molecular Diagnostics. Sequences of primers and probes are proprietary. Detailed methods are described to encourage standardized procedures that permit results to be compared across laboratories.
The analyte-specific reagents are provided in separate vials and include 1) Lightcycler EBV Primer/Hybridization Probes, 2) Lightcycler EBV Recovery Template, which is spiked into the sample before extraction, and 3) EBV Template DNA, which represents a series of prediluted standards by which to quantitate the virus. Reagents were stored frozen at −15 to −25°C until use, at which time the vials were thawed and gently flicked to dissolve any precipitate and then stored refrigerated at 4°C protected from light for up to 1 month.
Blood anticoagulated with EDTA was centrifuged at 3000 rpm for 10 minutes, and plasma was removed to a separate tube and frozen until analysis. A 200-μl aliquot of thawed plasma was spiked with 5 μl of recovery template (Roche Molecular Diagnostics), which is a synthetic plasmid mimic sequence having the same primer binding sites but a different internal probe recognition site from the natural EBV LMP2 sequence. Total DNA was extracted on a MagNaPure instrument (Roche Molecular Diagnostics) using the manufacturer’s external lysis protocol and extraction reagents (Total Nucleic Acid Isolation Kit; Roche Molecular Diagnostics) to yield 100 μl of eluate. EBV LMP2 DNA was amplified in duplicate by real-time PCR on the LightCycler (version 1.5). The spiked control sequence was coamplified in the same capillary. To test for false positive results due to reagent contamination, each run contained a control in which no template was added to the remaining reagents.
The LightCycler-FastStart DNA Master Hybridization Probes (vial 1b) was centrifuged, and then 60 μl of the reaction mix was pipetted into vial 1a of FastStart enzyme and mixed gently by pipetting up and down without vortexing. The mixture was relabeled using new labels provided by the manufacturer, one for the top of the cap and one for the side of the vial, to indicate that the mixture is a ready-to-use 10× “hot start” master mix. This reagent was aliquoted and stored refrigerated at 2 to 8°C for a maximum of 1 week. Each 20-μl reaction contained 5 μl of template DNA, 2 μl of 10× “hot start” master mix (prepared from vials 1a and 1b), 2 μl of Lightcycler EBV Primer/Hybridization Probes, and 11 μl of deionized distilled water (ddH2O).
The LightCycler was programed to automatically perform melt-curve analysis after amplification was complete. The cycling parameters included an initial denaturation step at 95°C for 10 minutes, followed by 45 cycles at 95°C for 10 seconds, annealing at 55°C for 15 seconds, and extension at 72°C for 15 seconds, with a 20°C/second transition time. The melt-curve parameters were 95°C for 0 seconds, 40°C for 60 seconds with a temperature transition rate of 20°C/second, and finally, a slow rise in the temperature to 80°C at a rate of 0.1°C/second with continuous acquisition of fluorescence data. LightCycler software (version 3.5) was used to evaluate amplification products in two fluorescence channels of the LightCycler. EBV product was visualized in the F2/back-F1 channel while the spiked control product was visualized in the F3/back-F1 channel. Calibration of the LightCycler must be done at least semiannually to diminish bleed through of fluorescence from one channel to another.
The stock standards (EBV Template DNA; Roche Molecular Diagnostics) represent serial 10-fold concentrations of 106 to 102 copies of EBV DNA per 5 μl, and 5 μl of each standard DNA was added as template to each of five reactions in every run. As the run progresses, Lightcycler software automatically displays an amplification curve in which accumulating fluorescence is visible. At the conclusion of the run, the software produces a graph of the standard curve of log concentration (x axis) versus crossing point (y axis) across the five standards. In this study, the slope of the standard curve was between −3.14 and −3.64, spanning the ideal slope value of −3.33.
The viral load for each experimental sample was extrapolated and automatically calculated by the LightCycler software, in copies per reaction. The duplicate values were averaged and then manually multiplied by the dilution factor of 100 to obtain an EBV viral load in copies of EBV DNA per ml of plasma. For purposes of data analysis, undetectable EBV DNA was reported as zero copies.
The assay described above, which relies on analyte-specific reagents from Roche, will heretofore be called assay 1. For comparison purposes, EBV viral load was also measured using assay 2 and assay 3, which are both in-lab-developed real-time PCR assays in which primers and a TaqMan probe target the BamH1W reiterated EBV sequence in 96-well plates on an Applied Biosystems platform, namely the ABI Prism 7900 instrument for assay 2 and the ABI Prism 7500 instrument for Assay 3 [Applied Biosystems, Inc. (ABI), Foster City, CA]. Reaction content and cycling parameters are as previously described,25 except that assay 2 used 10 μl of template DNA per 50 μl PCR, whereas assay 3 used 5 μl of template DNA per 25 μl of PCR and did not coamplify a spiked control sequence. Assay 3 was performed on an aliquot of the same MagNaPure-extracted eluate as assay 1, whereas assay 2 had to be performed on a separate plasma aliquot that had been spiked with a different control sequence (TaqMan Exogenous Internal Positive Control DNA; Applied BioSystems) before MagNaPure extraction to allow for multiplex amplification of the control sequence. Assay 2 and assay 3 both relied on standards prepared from the Namalwa Burkitt lymphoma cell line.25 Both assay 2 and assay 3 were run in duplicate, and viral load was calculated from the average copies measured per PCR.
Assay Performance Characteristics
EBV DNA prepared from three different cell lines was used to test assay sensitivity, accuracy, and linearity. The Namalwa Burkitt lymphoma cell line DNA was 10-fold serially diluted in ddH2O, representing 0.582 copies to 58,200 copies of EBV genome. B95.8 viral DNA (Advanced Biotechnologies, Inc., Columbia, MD) was 10-fold serially diluted in ddH2O, representing 6.5 copies to 65,000 copies, and P3HR1 viral DNA (Advanced Biotechnologies, Inc.) was 10-fold serially diluted in ddH2O, representing 7.5 copies to 75,000 copies of the EBV genome. In addition, to mimic actual patient samples, Namalwa cell line DNA was spiked into EBV-negative plasma at serial 10-fold dilutions such that, assuming 100% recovery of spiked EBV DNA after extraction, the 5-μl template used in each PCR would represent input levels of 0.582 to 58,200 EBV DNA copies per PCR. For the Namalwa cell line DNA, EBV DNA concentration had been determined by spectrophotometric measurement of genomic DNA content in concert with prior research showing two integrated copies of EBV genome per cell.26 For the B95.8 and P3HR1 products, both of which are purified viral DNA, the manufacturer (Advanced Biotechnologies, Inc.) specified the concentration of EBV genome. Assay specificity was tested by analyzing other herpes family viruses: cytomegalovirus, herpes simplex virus, Kaposi’s sarcoma-associated herpesvirus, and varicella-zoster virus. For each virus, 5 μl of purified viral DNA (Advanced Biotechnologies, Inc.) was used as PCR template.
Clinical Samples
Residual plasma (n = 69) was retrieved from the archives of the University of North Carolina and affiliated hospitals. A 200-μl aliquot of this plasma had been previously assayed for EBV viral load for purposes of patient care using assay 2 on an ABI 7900 instrument, and residual plasma was frozen at −20°C until the time it was thawed, and a second 200-μl aliquot was spiked and extracted for analysis by both the LightCycler assay 1 and the ABI 7500 assay 3. Plasma samples were selected to represent a variety of benign and malignant disorders having high, medium, low, or undetectable EBV loads by assay 2. Clinical information was obtained from medical records. In addition, EDTA blood from healthy blood donors was retrieved and centrifuged to prepare plasma. This plasma was mixed with EBV-positive plasma to create high volume plasma pools that were used to test assay precision and reproducibility. This study was approved by our Institutional Review Board.
Results
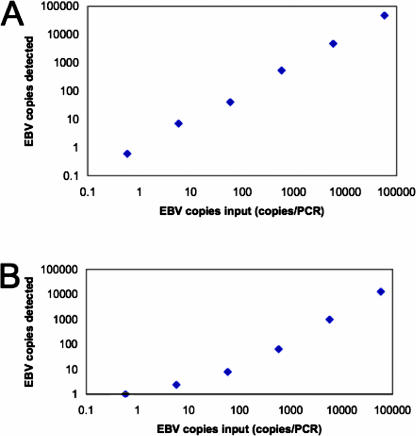
The performance characteristics of assay 1 were examined using a series of DNA samples prepared from cell lines. First, DNA from the Namalwa Burkitt lymphoma cell line was serially diluted in water and used as template for PCR to test assay sensitivity and linearity. Assay 1 was sensitive to the lowest tested level (0.58 copies of EBV DNA) and was linear over the entire 5-log range up to the highest available level of 58,200 copies of template (slope 1.2 versus ideal slope of 1.0, correlation coefficient 0.99 versus ideal of 1.0). We recognize that 0.58 copies of EBV DNA is an absurd value, because the targeted segment of the EBV genome was either present at one or more copies per reaction or not present at all; see below for further data and discussion on this point with regard to accuracy and reproducibility.
When Namalwa DNA was spiked into plasma and MagNaPure-extracted before being used as template, remarkably, the assay was still sensitive to 0.58 copies of template EBV DNA, although assay linearity was somewhat compromised (see Figure 1). On average, 79% of EBV DNA was lost in the extraction process (range, 59 to 88%; see Table 1). For viral DNA prepared from the P3HR1 Burkitt lymphoma cell line, the assay was sensitive to 7.5 copies per PCR. The assay was not quite as sensitive when tested on viral DNA prepared from the B95.8 lymphoid cell line, detecting 65 but not 6.5 template copies. It should be kept in mind that variation in assay sensitivity across the various cell lines may reflect inaccuracies in the assigned concentration of EBV DNA in each cell line sample.
Figure 1.
EBV assay 1 performed on the Roche Lightcycler is sensitive, accurate, and linear across a 5-log range. Genomic DNA from the Namalwa Burkitt lymphoma cell line was used as template in PCR (A) or was spiked into plasma and then extracted on the MagNaPure instrument before using as template in PCR (B). The amount of input EBV DNA is shown on the x axis, and the amount measured by the assay is shown on the y axis.
Table 1.
Assay Performance as Determined by Comparing Input Template Levels to Measured EBV Viral Load for Each of Four Serially Diluted DNA Samples
| Input EBV DNA, copies/PCR | LightCycler assay 1, EBV viral load, copies per PCR | Column 2 to column 1 ratio, % | ABI 7500 assay 3, EBV viral load, copies per PCR |
|---|---|---|---|
| Namalwa Burkitt Lymphoma Cell Line Genomic DNA | |||
| 58,200 | 48,750 | 84% | Not done* |
| 5820 | 4896 | 84 | Not done* |
| 582 | 543 | 93 | Not done* |
| 58.2 | 41 | 70 | Not done* |
| 5.82 | 7 | 120 | Not done* |
| 0.58 | 0.6 | 103 | Not done* |
| 0 | 0 | 0 | |
| Purified P3HR1 EBV DNA | |||
| 75,000 | 4424 | 6% | 2299 |
| 7500 | 470 | 6 | 326 |
| 750 | 41 | 5 | 26 |
| 75 | 2.6 | 3 | 3 |
| 7.5 | 1.4 | 19 | 0.4 |
| 0.75 | 0 | 0 | |
| 0 | 0 | 0 | |
| Purified B95.8 EBV DNA | |||
| 65,000 | 5630 | 9% | 3621 |
| 6500 | 573 | 9 | 565 |
| 650 | 61 | 9 | 60 |
| 65 | 2.9 | 4 | 0.4 |
| 6.5 | 0 | 0 | |
| 0.65 | 0 | 0 | |
| 0 | 0 | 0 | |
| Namalwa Burkitt cell line genomic DNA spiked into plasma | |||
| 58,200† | 12,930 | 22% | 15,169 |
| 5820† | 1003 | 17 | 1733 |
| 582† | 65 | 11 | 210 |
| 58.2† | 8 | 14 | 28 |
| 5.82† | 2.4 | 41 | 6 |
| 0.58† | 1 | 172 | 0.5 |
| 0 (plasma only) | 0 | 0 | Not done |
Serial dilutions of Namalwa DNA were used as the standard for assay 3, and therefore, it was inappropriate to use serial dilutions of Namalwa DNA to test the performance characteristics of assay 3.
For Namalwa DNA spiked into plasma and extracted before PCR, input EBV DNA levels are shown assuming that no EBV DNA was lost during extraction.
Our assessment of accuracy varied by the source of the EBV DNA (see Table 1). The results in the Namalwa cell line, when used directly as template for PCR, were remarkably accurate, with an average correlation of 93% between the amount of EBV template put into the reaction and the viral load measured by assay 1. For the P3HR1 cell line virus, there was a much greater difference between input and measured levels of EBV DNA, with the proportion of measured to expected levels averaging 6%. For the B95.8 cell line virus, there was also a substantial difference, averaging 8% of expected levels. Despite the magnitude of the differences, there was a fairly consistent proportional difference across each dilution series of a given stock DNA, suggesting that the “gold standard” values for viral DNA concentration that had been assigned to each DNA stock were not always accurate. We had no way of determining which stock had the most accurate assigned EBV concentration.
The viral loads measured by assay 1 and assay 3 were quite similar to each other, both in terms of absolute copies detected per PCR and in terms of sensitivity to low levels of input DNA (see Table 1). This correlation seems remarkable given that different standards were used in each of the two assays.
Specificity was tested by analyzing purified DNA from four other herpesvirus family members. Assay 1 did not cross-react with any of the other viruses tested including cytomegalovirus, herpes simplex virus, Kaposi’s sarcoma-associated herpesvirus, and varicella-zoster virus.
Precision and Reproducibility
Intra-assay precision and interassay reproducibility were assessed by first creating an EBV-positive plasma pool and then making 1:10 and 1:100 dilutions in normal plasma. Samples of the original pool and each dilution were aliquoted into three vials each and extracted on the MagNaPure for a total of nine extractions and then analyzed on the LightCycler in duplicate on 3 separate days for a total of 54 PCR results. Extracted DNA was stored at 4°C before analysis. Intra-assay coefficient of variation ranged from 4 to 8% in the pool with the highest EBV load, 15 to 20% in middle pool, and 22 to 47% in lowest pool, which contained EBV DNA at a level approaching the lower limit of detection. EBV DNA was detectable in all 18 replicates, even in the lowest pool. This result implies that the analytic sensitivity of the LightCycler assay is no worse than the level present in the lowest pool (12 copies per reaction, which is equivalent to 1200 copies per ml of plasma). Interassay coefficient of variation between run 1, run 2, and run 3 was 8, 20, and 40% in the highest, middle, and lowest sample pools, respectively (see Table 2).
Table 2.
Intra-Assay Precision and Inter-Assay Reproducibility
| Assay type | Run no. | Concentration | EBV viral load (copies/PCR)
|
|||
|---|---|---|---|---|---|---|
| Mean | Range | SD | CV% | |||
| Intra-assay* | Run 1 | Original | 881 | 761 to 944 | 70 | 8 |
| 10-fold dilution | 107 | 75 to 124 | 19 | 18 | ||
| 100-fold dilution | 12 | 8 to 23 | 5 | 47 | ||
| Run 2 | Original | 771 | 722 to 802 | 29 | 4 | |
| 10-fold dilution | 83 | 60 to 106 | 16 | 20 | ||
| 100-fold dilution | 9 | 5 to 12 | 3 | 31 | ||
| Run 3 | Original | 834 | 782 to 888 | 46 | 6 | |
| 10-fold dilution | 103 | 83 to 126 | 15 | 15 | ||
| 100-fold dilution | 16 | 12 to 21 | 4 | 22 | ||
| Interassay† | Run 1–3 | Original | 829 | 722 to 944 | 67 | 8 |
| 10-fold dilution | 98 | 60 to 126 | 19 | 20 | ||
| 100-fold dilution | 12 | 5 to 23 | 5 | 40 | ||
SD, standard deviation; CV, coefficient of variation.
Means of six PCR replicates.
Means of 18 PCR replicates.
Application to Human Plasma Specimens
A total of 69 human plasma samples were assayed by LightCycler assay 1. Twenty-one of these plasma samples were from 21 patients or donors who were thought to be free of EBV-related disease based on clinical findings and on absence of viremia as shown by undetectable EBV DNA by the ABI 7900 assay 2. The LightCycler assay 1 revealed undetectable EBV DNA in all 21 samples. Thus, the two assays were in complete agreement in all 21 specimens lacking evidence of EBV-related disease.
Forty-eight residual plasma samples were evaluated from 35 patients who were thought to have EBV-related disease based on clinical findings and viremia as shown by measurable EBV viral load by the ABI 7900 assay 2 (see Table 3). When the LightCycler assay 1 was applied to these plasma specimens, EBV DNA was detected in 42 of 48 samples. The remaining six specimens (two infectious mononucleosis, two transplant, and two nasopharyngeal carcinoma plasma samples) had undetectable EBV DNA by assay 1 despite detectable EBV by assay 2 and assay 3, albeit at a low level (range, 350 to 650 copies per ml of plasma by assay 2, and 144 to 756 copies per ml of plasma by assay 3). In nine specimens, only one of the two replicates was positive by assay 1, again in conjunction with a low viral load in the one positive test (range, 184 to 1101 copies/ml of plasma). In 33 samples with measurable viral loads, the duplicate values by assay 1 were very close to each other, never differing by more than twofold and averaging a difference of 20%.
Table 3.
EBV Load in 48 Plasma Samples from Patients with EBV-Related Disease
| Case number | Diagnosis | EBV viral load, copies per ml
|
||
|---|---|---|---|---|
| LightCycler assay 1 | ABI 7900 assay 2 | ABI 7500 assay 3 | ||
| 47 | AIDS lymphoma | 170,900 | 220,000 | 148,224 |
| 46 | AIDS lymphoma | 696,000 | 1029,200 | 513,897 |
| 48 | AIDS lymphoma | 369,750 | 1500,000 | 817,884 |
| 17 | AIDS lymphoma | 254 | 1280 | 2100 |
| 7 | AIDS | 92 | 1300 | 1409 |
| 20 | AIDS | 270 | 420 | 1096 |
| 31 | AIDS | 663 | 9000 | 12,554 |
| 33 | AIDS | 610 | 580 | 2573 |
| 21 | Aplastic anemia | 356 | 1400 | 1536 |
| 11 | Autoimmune lymphoproliferative syndrome | 127 | 630 | 1259 |
| 44 | Terminal multi-organ failure after drug overdose | 13,545 | 14,134 | 11,958 |
| 30 | Infectious mononucleosis | 570 | 1040 | 2067 |
| 1 | Infectious mononucleosis | 0 | 350 | 710 |
| 25 | Infectious mononucleosis | 455 | 3741 | 3242 |
| 26 | Infectious mononucleosis | 473 | 3140 | 4105 |
| 43 | Infectious mononucleosis | 2592 | 133,003 | 6232 |
| 41 | Infectious mononucleosis | 1844 | 912 | 812 |
| 42 | Infectious mononucleosis | 1416 | 399 | 1924 |
| 2 | Infectious mononucleosis | 0 | 351 | 177 |
| 37 | Infectious mononucleosis | 1261 | 654 | 484 |
| 27 | Leukemia | 476 | 11,500 | 10,286 |
| 45 | Heart transplant | 144,000 | 20,000 | 30,133 |
| 23 | Heart transplant | 411 | 2030 | 3822 |
| 3 | Liver transplant | 0 | 454 | 425 |
| 16 | Liver transplant | 253 | 1530 | 1230 |
| 28 | Liver transplant | 595 | 520 | 1684 |
| 9 | Liver transplant | 124 | 1200 | 985 |
| 14 | Liver transplant | 244 | 1400 | 1250 |
| 13 | Liver transplant | 211 | 990 | 850 |
| 34 | Liver transplant | 736 | 2018 | 1661 |
| 39 | Liver transplant | 1232 | 710 | 4106 |
| 24 | Liver transplant | 391 | 3170 | 619 |
| 36 | Liver transplant | 966 | 450 | 1042 |
| 29 | Liver transplant | 551 | 1265 | 1794 |
| 18 | Liver transplant | 266 | 819 | 1476 |
| 12 | Liver transplant | 151 | 413 | 423 |
| 8 | Lung transplant | 113 | 560 | 644 |
| 32 | Lung transplant | 711 | 2000 | 3934 |
| 15 | Lung transplant | 191 | 1500 | 3084 |
| 4 | Lung transplant | 0 | 400 | 380 |
| 10 | Lung transplant | 166 | 910 | 2793 |
| 35 | Lung transplant | 777 | 1770 | 1900 |
| 38 | Marrow transplant | 991 | 1143 | 1450 |
| 22 | Nasopharyngeal carcinoma | 401 | 311 | 652 |
| 40 | Nasopharyngeal carcinoma | 1752 | 2643 | 3455 |
| 5 | Nasopharyngeal carcinoma | 0 | 650 | 144 |
| 19 | Nasopharyngeal carcinoma | 388 | 3600 | 9288 |
| 6 | Nasopharyngeal carcinoma | 0 | 410 | 756 |
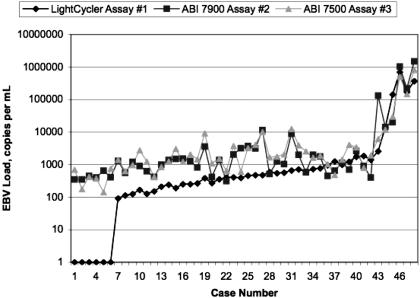
Results of the LightCycler assay 1 were evaluated in comparison with results of the ABI assay 2 and the ABI 7500 assay 3 (see Figure 2). There was strong correlation between the EBV viral loads measured by assay 1 compared with assay 2 (correlation, 0.86) or assay 3 (correlation, 0.85). Failure to detect EBV DNA in six specimens implied that the Lightcycler assay 1 is slightly less sensitive than the other assays when applied to patient plasma samples.
Figure 2.
EBV levels are shown as measured by three different viral load assays in 48 plasma samples from patients with EBV-related disease. EBV DNA was undetectable by LightCycler assay 1 in six patients who had low-level EBV by the other two assays. In the remaining 42 samples, Lightcycler assay 1 levels tended to be lower than those of the other two assays at levels below 1000 copies per ml, whereas the values were quite similar across the three assays for samples above 1000 copies per ml. It is not clear why the values of all three assays correlated most closely at higher rather than lower viral loads.
To examine technical issues that could potentially contribute to false negative EBV results by assay 1, we tested for efficacy of extraction and for presence of amplification inhibitors by first spiking a synthetic control sequence into patient plasma before extraction. The spiked plasmid DNA has been engineered to have the same primer recognition sequences but a different internal probe sequence compared with natural EBV LMP2. On subsequent real-time amplification of the spiked sequence, all of the patient samples had nearly identical amplification profiles, suggesting that the extraction and amplification steps were quite consistent and reproducible. Even when varying levels of EBV DNA resulted in competition for reagents in this multiplex PCR, the crossing points for the spiked control assay remained around 32 to 33 cycles. Neither failed extraction nor substantial inhibition of PCR occurred in this study.
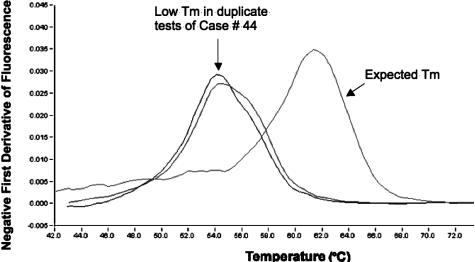
Melt-curve analysis of the hybridization probe-binding region showed that all but one of the EBV products had a dissociation temperature of 61°C ± −2°C. The single outlier had a dissociation temperature of only 54°C in both replicates, suggesting the likelihood of a sequence variant in the probe-binding region (see Figure 3). This sample was from a patient with terminal multiorgan failure having an EBV viral load of 13,545 copies per ml of plasma. Further testing was not done to characterize the nature of the genetic defect that altered the melt temperature. (The precise location of probe hybridization in the LMP2 gene is proprietary information not disclosed by the probe manufacturer.)
Figure 3.
Melt-curve analysis reveals an abnormal melt temperature (Tm) in duplicate EBV PCR assays on sample 44. This plasma sample was taken from a woman with terminal multiorgan failure following a drug overdose. A typical melt curve on the right has probe dissociation at approximately 61°C, whereas the atypical melt curves on this particular sample have probe dissociation at approximately 54°C.
Discussion
This study represents the first comprehensive evaluation of the performance characteristics of an assay that uses commercial analyte-specific reagents for measuring EBV viral load. The assay was optimized for implementation in our clinical laboratory where automated instruments and software are used to facilitate extraction and detection of amplification products. When the EBV-specific primers and hybridization probes were applied to samples containing known amounts of EBV DNA from any of three cell lines, the assay was shown to be sensitive to approximately 7.5 copies of EBV per PCR reaction, which translates to 750 copies per ml of plasma. The assay was linear across an average of four orders of magnitude, although the linear range may have been underestimated because the upper boundary was not studied. When applied to residual plasma samples from patients with or without viremia, the assay performed very well from a clinical standpoint. All of the medically significant EBV levels were identified in immunosuppressed individuals where the LightCycler assay 1 was considered equivalent to two other in-laboratory-developed assays.
A total of six samples had undetectable EBV by LightCycler assay 1 in contrast to low EBV loads by the ABI 7900 assay 2 and by ABI 7500 assay 3 (up to 756 copies/ml), implying that low-level EBV DNA was missed by the Roche assay. False negative results were seen in two of nine infectious mononucleosis patients, reinforcing prior studies suggesting that serology rather than PCR be used as the first line test to confirm a diagnosis of infectious mononucleosis.1,2,3 Undetectable EBV results were seen in 2 of 22 transplant recipients; however, in both instances, the level of EBV DNA by the other two assays was below our threshold of 700 copies per milliliter of plasma, a threshold that was validated to distinguish clinically significant EBV infection from the insignificant levels of plasma EBV that are frequently found in healthy allogeneic transplant recipients.27 In nasopharyngeal carcinoma subjects, the LightCycler assay 1 failed to detect low-level EBV DNA in two of five plasma samples. Further studies of this patient group are warranted to determine whether this difference is clinically significant. Assay sensitivity may be critical in screening individuals at high risk for nasopharyngeal carcinoma, especially those with early stage disease.12,17,24,28 In addition, carcinoma patients being followed for residual disease after therapy may benefit from a sensitive assay to detect early relapse.13,17,24
Contributing factors to assay sensitivity include 1) targeting a single-copy gene versus a reiterated viral sequence, 2) signal-to-noise ratio in the detection system, 3) primer and probe design with associated optimization of reaction components and cycling parameters, 4) multiplex versus single-target amplification, and 5) input DNA volume. In the current study, the LightCycler assay 1 targeted the unique LMP2 gene, whereas the ABI assays 2 and 3 targeted the reiterated BamH1W sequence. Assays 1 and 2 were both multiplexed with a spiked control coamplification, whereas assays 1 and 3 both used the same input DNA volume (5 μl) versus 10 μl for assay 2. Despite all of these technical variables, the assays performed remarkably similarly when applied to either patient plasmas or to mock samples prepared from cell lines.
False negative test results may also be a consequence of failed extraction or PCR inhibition. Our study revealed no evidence of either problem as shown by consistent recovery of a spiked control sequence. Despite the added expense of this control assay, we recommend its use on a regular and ongoing basis to provide assurance that extraction and amplification function as expected.
Another potential cause of false negative results is sampling error, which is most evident clinically when testing for low level disease or early relapse. In the current study, duplicate testing sometimes revealed negative results in one of the replicates with a low viral load measured in the other replicate, suggesting that sampling error contributed to a false negative result in one of the replicate wells. For purposes of patient care, one should consider replicate analysis whenever clinical circumstances suggest that low positive versus negative results would make a difference inpatient care. Further studies are needed to determine whether replicate testing would benefit nasopharyngeal carcinoma patients for whom assay sensitivity seems to be important for screening high-risk individuals and for early detection of relapse.13,28 On the other hand, replicate testing is probably not required when monitoring allogeneic transplant recipients because there is a low likelihood of missing a clinically significant EBV-related posttransplant lymphoproliferation as a consequence of sampling error. In this regard, the Lightcycler assay is well suited for monitoring transplant recipients.
Prior studies showed that most AIDS lymphoma patients whose tumors are EBV-related (as proven by in situ hybridization) have moderate to high levels of circulating EBV, but a few have low or undetectable levels in plasma, and some with primary brain lymphoma had EBV DNA detectable only in cerebrospinal fluid.14,29,30 In the current study, we did not assess cerebrospinal fluid, but we did show that the Lightcycler assay detected plasma EBV DNA in all four samples from AIDS patients who had EBV-related lymphoma.
Polymorphisms of EBV DNA can interfere with primer or probe binding and thus alter viral load measurement. Therefore, it is important to target highly conserved sequences within the viral genome. The LMP2 gene targeted by LightCycler assay 1 is a highly conserved gene that prevents antigen-mediated B lymphocyte activation, thus promoting latent viral persistence by inhibiting the cell from undergoing terminal differentiation. Our study showed an abnormal melt curve in one patient sample, which suggests there is a mutation in the EBV genome of that patient’s strain of virus. The nature of the putative polymorphism and its potential impact on viral pathogenicity is beyond the scope of this study. In routine laboratory practice, melt-curve analysis is an added benefit of the hybridization probe format of the LightCycler assay 1 because it provides extra confidence in the specificity of the assay.
A major advantage of a commercial assay is that all testing laboratories have access to the same reagents and equipment, thus facilitating standardization of testing across many clinical laboratories. Standardization is especially important when patients are tested at more than one laboratory, because serial monitoring of disease requires use of equivalent tests to assure that trends are interpreted correctly. Another advantage of standardization is improved ability to compare proficiency survey results across testing laboratories, whereas interlaboratory comparisons are difficult to interpret when each testing laboratory uses different procedures, standards, and units of reporting. Finally, commercial manufacturers offer many different molecular products that complement each other, thus facilitating use of their instruments and reagents for many different applications merely by switching out the analyte specific reagents. In this regard, multiple patient samples may be extracted simultaneously and amplified simultaneously because the extraction and cycling parameters are designed to be the same for multiple different analytic tests. A downside of commercial systems, beyond their expense, is that the laboratory is at the mercy of the manufacturer to provide a continuous pipeline of a high quality product. If the manufacturer fails to deliver such product when needed, the testing laboratory is faced with validating a back-up assay or else sending samples to another laboratory that may use a different test procedure, either of which can confound interpretation of serial test results.
It was interesting to note that LightCycler assay 1 seemed to be as sensitive as assay 3 when tested on cell line DNA, whereas it seemed to be less sensitive when tested on actual patient samples. Even if true, this did not appear to adversely impact the measurement of clinically important levels of EBV in immunocompromised patients. Furthermore, accuracy of assigned concentration values is questionable in each of the three cell line DNA samples that we tested. Two major factors are likely to contribute to the variation in findings across the three cell lines. First, Namalwa represents total DNA from an infected cell line, so EBV DNA is mixed with abundant human DNA, whereas the B95.8 and P3HR1 samples represent pure viral DNA with little or no human DNA. The human DNA may act as a blocker that nonspecifically binds to the pipette tip and the reaction well, thus preventing viral DNA from binding and thereby enhancing amplifiability of viral sequences. Second, different methods were used to determine the “correct” amount of EBV DNA in Namalwa compared with the B95.8 and P3HR1 samples, with the latter two samples coming from the same commercial source and yielding results similar to each other. We cannot determine which, if any, of the cell lines had the more accurate assigned EBV concentration because there is currently no “gold standard” by which to quantitate EBV DNA. International effort is required to gain agreement on a single, stable standard that can be used by all testing laboratories to calibrate EBV viral load assays.
Acknowledgments
We thank the staff of the University of North Carolina Hospitals Molecular Genetics Laboratory including Kristin Pittman, Gwen Evans, Karen Weck, and Michael Langley. We are grateful to Roche Diagnostics Corporation for funding this study, and we acknowledge discussions with corporate staff including Bryan Moore, Joel Cox, James Floberg, Robert Gibbs, Vivian Cintron, Jarman Smith, and Deborah Greene. Roche Diagnostics Corporation had no role in data analysis, manuscript content, or the decision to submit this manuscript for publication.
Footnotes
Current address for H.F.: Department of Pathology, University of Texas Health Science Center at San Antonio, San Antonio, TX.
References
- Ryan JL, Fan H, Swinnen LJ, Schichman SA, Raab-Traub N, Covington M, Elmore S, Gulley ML. Epstein-Barr Virus (EBV) DNA in plasma is not encapsidated in patients with EBV-related malignancies. Diagn Mol Pathol. 2004;13:61–68. doi: 10.1097/00019606-200406000-00001. [DOI] [PubMed] [Google Scholar]
- Fafi-Kremer S, Morand P, Brion JP, Pavese P, Baccard M, Germi R, Genoulaz O, Nicod S, Jolivet M, Ruigrok RW, Stahl JP, Seigneurin JM. Long-term shedding of infectious epstein-barr virus after infectious mononucleosis. J Infect Dis. 2005;191:985–989. doi: 10.1086/428097. [DOI] [PubMed] [Google Scholar]
- Balfour HH, Jr, Holman CJ, Hokanson KM, Lelonek MM, Giesbrecht JE, White DR, Schmeling DO, Webb CH, Cavert W, Wang DH, Brundage RC. A prospective clinical study of Epstein-Barr virus and host interactions during acute infectious mononucleosis. J Infect Dis. 2005;192:1505–1512. doi: 10.1086/491740. [DOI] [PubMed] [Google Scholar]
- Maurmann S, Fricke L, Wagner HJ, Schlenke P, Hennig H, Steinhoff J, Jabs WJ. Molecular parameters for precise diagnosis of asymptomatic epstein-barr virus reactivation in healthy carriers. J Clin Microbiol. 2003;41:5419–5428. doi: 10.1128/JCM.41.12.5419-5428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Gulley ML. Epstein-Barr viral load measurement as a marker of EBV-related disease. Mol Diagn. 2001;6:279–289. doi: 10.1054/modi.2001.29161. [DOI] [PubMed] [Google Scholar]
- Gulley ML. Molecular diagnosis of Epstein-Barr virus-related diseases. J Mol Diagn. 2001;3:1–10. doi: 10.1016/S1525-1578(10)60642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lit LC, Chan KC, Leung SF, Lei KI, Chan LY, Chow KC, Chan AT, Lo YM. Distribution of cell-free and cell-associated Epstein-Barr virus (EBV) DNA in the blood of patients with nasopharyngeal carcinoma and EBV-associated lymphoma. Clin Chem. 2004;50:1842–1845. doi: 10.1373/clinchem.2004.036640. [DOI] [PubMed] [Google Scholar]
- Au WY, Pang A, Choy C, Chim CS, Kwong YL. Quantification of circulating Epstein-Barr virus (EBV) DNA in the diagnosis and monitoring of natural killer cell and EBV-positive lymphomas in immunocompetent patients. Blood. 2004;104:243–249. doi: 10.1182/blood-2003-12-4197. [DOI] [PubMed] [Google Scholar]
- Allen UD, Farkas G, Hebert D, Weitzman S, Stephens D, Petric M, Tellier R, Ngan B, Fecteau A, West L, Wasfy S. Risk factors for post-transplant lymphoproliferative disorder in pediatric patients: a case-control study. Pediatr Transplant. 2005;9:450–455. doi: 10.1111/j.1399-3046.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- Lee TC, Savoldo B, Rooney CM, Heslop HE, Gee AP, Caldwell Y, Barshes NR, Scott JD, Bristow LJ, O’Mahony CA, Goss JA. Quantitative EBV viral loads and immunosuppression alterations can decrease PTLD incidence in pediatric liver transplant recipients. Am J Transplant. 2005;5:2222–2228. doi: 10.1111/j.1600-6143.2005.01002.x. [DOI] [PubMed] [Google Scholar]
- Greenfield HM, Gharib MI, Turner AJ, Guiver M, Carr T, Will AM, Wynn RF. The impact of monitoring Epstein-Barr virus PCR in paediatric bone marrow transplant patients: Can it successfully predict outcome and guide intervention? Pediatr Blood Cancer. 2006;47:200–205. doi: 10.1002/pbc.20604. [DOI] [PubMed] [Google Scholar]
- Leung SF, Chan AT, Zee B, Ma B, Chan LY, Johnson PJ, Lo YM. Pretherapy quantitative measurement of circulating Epstein-Barr virus DNA is predictive of posttherapy distant failure in patients with early-stage nasopharyngeal carcinoma of undifferentiated type. Cancer. 2003;98:288–291. doi: 10.1002/cncr.11496. [DOI] [PubMed] [Google Scholar]
- Kalpoe JS, Douwes Dekker PB, van Krieken JH, Baatenburg de Jong RJ, Kroes AC. The role of Epstein-Barr virus DNA measurement in plasma in the clinical management of nasopharyngeal carcinoma (NPC) patients in a low risk population. J Clin Pathol. 2006;59:537–541. doi: 10.1136/jcp.2005.030544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Kim SC, Chima CO, Israel BF, Lawless KM, Eagan PA, Elmore S, Moore DT, Schichman SA, Swinnen LJ, Gulley ML. Epstein-Barr viral load as a marker of lymphoma in AIDS patients. J Med Virol. 2005;75:59–69. doi: 10.1002/jmv.20238. [DOI] [PubMed] [Google Scholar]
- Bauer CC, Aberle SW, Popow-Kraupp T, Kapitan M, Hofmann H, Puchhammer-Stockl E. Serum Epstein-Barr virus DNA load in primary Epstein-Barr virus infection. J Med Virol. 2005;75:54–58. doi: 10.1002/jmv.20237. [DOI] [PubMed] [Google Scholar]
- Chan KC, Zhang J, Chan AT, Lei KI, Leung SF, Chan LY, Chow KC, Lo YM. Molecular characterization of circulating EBV DNA in the plasma of nasopharyngeal carcinoma and lymphoma patients. Cancer Res. 2003;63:2028–2032. [PubMed] [Google Scholar]
- Shao JY, Li YH, Gao HY, Wu QL, Cui NJ, Zhang L, Cheng G, Hu LF, Ernberg I, Zeng YX. Comparison of plasma Epstein-Barr virus (EBV) DNA levels and serum EBV immunoglobulin A/virus capsid antigen antibody titers in patients with nasopharyngeal carcinoma. Cancer. 2004;100:1162–1170. doi: 10.1002/cncr.20099. [DOI] [PubMed] [Google Scholar]
- Wang WY, Chien YC, Jan JS, Chueh CM, Lin JC. Consistent sequence variation of Epstein-Barr virus nuclear antigen 1 in primary tumor and peripheral blood cells of patients with nasopharyngeal carcinoma. Clin Cancer Res. 2002;8:2586–2590. [PubMed] [Google Scholar]
- Tierney RJ, Edwards RH, Sitki-Green D, Croom-Carter D, Roy S, Yao QY, Raab-Traub N, Rickinson AB. Multiple Epstein-Barr virus strains in patients with infectious mononucleosis: comparison of ex vivo samples with in vitro isolates by use of heteroduplex tracking assays. J Infect Dis. 2006;193:287–297. doi: 10.1086/498913. [DOI] [PubMed] [Google Scholar]
- Edwards RH, Sitki-Green D, Moore DT, Raab-Traub N. Potential selection of LMP1 variants in nasopharyngeal carcinoma. J Virol. 2004;78:868–881. doi: 10.1128/JVI.78.2.868-881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung E, Shenton BK, Green K, Jackson G, Gould FK, Yap C, Talbot D. Dynamic EBV gene loads in renal, hepatic, and cardiothoracic transplant recipients as determined by real-time PCR light cycler. Transpl Infect Dis. 2004;6:156–164. doi: 10.1111/j.1399-3062.2004.00073.x. [DOI] [PubMed] [Google Scholar]
- Jebbink J, Bai X, Rogers BB, Dawson DB, Scheuermann RH, Domiati-Saad R. Development of real-time PCR assays for the quantitative detection of Epstein-Barr virus and cytomegalovirus, comparison of TaqMan probes, and molecular beacons. J Mol Diagn. 2003;5:15–20. doi: 10.1016/S1525-1578(10)60446-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz G, Pena P, de Ory F, Echevarria JE. Comparison of commercial real-time PCR assays for quantification of Epstein-Barr virus DNA. J Clin Microbiol. 2005;43:2053–2057. doi: 10.1128/JCM.43.5.2053-2057.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le QT, Jones CD, Yau TK, Shirazi HA, Wong PH, Thomas EN, Patterson BK, Lee AW, Zehnder JL. A comparison study of different PCR assays in measuring circulating plasma epstein-barr virus DNA levels in patients with nasopharyngeal carcinoma. Clin Cancer Res. 2005;11:5700–5707. doi: 10.1158/1078-0432.CCR-05-0648. [DOI] [PubMed] [Google Scholar]
- Ryan JL, Fan H, Glaser SL, Schichman SA, Raab-Traub N, Gulley ML. Epstein-Barr virus quantitation by real-time PCR targeting multiple gene segments: a novel approach to screen for the virus in paraffin-embedded tissue and plasma. J Mol Diagn. 2004;6:378–385. doi: 10.1016/S1525-1578(10)60535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Heller M, Petti L, O’Shiro E, Kieff E. Persistence of the entire Epstein-Barr virus genome integrated into human lymphocyte DNA. Science. 1984;226:1322–1325. doi: 10.1126/science.6095452. [DOI] [PubMed] [Google Scholar]
- Swinnen LJ, Gulley ML, Hamilton E, Schichman SA. EBV DNA quantitation in serum is highly correlated with the development and regression of post-transplant lymphoproliferative disorder (PTLD) in solid organ transplant recipients. Blood. 1998;92(Suppl 1):314a. [Google Scholar]
- Stevens SJ, Verkuijlen SA, Hariwiyanto B, Harijadi , Fachiroh J, Paramita DK, Tan IB, Haryana SM, Middeldorp JM. Diagnostic value of measuring Epstein-Barr virus (EBV) DNA load and carcinoma-specific viral mRNA in relation to anti-EBV immunoglobulin A (IgA) and IgG antibody levels in blood of nasopharyngeal carcinoma patients from Indonesia. J Clin Microbiol. 2005;43:3066–3073. doi: 10.1128/JCM.43.7.3066-3073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossolasco S, Cinque P, Ponzoni M, Vigano MG, Lazzarin A, Linde A, Falk KI. Epstein-Barr virus DNA load in cerebrospinal fluid and plasma of patients with AIDS-related lymphoma. J Neurovirol. 2002;8:432–438. doi: 10.1080/13550280260422730. [DOI] [PubMed] [Google Scholar]
- Ivers LC, Kim AY, Sax PE. Predictive value of polymerase chain reaction of cerebrospinal fluid for detection of Epstein-Barr virus to establish the diagnosis of HIV-related primary central nervous system lymphoma. Clin Infect Dis. 2004;38:1629–1632. doi: 10.1086/420934. [DOI] [PubMed] [Google Scholar]