Abstract
Nasal carriage of Staphylococcus aureus is considered a source of subsequent infection in health care settings. Utilization of real-time polymerase chain reaction (PCR) for detection of S. aureus has the potential to dramatically affect infection control practice by rapidly identifying S. aureus-colonized patients. We developed and validated the use of real-time PCR for detection of S. aureus colonization in two patient populations. Paired nasal swabs were collected from 299 neonates and from 151 adult patients at Evanston Hospital. One swab was used for culture and the other placed into a bacterial lysis solution containing achromopeptidase. The DNA liberated was used as the template for real-time PCR with primers for the femA gene. SYBR Green was used for amplicon detection. In the neonatal population the sensitivity, specificity, predictive value positive and predictive value negative for culture and PCR was 92% versus 96%, 100% versus 100%, 100% versus 100%, and 98% versus 99%, respectively. In the adults the results were 90% versus 100%, 100% versus 98%, 100% versus 96%, and 95% versus 100%, respectively. Real-time PCR was able to detect S. aureus in 2 hours compared to 1 to 4 days for culture and provided sensitivity equal to or greater than culture.
Staphylococcus aureus is an important cause of skin, soft tissue, and bloodstream infections that can be rapidly fatal if not treated effectively and is the single leading pathogen in health care-associated infections.1 Nasal carriage of S. aureus has been suggested as the source of bacteremia, surgical-site, and other infections and a reservoir of S. aureus in hospitals.2 Early detection followed by decolonization with topical mupirocin may prevent infections and reduce transmission.3
Screening patients for S. aureus colonization using culture methods requires 1 to 4 or more days for accurate detection and identification of S. aureus.4 However, the results of a real-time polymerase chain reaction (PCR) assay to detect S. aureus could be obtained within 2 hours. Thus, an effective real-time PCR strategy to screen patients would theoretically enable earlier identification of S. aureus colonization.
The purpose of this investigation was to compare culture using selective media to real-time PCR using femA primers specific for detecting S. aureus colonization directly in nasal swab specimens collected from hospitalized neonatal and adult patients.
Materials and Methods
Patient Population and Specimen Collection
The study site was Evanston Hospital, which is a 420-bed academic facility in Evanston, Illinois that is affiliated with Northwestern University’s Feinberg School of Medicine. The hospital contains both neonatal and adult intensive care units, with nasal swab surveillance being part of routine infection control practice. Starting in December 2002 and continuing through March 2003, we collected nasal swabs from neonatal and adult patients. Specimens were obtained weekly from all neonates in the Level III Infant Special Care Unit (ISCU) caring for premature neonates and young infants (with a median length of stay of 16 days) and from adults on admission and discharge from the Surgical Intensive Care Unit (ICU) at Evanston Hospital. Nasal samples were collected using pre-moistened, double-headed rayon tipped swabs (CultureSwab, BBL, Becton Dickinson Inc., Cockeysville, MD). Both swabs were inserted into one nostril followed by the other, which yielded paired swabs, with one swab to be used for culture analysis and the other processed for real-time PCR. Institutional Review Board approval for comparison of these test methodologies was obtained from Evanston Northwestern Healthcare.
Culture
Before this study, a comparison of selective media was performed. The nasal swab for culture was plated to Columbia colistin-nalidixic agar (CNA) with 5% sheep blood (Remel, Inc., Lenexa, KS) and to mannitol salt agar (Remel), then immersed and left in phenol red mannitol broth with 75 μg/ml aztreonam and 5 μg/ml ceftazidime. There was no statistical difference in the recovery of S. aureus from CNA, mannitol salt agar, or the mannitol broth.4 Thus, one of the paired swabs from the nasal specimen was plated to CNA with 5% sheep blood and incubated in 5% CO2 at 35°C for 24 to 48 hours to recover S. aureus.
S. aureus was identified by colony morphology and Staphaurex latex agglutination test (Murex Biotech Limited, Dartford, Kent, UK). Oxacillin susceptibility testing using oxacillin disk diffusion and oxacillin agar screen was performed following National Committee for Clinical Laboratory Standards guidelines.5 Susceptibility results were read and interpreted after 24 hours of incubation at 35°C.
Cell Lysis
The second swab from the nasal specimen was broken off into a microcentrifuge tube containing 200 μl of lysis buffer. The lysis buffer was a solution containing 1 U/μl achromopeptidase (Sigma-Aldrich, St. Louis, MO)6 in 10 mmol/L Tris-HCl pH 8.0, 1 mmol/L EDTA (Sigma-Aldrich). Swabs in the achromopeptidase solution were vortexed for 5 seconds, incubated at 37°C for 15 minutes, and then boiled for 5 minutes. Subsequently, the sample was centrifuged at >10000 × g for 1 minute and the swab was removed. The remaining supernatant was used directly in real-time PCR.
Real-Time PCR Method
Real-time PCR was accomplished using the LightCycler (Roche Applied Science, Indianapolis, IN). Oligonucleotide primers were designed to amplify a unique conserved region of the femA gene in S. aureus only (femA-2F: 5′ AACTGTTGGCCACTATGAGT 3′ and femA-2R:5′ CCAGCATTACCTGTAATCTCG 3′ yielding a 306-bp product), which is not present in other Staphylococcus species. Primers were synthesized by MWG Biotech (High Point, NJ). Lysis supernatant (2 μl) was added to a hot-start reaction mixture. The final 20 μl real-time PCR reaction contained 1X LightCycler FastSart DNA Master SYBR Green I (containing a modified Taq polymerase with heat-labile blocking groups), 2% DMSO (Sigma), 5 mmol/L MgCl2 and 0.25 μmol/L of each primer. Included in each run were blank (water), negative (S. epidermidis), and positive (methicillin-resistant S. aureus; MRSA) controls. The real-time PCR conditions consisted of an initial step of 95°C for 10 minutes followed by an amplification program for 40 cycles of 3 seconds at 95°C, 5 seconds at 61°C, 20 seconds at 72°C with fluorescence acquisition at the end of each extension. The amplification program was immediately followed by a melt program consisting of 60 seconds at 95°C, 60 seconds at 65°C, and a gradual increase to 90°C at a rate of 0.2°C/sec with fluorescence acquisition at each temperature transition.
Laboratory Evaluation of Real-Time PCR Detection Sensitivity
The positive control strain of MRSA was suspended in sterile saline and adjusted to a No. 3 McFarland standard (5 × 108 to 1 × 109 colony forming unit (CFU)/ml).7 Ten-fold serial dilutions of this suspension were made in saline. A 10-μl aliquot of each dilution was spotted onto a fresh blood agar plate and another 20-μl aliquot into a microcentrifuge tube containing 180 μl of lysis solution for DNA extraction. Cell lysis and real-time PCR were performed as earlier described, and results were compared to the colony counts to determine the detection sensitivity of the assay. Blood agar plates were incubated for 24 hours and the CFUs were counted. This procedure was done in triplicate. A similar evaluation was done to determine the efficiency of achromopeptidase lysis from swab material except that the 20-μl aliquot was placed on a pre-moistened swab and that swab was then placed in 200-μl achromopeptidase solution for processing.
Stability of S. aureus Density in Swab Transport Containers
The stability of colony forming units in transport swabs was assessed over seven days using simulated nasal surveillance swabs with growth measured semi-quantitatively. To accomplish this, a cotton ball was placed in a petri dish and inoculated with organism suspended in nutrient broth until saturated. Next each test swab was rolled over the cotton ball, then returned to the swab holder and placed in contact with the transport material. These swabs were held at room temperature (22°C) for 7 days and assessed for growth. The swabs were rolled onto blood agar plates that were subsequently incubated for 48 hours. Growth was graded as rare, few, moderate, or many. Nine isolates of S. aureus were tested in duplicate, eight of the isolates had been recovered from individual patient samples, and one strain was an ATCC control (American Type Culture Collection, Manassas, VA, ATCC strain 29213).
Results
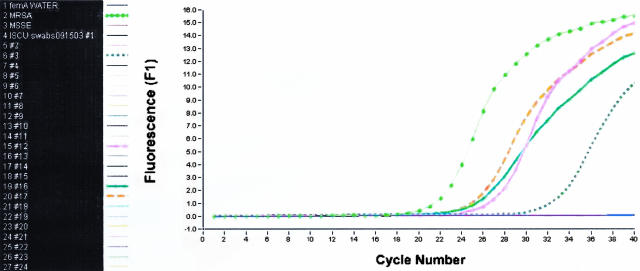
There was no difference in the real-time PCR amplification curves between adult and neonate specimens. An example of a real-time PCR assay run is in Figure 1. A total of 299 paired nasal swabs were collected from neonates in the ISCU and 151 paired nasal swabs were collected from adult patients in the ICU. In stability testing, all nine duplicate swabs showed growth of S. aureus equal or greater on day 7 of storage compared to the amount detected at day 0. Two strains had a single category reduction in one of their respective paired samples on day 5, and both of these swabs equaled (1) or exceeded (1) the amount of baseline growth following that to day 7, when the study was terminated.
Figure 1.
Representative real-time PCR run that included both positive and negative samples. ISCU nasal samples real-time PCR run with femA primers and SYBR Green detection of amplicon products. Blank and S. epidermidis were negative controls; MRSA (light green dotted line) was the positive control; and samples #3 (dark green dashed line), #12 (pink line), #16 (green hashed line), and #17 (brown line) were positive. The remaining samples were negative and overlap in the flat line across the bottom of the graph.
Neonates
In this population, 254 swab samples were negative by both methods (84.9%), 39 were positive by both methods (13%), two were positive by culture only (0.7%), and four were positive by real-time PCR only (1.3%). For this set of samples, the technologists were blinded from the other method’s results until both tests were considered final. The original culture plates were re-examined for all specimens with discrepant results. Review of results for the two samples that were positive by culture and negative by real-time PCR found that only one and two colonies of S. aureus were present on each culture plate, indicating very low density colonization. With the four samples that were real-time PCR positive and culture negative, routine processing provided a final culture result at 48 hours as negative for S. aureus, yet an extended investigation of the culture plates recovered S. aureus in all cases. Melt curve analysis also confirmed that the correct femA target was amplified. Thus, there were four false-negative cultures and two false-negative PCR tests. The sensitivity, specificity, positive, and negative predictive values are in Table 1.
Table 1.
Sensitivity, Specificity, Predictive Value Positive, and Predictive Value Negative of the Culture and Real-Time PCR Assays for S. aureus Directly from Nasal Swab Specimens
| Population (n) | Sensitivity | Specificity | Predictive value of a positive test | Predictive value of a negative test |
|---|---|---|---|---|
| Neonate culture (299) | 92% | 100% | 100% | 98% |
| Neonate PCR (299) | 96% | 100% | 100% | 99% |
| Adult culture (151) | 90% | 100% | 100% | 95% |
| Adult PCR (151) | 100% | 98% | 96% | 100% |
| Total subjects culture (450) | 90% | 100% | 100% | 98% |
| Total subjects PCR (450) | 98% | 99% | 98% | 99% |
Adults
In this population, 101 nasal swabs were negative by both methods (66.9%), 43 were positive by both methods (28.5%), none were positive by culture only (0%), and seven were positive by real-time PCR only (4.6%). For this set of samples, the technologists were not blinded to the other method’s results. After 48 hours of culture incubation any discrepant results were investigated thoroughly, which involved extended incubation and occasionally placing the original swabs into a broth. For the seven samples that were positive by real-time PCR only, all indicated that the femA target was amplified on melt curve review. Additional work-up of the original culture specimen found S. aureus in three of the swabs, another two swab specimens were from patients that had S. aureus identified from a different nasal culture or a clinical specimen sent for culture (one each) as part of an investigation for symptomatic infection. Two swabs were from patients with no clinical cultures and no S. aureus identified from a second nasal culture. We believe that clinical specimens positive for S. aureus are good evidence that these patients carried this pathogen in their nose since the ecological niche of S. aureus is the anterior nares, from where the organism spreads to other parts of the body.8 Nasal colonization with S. aureus antedates bacteremic as well as non-bacteremic infection, 9 so it is extremely likely that these patients harbored the microbe, even if it was not viable at the time of our sampling. Therefore, these seven discrepant results were interpreted to indicate that there were two false-positive real-time PCR reactions and five false-negative cultures. A summary of these results also is in Table 1.
Assay Performance
The detection limit for the real-time PCR test using the achromopeptidase solution for cell lysis from the nasal swabs was 2 CFU/PCR reaction. Comparison studies had demonstrated that this cell lysis method was approximately 100-fold more sensitive than one using a lysis solution consisting of 1% Triton X-100, 0.5% Tween 20, 1 mmol/L Tris-HCl pH 8.0, and 10 mmol/L EDTA (data not shown). The total time for completion of this assay, including cell lysis, was about 2 hours.
Discussion
We found that among neonatal and adult patients, real-time PCR performed directly on nasal specimens yielded positive results more often than did culture (Table 1). One key aspect of a study such as this is the comparative media used. Other investigators have reported varied levels of sensitivity for different selective media. Wertheim and colleagues10 compared selective broth with routine methods and found the broth superior. However, we compared this methodology to CNA medium and did not find the broth-based system superior, and concluded that CNA medium was the easiest to read for detection of S. aureus as well as the least expensive of the media tested.4 More recently, Safdar and co-workers11 reported their study of 32 culture-based methods for detecting MRSA. While they did not use a molecular-based test as part of their comparison, the best single agar evaluated was only able to recover approximately 90% of the MRSA strains colonizing their patient sample,11 which is similar to the performance of our agar test method when compared to PCR.
In recent years, applications of PCR have been used to increase the rapidity and accuracy for identification of S. aureus and confirmation of MRSA by detection of mecA.12 Multiplex PCR using detection of the mec and fem genes has been suggested as one approach for the routine diagnostic laboratory to rapidly identify MRSA from cultures,13 or following enrichment of patient screening swabs.14 This approach was found to be more accurate and sensitive than routine, culture-based analysis, with results available in 24 hours.13 Additional reports have appeared with similar recommendations for the routine diagnostic laboratory to speed the accurate detection of MRSA in blood cultures,15 including the use of real-time PCR for this application.16,17 Even direct detection of MRSA (targeting mecA and femA) in tracheal aspirates has been suggested as a management tool, based on conventional, gel-based PCR.18 However, only rare reports have discussed the use of real-time PCR directly from nasal swab specimens for rapid detection of S. aureus carriers,19 and no one has reported the level of sensitivity achieved with our lysis and amplification protocol. This high level of sensitivity and specificity obtained with our primers in a simple SYBR Green assay indicated no need for developing a hybridization probe-based assay. In addition, the melt curve analysis provided a secondary confirmation that the correct target was amplified.
In a research project on the economic feasibility of a direct real-time-PCR test for S. aureus detection, Shrestha and colleagues19 found this technology might be cost effective. While their initial test appeared slightly lower in sensitivity (97.0%) and specificity (97.1%) compared to our results, in a population with a MRSA prevalence up to 50% the predictive value of a negative test using their assay exceeded 97%.19 Our study validates this concept as a viable option, particularly with the development of a real-time PCR that detected more S. aureus than did culture, which raises the predictive value of a negative test to 99%.
The eventual goal is to not only rapidly detect S. aureus nasal carriers, but also those harboring MRSA, as well as determine whether the isolate is susceptible to the decolonizing antibiotic mupirocin. Nasal swabs are excellent and easily collected samples for surveillance of S. aureus colonization, which can be readily detected using the assay we describe.20 Importantly, methicillin resistance can be detected by amplifying the mecA gene. In addition, and particularly relevant to the identification of high-level mupirocin resistance, is the detection of ileS-2, the plasmid-borne gene responsible for clinically relevant mupirocin resistance.21,22,23 Detection of this high-level resistance is important to detect since it correlates with failure of mupirocin therapy.24,25 Because both of these resistance genes are carried in coagulase-negative staphylococci as well as S. aureus, it is not practical to amplify the DNA extracted directly from nasal swabs for false-positive results will occur with both organisms potentially present. Our current protocol is to determine and report the presence or absence of S. aureus by real-time PCR for the femA gene. Since we are interested in MRSA detection and decolonization, all of the PCR-positive swabs are plated for culture. The next morning isolated S. aureus colonies are tested for methicillin and mupirocin resistance by real-time PCR amplification for mecA and ileS-2 genes, respectively. This testing strategy can provide all of the relevant information on nasal carriers. Therefore, the presence of S. aureus is detected in a few hairs and followed by methicillin and mupirocin susceptibility within 24 hours. Such testing provides the needed speed for appropriate isolation of MRSA-colonized patients, and offers reliable susceptibility testing for mupirocin in a setting where decolonization therapy is contemplated.
Since implementing this protocol as a routine practice, we have tested 434 additional adult patient swabs. A total of 115 swabs were positive for S. aureus by real-time PCR (26.5%). For 108 of those swabs real-time PCR for mecA and ileS-2 was done on S. aureus that were isolated from culture while seven swabs (from six patients) failed to grow S. aureus. One of these six patients subsequently developed an infection from S. aureus. We also have tested 1340 additional swabs from neonates. A total of 258 swabs were positive by real-time PCR (19.3%). For 252 of those swabs, real-time PCR for mecA was done on S. aureus isolated from culture while six swabs failed to grow S. aureus. All six of these latter swabs were from patients with prior or subsequent nasal swabs positive by real-time PCR that did grow S. aureus in culture. While sampling error is always possible when using paired swabs, the data supports our belief that a positive real-time PCR result from a swab that did not grow S. aureus, which is from a patient with a concurrent culture positive for S. aureus, does represent a true-positive PCR result, even if it is detecting DNA from a non-viable S. aureus. All our false-negative PCR results seemed to be due to a very low quantity of S. aureus in the sample that was below our level of detection. With increased test sensitivity and the benefit of results in a few hours as opposed to days, real-time PCR can be used for early detection of S. aureus nasal colonization, which can lead to prevention of S. aureus infection and reduced spread of MRSA.
Recently, several papers have appeared indicating the need for more active surveillance and control of MRSA to reduce the medical and economic burden from this aggressive pathogen.26,27,28 Novel molecular methods continue to be explored that can aid in following through on these recommendations.29,30,31 We believe that real-time PCR, using a protocol consistent with that described in our report is readily applicable now to begin better management and control of staphylococcal infections. In conclusion, we have developed the first real-time PCR test that is equal to or better in sensitivity than culture for detection of S. aureus directly from nasal swabs in neonates and adults. The test is rapidly performed (2-hour assay time) with a simple cell lysis technique. We found that real-time PCR was a sensitive, accurate, and rapid strategy for detection of S. aureus nasal colonization from direct surveillance swab specimens.
Footnotes
Supported by the Department of Pathology and Laboratory Medicine, Evanston Northwestern Healthcare.
Presented in part at the 43rd Meeting of the Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, September 14–17, 2003.
References
- Jarvis WR. Infection control and changing health care delivery systems. Emerg Infect Dis. 2001;7:170–173. doi: 10.3201/eid0702.010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group N Engl J Med. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- Hudson IRR. The efficacy of intranasal mupirocin in the prevention of staphylococcal infections: a review of recent experience. J Hosp Infect. 1994;27:81–98. doi: 10.1016/0195-6701(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Hacek D, Paule S, Small M, Gottschall R, Thomson R, Peterson L. Comparison of colistin nalidixic agar (CNA), mannitol salt agar (MS), and phenol mannitol broth with antibiotics (PMB) for the recovery of Staphylococcus aureus (SA) from nasal swabs. Washington, DC: (Abstract C-323); Abstracts of the 103rd Annual Meeting of the American Society of Microbiology. 2003 [Google Scholar]
- National Committee for Clinical Laboratory Standards National Committee for Laboratory Standards; Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. (ed 6.) 2003 NCCLS document M7–A6. Wayne, PA, [Google Scholar]
- Leonard RB, Carroll KC. Rapid lysis of gram-positive cocci for pulsed-field gel electrophoresis using achromopeptidase. Diagn Mol Pathol. 1997;6:288–291. doi: 10.1097/00019606-199710000-00007. [DOI] [PubMed] [Google Scholar]
- Chapin KC, Lauderdale TL. Reagents, stains and media: bacteriology. Murray PR, Baron EJ, Jorgense JH, Pfaller MA, Yolken RH, editors. Washington, DC: ASM Press, American Society for Microbiology; Manual of Clinical Microbiology. 2003:358. [Google Scholar]
- Kluytmans J, Van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10:505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Eiff C, Karsten B, Konstanze M, Holger S, Georg P. Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- Wertheim H, Verbrugh HA, van Pelt C, de Man P, van Belkum A, Voss MC. Improved detection of methicillin-resistant Staphylococcus aureus using phenyl mannitol broth containing aztreonam and ceftizoxime. J Clin Microbiol. 2001;39:2660–2662. doi: 10.1128/JCM.39.7.2660-2662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar N, Narans L, Gordon B, Maki DG. Comparison of culture screening methods for detection of nasal carriage of methicillin-resistant Staphylococcus aureus: a prospective study comparing 32 methods. J Clin Microbiol. 2003;41:3163–3166. doi: 10.1128/JCM.41.7.3163-3166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisold AJ, Leitner E, Muhlbauer G, Marth E, Kessler HH. Detection of methicillin-resistant Staphylococcus aureus and simultaneous confirmation by automated nucleic acid extraction and real-time PCR. J Clin Microbiol. 2002;40:2392–2397. doi: 10.1128/JCM.40.7.2392-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner KJ, Talbot DC, Curran R, Webster CA, Humphreys H. Development and evaluation of a PCR-based immunoassay for the rapid detection of methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1998;47:607–613. doi: 10.1099/00222615-47-7-607. [DOI] [PubMed] [Google Scholar]
- Jonas D, Speck M, Daschner FD, Grundmann H. Rapid PCR-based identification of methicillin-resistant Staphylococcus aureus from screening swabs. J Clin Microbiol. 2002;40:1821–1823. doi: 10.1128/JCM.40.5.1821-1823.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie L, Goodfellow J, Mathieu P, Glatt A, Louie M, Simor AE. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J Clin Microbiol. 2002;40:2786–2790. doi: 10.1128/JCM.40.8.2786-2790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha NK, Tuohy MJ, Hall GS, Isada CM, Procop GW. Rapid identification of Staphylococcus aureus and the mecA gene from BacT/ALERT blood culture bottles by using the LightCycler system. J Clin Microbiol. 2002;40:2659–2661. doi: 10.1128/JCM.40.7.2659-2661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TY, Corden S, Barnes R, Cookson B. Rapid identification of methicillin-resistant Staphylococcus aureus from positive blood cultures by real-time fluorescence PCR. J Clin Microbiol. 2001;39:4529–4531. doi: 10.1128/JCM.39.12.4529-4531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannuffel P, Laterre PF, Bouyer M, Gigi J, Vandercam B, Reynaert M, Gala JL. Rapid and specific molecular identification of methicillin-resistant Staphylococcus aureus in endotracheal aspirates from mechanically ventilated patients. J Clin Microbiol. 1998;36:2366–2368. doi: 10.1128/jcm.36.8.2366-2368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha NK, Shermock KM, Gordon SM, Tuohy MJ, Wilson DA, Cwynar RE, Banbury MK, Longworth DL, Isada CM, Mawhorter SD, Procop GW. Predictive value and cost-effectiveness analysis of a rapid polymerase chain reaction for preoperative detection of nasal carriage of Staphylococcus aureus. Infect Cont Hosp Epidemiol. 2003;24:327–333. doi: 10.1086/502219. [DOI] [PubMed] [Google Scholar]
- Singh K, Gavin PJ, Vescio T, Thomson RB, Jr, Deddish RB, Fisher A, Noskin GA, Peterson LR. Microbiologic surveillance using nasal cultures alone is sufficient for detection of methicillin-resistant Staphylococcus aureus isolates in neonates. J Clin Microbiol. 2003;41:2755–2757. doi: 10.1128/JCM.41.6.2755-2757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Roth E, Claverie-Martin F, Batista N, Moreno A, Mendez-Alvarez S. Mupirocin resistance in methicillin-resistant Staphylococcus aureus clinical isolates in a Spanish hospital: co-application of multiplex PCR assay and conventional microbiology methods. Diagn Microbiol Infect Dis. 2002;43:123–128. doi: 10.1016/s0732-8893(02)00388-7. [DOI] [PubMed] [Google Scholar]
- Perez-Roth E, Claverie-Martin F, Villar J, Mendez-Alvarez S. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J Clin Microbiol. 2001;39:4037–4041. doi: 10.1128/JCM.39.11.4037-4041.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo EE, Jacob LE, Mathew B. Genetic analysis of methicillin-resistant Staphylococcus aureus expressing high- and low-level mupirocin resistance. J Med Microbiol. 2001;50:909–915. doi: 10.1099/0022-1317-50-10-909. [DOI] [PubMed] [Google Scholar]
- Harbarth S, Liassine N, Dharan S, Herrault P, Auckenthaler R, Pittet D. Risk factors for persistent carriage of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2000;31:1380–1385. doi: 10.1086/317484. [DOI] [PubMed] [Google Scholar]
- Fung S, O’Grady S, Kennedy C, Dedier H, Campbell I, Conly J. The utility of polysporin ointment in the eradication of methicillin-resistant Staphylococcus aureus colonization: a pilot study. Infect Control Hosp Epidemiol. 2000;21:653–655. doi: 10.1086/501709. [DOI] [PubMed] [Google Scholar]
- Arnold MS, Dempsey JM, Fishman M, McAuley PJ, Tibert C, Vallande NC. The best hospital practices for controlling methicillin-resistant Staphylococcus aureus: on the cutting edge. Infect Control Hosp Epidemiol. 2002;23:69–76. doi: 10.1086/502009. [DOI] [PubMed] [Google Scholar]
- LeDell K, Muto CA, Jarvis WR, Farr BM. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and Enterococcus. Infect Control Hosp Epidemiol. 2003;24:639–641. doi: 10.1086/502924. [DOI] [PubMed] [Google Scholar]
- Farr BM, Jarvis WR. Would active surveillance cultures help control healthcare-related methicillin-resistant Staphylococcus aureus infections? Infect Control Hosp Epidemiol. 2002;23:65–68. doi: 10.1086/502008. [DOI] [PubMed] [Google Scholar]
- Francois P, Pittet D, Bento M, Pepey B, Vaudaux P, Lew D, Schrenzel J. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or non-sterile clinical samples by a new molecular assay. J Clin Microbiol. 2003;41:254–260. doi: 10.1128/JCM.41.1.254-260.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi K, Bailey C, Bennett A, Marsh P, Cardy DLN, Towner KJ. Evaluation of an isothermal signal amplification method for rapid detection of methicillin-resistant Staphylococcus aureus from patient-screening swabs. J Clin Microbiol. 2003;41:3187–3191. doi: 10.1128/JCM.41.7.3187-3191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmeier R, Stuhlmeier KM. Fast, simultaneous, and sensitive detection of staphylococci. J Clin Pathol. 2003;56:782–785. doi: 10.1136/jcp.56.10.782. [DOI] [PMC free article] [PubMed] [Google Scholar]