Abstract
Aldolases are enzymes with potential applications in biosynthesis, depending on their activity, specificity and stability. In the present study, the genomes of Sulfolobus species were screened for aldolases. Two new KDGA [2-keto-3-deoxygluconate (2-oxo-3-deoxygluconate) aldolases] from Sulfolobus acidocaldarius and Sulfolobus tokodaii were identified, overexpressed in Escherichia coli and characterized. Both enzymes were found to have biochemical properties similar to the previously characterized S. solfataricus KDGA, including the condensation of pyruvate and either D,L-glyceraldehyde or D,L-glyceraldehyde 3-phosphate. The crystal structure of S. acidocaldarius KDGA revealed the presence of a novel phosphate-binding motif that allows the formation of multiple hydrogen-bonding interactions with the acceptor substrate, and enables high activity with glyceraldehyde 3-phosphate. Activity analyses with unnatural substrates revealed that these three KDGAs readily accept aldehydes with two to four carbon atoms, and that even aldoses with five carbon atoms are accepted to some extent. Water-mediated interactions permit binding of substrates in multiple conformations in the spacious hydrophilic binding site, and correlate with the observed broad substrate specificity.
Keywords: carbon–carbon bond formation, 2-keto-3-deoxygluconate aldolase (KDGA), substrate specificity, Sulfolobus, thermophilic enzyme
Abbreviations: DHAP, dihydroxyacetone phosphate; DHDPS, dihydrodipicolinate synthase; GA, glyceraldehyde; GAP, glyceraldehyde 3-phosphate; IPTG, isopropyl β-D-thiogalactoside; KDG, 2-keto-3-deoxygluconate; KDGA, 2-keto-3-deoxygluconate aldolase; KDGal, 2-keto-3-deoxygalactonate; KDO, 2-keto-3-deoxyoctonate; KDPG, 2-keto-3-deoxy-6-phosphogluconate; LDH, lactate dehydrogenase; NAL, N-acetylneuraminate lyase; PEG, poly(ethylene glycol); rmsd, root-mean-square deviation; Sac-KDGA, Sulfolobus acidocaldarius KDGA; Sso-KDGA, Sulfolobus solfataricus KDGA; Sto-KDGA, Sulfolobus tokodaii KDGA; TBA, thiobarbituric acid; Tte-KDGA, Thermoproteus tenax KDGA
INTRODUCTION
Aldolases are among the few classes of enzymes capable of enlarging the carbon skeleton of molecules in a specific way by catalysing the condensation of a ketone donor and an aldehyde acceptor. These enzymes can be a tool for the specific production of carbon–carbon bonds in organic synthesis, since it is difficult to achieve stereochemical control with the widely used chemical aldol condensation reaction [1]. Aldolases have already successfully been used for the biosynthesis of a limited number of specialty compounds [2]. However, many of the currently used aldolases are expensive, unstable, limited in their catalytic range and often require expensive substrates. The discovery of new aldolases or the engineering of known aldolases is therefore of continuing importance [2]. The availability of many sequenced genomes opens up new opportunities for the identification of potentially suitable aldolases.
In Nature, many different aldolases are encountered. Some aldolases play a vital physiological role in the degradation of metabolites, for instance during glycolysis; other aldolases are employed for carbon backbone assembly, e.g. in amino acid synthesis [3]. Two types of aldolase enzymes are distinguished based on their mechanisms. Type-I aldolases form a Schiff-base intermediate with the donor substrate, whereas type-II aldolases are metal-dependent enzymes [1,4]. Although both types can usually accept different acceptor substrates, they are rather specific for the donor substrate. Using a different classification, synthesizing aldolases are therefore also grouped according to their donor specificity: (1) DHAP (dihydroxyacetone phosphate)-dependent aldolases; (2) DERA (2-deoxyribose-5-phosphate aldolase), which is acetaldehyde-dependent; (3) phosphoenolpyruvate-dependent aldolases; and (4) glycine-dependent aldolases [4]. Novel aldolases belonging to these classes may be found in organisms with divergent sugar metabolic pathways or degradation pathways of xenobiotics [2]. The thermostability of their proteins make extremophiles such as Sulfolobus particularly interesting sources of new enzymes.
Sulfolobus species are thermoacidophilic archaea that typically grow optimally at high temperatures (75–85 °C) and at a low pH (pH 2–4) [5–7]. Because of their global abundance [8] and ease of cultivation [6], they have become model archaea; genetic tools have been established [9–11] and the complete genome sequences of Sulfolobus solfataricus, Sulfolobus tokodaii and Sulfolobus acidocaldarius have recently been unravelled [12–14]. In particular, because of their resemblance to eukaryotic counterparts, several protein complexes from Sulfolobus have been selected for the analyses of fundamental processes, such as replication [15–17], transcription and translation [18–20]. In addition, potential applications have been described for some of the thermostable Sulfolobus enzymes, including an unusual KDGA {KDG [2-keto-3-deoxygluconate (2-oxo-3-deoxygluconate)] aldolase} [21].
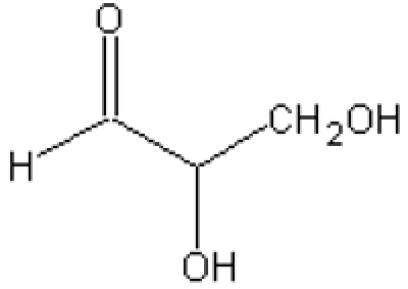
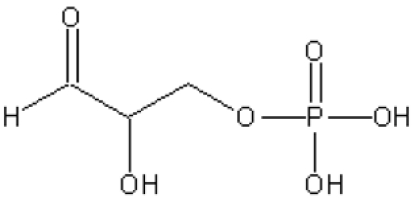
KDGA from S. solfataricus is a type-I aldolase and is a member of the NAL (N-acetylneuraminate lyase) subfamily. This NAL subfamily consists of tetrameric enzymes that are specific for pyruvate as a donor substrate, but use different aldehydes as acceptor substrates. The S. solfataricus KDGA was first characterized as an enzyme that catalyses the reversible conversion D,L-KDG⇋pyruvate+D,L-GA (D,L-glyceraldehyde). It was later discovered that the enzyme was not only able to synthesize KDG, but that it could also synthesize KDGal [2-keto-3-deoxygalactonate (2-oxo-3-deoxygalactonate)] from pyruvate and D-GA [22]. Recently, Ahmed et al. [23] have shown that the enzyme also catalyses the interconversion KDPG [D,L-2-keto-3-deoxy-6-phosphogluconate (D,L-2-oxo-3-deoxy-6-phosphogluconate)]⇋pyruvate+D,L-GAP (glyceraldehyde 3-phosphate) [23], suggesting that this bifunctional enzyme is part of a semi-phosphorylated Entner–Doudoroff pathway [23,24]. Previously, the crystal structure of S. solfataricus KDGA (Sso-KDGA) was solved; in agreement with the abovementioned specificity studies, substrate-soaking studies with KDG and KDGal revealed promiscuous binding with multiple conformations of these substrates owing to different water-mediated interactions of the O-5 and O-6 hydroxy groups in the rather spacious binding cavity [25].
In the present study, two hypothetical KDGA genes from S. acidocaldarius and S. tokodaii were cloned, and, after expression in Escherichia coli, their biochemical properties were assessed, together with the previously characterized S. solfataricus KDGA. In addition, the crystal structure of S. acidocaldarius KDGA was determined, providing a molecular basis for the observed substrate specificity and stereoselectivity. Furthermore, we have identified a novel phosphate-binding motif in the KDGAs that explains their high activity with phosphorylated substrates.
MATERIALS AND METHODS
Cloning and expression
The hypothetical KDGA genes from S. acidocaldarius (Saci_0225; GenBank® Identifier 70606067) and S. tokodaii (ST2479; GenBank® Identifier 15623602) were PCR-amplified from genomic DNA with primer pairs BG 1816 and BG 1817, and BG 1783 and 1784 (Table 1). The products obtained were restricted with NcoI and SalI and ligated into NcoI/SalI-restricted pET24d (Novagen). These plasmids, designated pWUR193 (containing Saci_0225) and pWUR192 (containing ST2479) were transformed into E. coli BL21(DE3) and E. coli BL21(DE3)/pRIL cells. The clone pWUR122, containing the KDGA gene from S. solfataricus (SSO3197; GenBank® identifier 2879782), described previously [23], was also transformed into E. coli BL21(DE3) cells.
Table 1. Primers used in the present study.
| Primer | Sequence |
|---|---|
| BG1784 | 5′-CGCGCCCATGGTGTTCAAAATTTTAAGTATGGATATTG-3′ |
| BG1816 | 5′-CGCGCGTCGACTTTACGAAACAGCTCTTTCTATTTC-3′ |
| BG1817 | 5′-CGCGCCATGGAAATAATTTCACCTATCATTAC-3′ |
| BG1852 | 5′-GCGCGTCGACTTAATGTACCAGTTCTTGAATCTTTC-3′ |
Purification
S. acidocaldarius KDGA (Sac-KDGA) was produced by inoculating 2 litres of LB (Luria–Bertani) medium supplemented with 50 μg/ml kanamycin with E. coli BL21(DE3) cells containing pWUR193. Subsequently, after 4 h of growth at 37 °C, the culture was induced with 0.1 mM IPTG (isopropyl β-D-thiogalactoside) and growth was continued overnight. The cells were harvested by centrifugation at 3800 g for 10 min, resuspended in 50 mM Hepes, pH 7, and 20 mM KCl, and lysed using a French press. After centrifugation at 27000 g for 20 min, the soluble protein fraction was incubated at 75 °C for 30 min. This was centrifuged at 48000 g for 30 min to obtain the heat-stable cell-free extract. The Sac-KDGA was purified further using anion-exchange chromatography (Q-Sepharose; 20 mM Tris/HCl, pH 8.5, 0–1 M gradient NaCl), eluting at 0.30 M NaCl and gel filtration (Superdex™ 200 10/300 GL; 50 mM Tris/HCl, pH 7.8, 100 mM NaCl). S. tokodaii KDGA (Sto-KDGA) was produced in the same way, except that the culture, inoculated with E. coli BL21(DE3)/pRIL cells containing pWUR192, was induced with 0.02 mM IPTG on ice and grown for a further 2 days at 20 °C (Q-Sepharose elution at 0.21 M NaCl). Sso-KDGA was also produced in a similar way to that for Sac-KDGA, but the cell pellet was resuspended in 90 mM Hepes, pH 7, and 160 mM KCl and purified only by heat incubation and anion-exchange chromatography, eluting at 0.32 M NaCl. The enzyme purity was checked by SDS/PAGE (10% gels), staining the proteins with Coomassie Brilliant Blue. Protein concentrations were determined with the Bio-Rad Bradford method.
Activity measurements
KDGA activity was measured with the TBA (thiobarbituric acid) assay as described by Buchanan et al. [26]. In a standard aldolase assay, the aldolase reaction was carried out using the following conditions: 50 mM sodium phosphate buffer, pH 6, 50 mM sodium pyruvate, 20 mM D,L-GA and an appropriate amount of enzyme (∼1 μg) were incubated at 70 °C for 10 min. The TBA assay was then performed as described previously [26]. Vmax and Km determinations were performed at optimal pH at 70 °C.
Aldolase activities with different aldehydes were measured by assaying the remaining pyruvate in a standardized LDH (lactate dehydrogenase) assay in triplicate. Reaction mixtures (100 μl volumes) containing 50 mM sodium phosphate buffer, pH 6, 10 mM pyruvate and 20 mM aldehyde and 1–100 μg of enzyme were incubated at 70 °C for 10–60 min. The reaction was stopped by adding 6 μl of 20% TCA (trichloroacetic acid), and precipitated protein was removed by centrifugation at 16000 g for 10 min. From 984 μl of assay buffer (containing 100 mM Tris/HCl, pH 7.5, 0.16 mM NADH and ∼3 units of LDH), the starting absorbance at 340 nm was read. After adding 16 μl of sample and incubating for ∼5 min, the absorbance was read again. The absorbance difference in NADH (molecular absorption coefficient of 6.22 mM−1·cm−1; [27]) was taken to be the amount of pyruvate remaining. When aldose substrates were used, the TBA assay was also used.
Thermostability of the enzymes at 100 °C was checked as follows. KDGA was diluted to 40 μg/ml in 50 mM sodium phosphate, pH 6, divided into 175 μl aliquots and closed into HPLC vials, extra sealed with nylon. Vials were immersed in a 100 °C oil bath and sampled at different time points and immediately cooled on ice. Residual activity was measured with the standard TBA assay.
Crystallization
Sac-KDGA was crystallized using the hanging-drop method. Hexagonal crystals of Sac-KDGA with typical dimensions of 0.3 mm×0.3 mm×0.2 mm were grown at 18 °C. Drops were a mixture of 1 μl of a 19 mg/ml protein solution in water and 1 μl of reservoir solution {0.1 M Hepes, pH 7.5, 30% (v/v) PEG [poly(ethylene glycol)] 400 and 0.2 M MgCl2}. Diffraction data of native crystals were collected at the ID14-1 beam line at the ESRF (European Synchrotron Radiation Facility; Grenoble, France) at 100 K using the reservoir solution as a cryoprotectant. Data were processed with XDS [28] and programs from the CCP4 package [29], with different crystals being indexed either in space group P3121 or in a P6522 cell with related dimensions (see Table 5). MOLREP [30] was first used to find a molecular replacement solution for the P3121 data, using the co-ordinates of one Sso-KDGA subunit (PDB code 1W3I [25]) as a search model. Afterwards, a molecular replacement solution was found in the second space group, P6522, using the preliminary model of Sac-KDGA as a search model. The prime-and-switch protocol of RESOLVE [31] was used for density modification and removal of model bias, after which final models were obtained by iterative cycles of refinement in Refmac5 [32] and model building in Xtalview [33]. To obtain a Schiff-base intermediate with pyruvate, a crystal was transferred to a solution of 0.1 M Mes buffer, pH 6.3, 32% (v/v) PEG 400, 0.2 M MgCl2 and 50 mM sodium pyruvate at 291 K and was flash-frozen after 20 min. Data were collected at the ID14-2 beamline at the ESRF, and processing was as before. Co-ordinates have been deposited with the Protein Data Bank for the native Sac-KDGA structure in P3121 (accession code 2NUW), in P6522 (accession code 2NUX) and the complex with pyruvate (accession code 2NUY).
Table 5. Crystallographic data collection and refinement statistics on Sac-KDGA.
Values in parentheses are for the highest resolution shell. Rsym=|I−〈I〉|/I, where I is the observed intensity and 〈I〉 is the average intensity. Rcryst=hkl, work ||Fobs|−k|Fcalc||/hkl|Fobs|×100%, where Fobs=observed structure factor and Fcalc=calculated structure factor. Rfree=R calculated with 5% of randomly selected data that were omitted from the refinement.
| Native, pH 7.5 | Native, pH 7.5 | Pyruvate, pH 6.3 | |
|---|---|---|---|
| Space group | P3121 | P6522 | P3121 |
| Unit cell dimensions (Å) | a=b=108.2 c=171.9 | a=b=109.5 c=319.6 | a=b=109.1 c=171.3 |
| Resolution (Å) | 30–1.8 (1.88–1.8) | 36–2.5 (2.59–2.5) | 50–2.5 (2.65–2.5) |
| Wavelength (Å) | 0.934 | 0.934 | 0.933 |
| Unique reflections | 107995 | 36273 | 39118 |
| Completeness (%) | 99.9 (99.8) | 90.1 (55.2) | 99.6 (97.1) |
| Redundancy | 10.9 (4.0) | 33.1(19.8) | 6.2 (4.2) |
| I/σ(I) | 14.1 (2.3) | 23.8 (8.4) | 17.6 (5.3) |
| Rsym (%) | 9.6 (55.9)* | 15.5 (37.8) | 5.3 (34.8) |
| Refinement | |||
| Rcryst (%) | 15.7 | 17.6 | 15.9 |
| Rfree (%) | 17.9 | 21.7 | 19.1 |
| Number of atoms | |||
| Protein | 4634 | 4598 | 4616 |
| Pyruvate | − | − | 10 |
| Solvent | 412 | 95 | 162 |
| Average B factor (Å2) protein | 35.5 | 52.0 | 52.9 |
| Pyruvate | − | − | 42.6 |
| Solvent | 44.2 | 43.6 | 50.8 |
| Rmsd bonds/angles (Å/°) | 0.012/1.5 | 0.008/1.3 | 0.008/1.3 |
*The value for the highest resolution shell is affected by anisotropy in diffraction.
RESULTS AND DISCUSSION
Sulfolobus genomes contain putative KDGAs
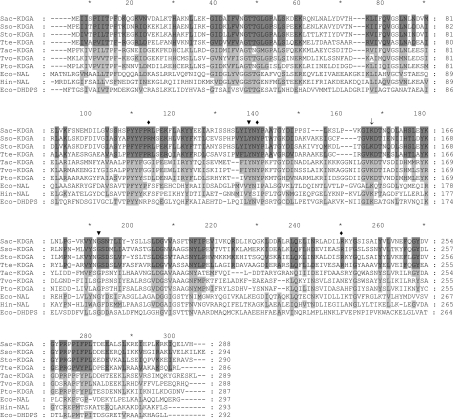
The three sequenced Sulfolobus genomes contain several open reading frames encoding hypothetical aldolases, i.e. enzymes that potentially catalyse aldol-cleavage or aldol-condensation reactions (Table 2). These include DHAP-dependent fructose-1,6-bisphosphate aldolases [34], pyruvate-dependent DHDPSs (dihydrodipicolinate synthases) [35] and KDGAs. DHDPSs and KDGAs do not require (expensive) phosphorylated substrates, and are thus potentially interesting for industrial applications. Previous studies on the KDGA from S. solfataricus suggested a broad substrate specificity [22,24] and therefore we focused on homologues of this enzyme. A BLAST analysis with the protein sequence of the S. solfataricus KDGA (SSO3197) revealed high homology with the KDGA from Thermoproteus tenax (Tte-KDGA) and hypothetical KDGAs from S. acidocaldarius, S. tokodaii, Thermoplasma and Picrophilus torridus. In particular, the active-site residues (Lys-153, Tyr-129, Gly-177, Val-193 and Pro-6) are conserved in these KDGAs, and also in some well-characterized enzymes of the NAL subfamily of type-I aldolases, such as the NALs from Haemophilus influenzae [36] and E. coli [37], and the DHDPS from E. coli [35] (Figure 1). On the basis of this high similarity and a well-conserved gene context, in which the genes for the putative KDGA, KDG kinase and non-phosphorylating-type GAP dehydrogenase are clustered, the hypothetical aldolases from S. acidocaldarius and S. tokodaii are most likely part of a semi-phosphorylated Entner–Doudoroff pathway as was demonstrated for Sso-KDGA and Tte-KDGA [23]. On the basis of that, the homologues analysed were expected to have a similar activity with pyruvate and D,L-GA (phosphorylated and non-phosphorylated) as the KDGA from S. solfataricus [23,24,26].
Table 2. Overview of possible enzymes in Sulfolobus species that hypothetically catalyse aldolase reactions.
| Enzyme | Function | Reaction |
|---|---|---|
| DAHP aldolase | Synthesis | Phosphoenolpyruvate+erythrose 4-phosphate→7-phospho-2-dehydro-3-deoxy-D-arabinoheptonate |
| Citrate lyase | Degradation | Citrate→oxaloacetate+acetate |
| DHDPS-like* | Synthesis | Pyruvate+aspartate-β-semialdehyde→dihydrodipicolinate |
| FBP-aldolase† | Degradation | D-Fructose 1,6-bisphosphate→DHAP+D-GAP |
| Isocitrate lyase‡ | Degradation | Isocitrate→succinate+glyoxylate |
| KDGA | Degradation | KDG→pyruvate+glyceraldehyde |
*Three paralogues in S. solfataricus; one in S. tokodaii and S. acidocaldarius.
†In S. solfataricus and S. tokodaii [34].
‡In S. solfataricus and S. acidocaldarius.
Figure 1. Protein sequence alignment of different members of the NAL subfamily.
Alignment of the three Sulfolobus KDGAs with Thermoproteus tenax KD(P)G aldolase, hypothetical KDGAs from Thermoplasma acidophilum (Tac-KDGA), Thermoplasma volcanium (Tvo-KDGA) and Picrophilus torridus (Pto-KDGA), NALs from E. coli (Eco-NAL) and H. influenzae (Hin-NAL), and the E. coli DHDPS (Eco-DHDPS). Residues that are conserved between the KD(P)G aldolases are indicated with dark-grey shading, while those residues that are also conserved in the hypotheticals and other aldolases are shaded in a lighter grey. The arrow indicates the conserved catalytic lysine residue. Catalytic residues (▼) and residues forming the putative phosphate-binding motif in Sac-KDGA (♦) are indicated.
Stability and activity of Sulfolobus KDGAs
For a thorough characterization of their properties, the hypothetical KDGA genes from S. acidocaldarius and S. tokodaii were cloned and expressed in E. coli using the pET expression system. Sso-KDGA was produced using a previously generated pET24d-derived construct [23]. The three recombinant KDGAs were readily purified by heat incubation and subsequent anion-exchange chromatography. KDGA activity was assayed in the synthesis direction with the substrates pyruvate and D,L-GA producing KDG or its C4-epimer, KDGal. The recombinant enzymes from S. acidocaldarius and S. tokodaii had KDGA activity in the same range as Sso-KDGA. For a thorough comparison between the three KDGAs, their pH and temperature optima, and Michaelis–Menten kinetics were determined. The biochemical properties of Sac-KDGA and Sto-KDGA are not very different from the previously studied Sso-KDGA (Table 3). All three enzymes had higher activity with GAP than with GA (Table 4), similar to reports by Ahmed et al. [23] and Lamble et al. [24] on Sso-KDGA activity. The stability of these enzymes at high temperature was also determined. The KDGAs show a high thermostability, although the half-life time of Sac-KDGA at 100 °C is relatively short, which may be related to the optimal growth temperature of S. acidocaldarius, which is lower than that of the other two species [6]. Taken together, the biochemical experiments and the conserved gene context indicate that these enzymes are involved in a modified Entner–Doudoroff pathway, similar to what has recently been reported for Sso-KDGA [23].
Table 3. Comparison of biochemical data of Sulfolobus KDGAs.
| S. acidocaldarius | S. solfataricus | S. tokodaii | |
|---|---|---|---|
| Optimum pH | 6.5 | 6* | 5.5 |
| Optimum temperature (°C) | ∼99 | 95 | 90 |
| Vmax(pyruvate) (units/mg) | 26.3 (±0.9) | 15.7 (±0.3)* | 17.8 (±0.7) |
| Km(pyruvate) (mM) | 1.1 (±0.2) | 1.0 (±0.1)* | 0.7 (±0.1) |
| Vmax(GA) (units/mg) | 33.1 (±1.3) | 17.1 (±0.4)* | 20.3 (±0.8) |
| Km(GA) (mM) | 6.3 (±0.5) | 5.2 (±0.1)* | 3.9 (±0.4) |
| ½ (min at 100 °C) | 18.9 (±2.1) | 120 (150*) | 110 |
*Data from Buchanan et al. [26].
Table 4. Substrate specificity of Sulfolobus KDGAs based on the rate of condensation with pyruvate as donor substrate (units/mg).
Similar results were obtained with Sto-KDGA. S.D. values are given in parentheses.
Crystal structure of Sac-KDGA
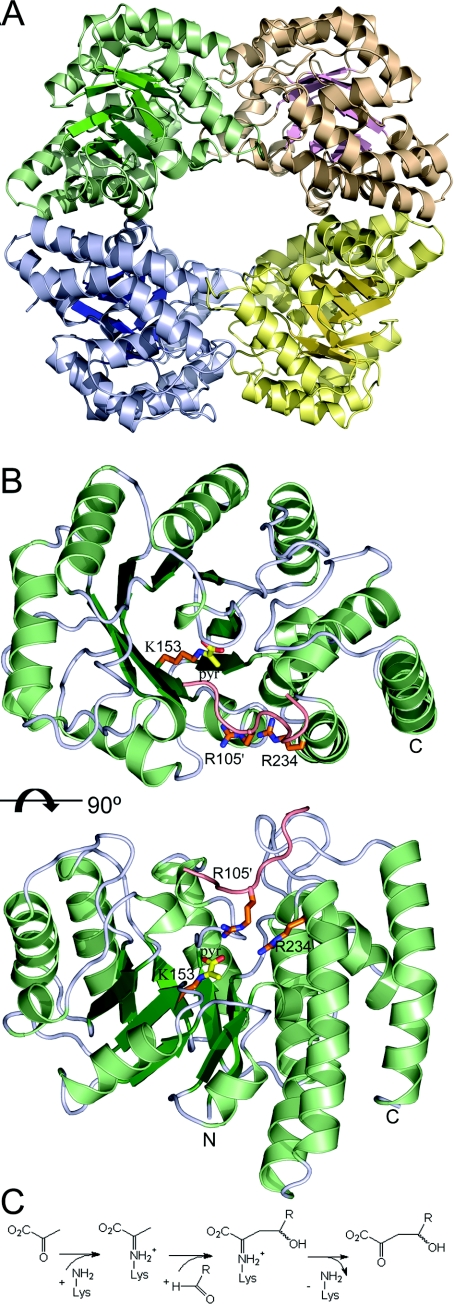
Structural information can provide detailed insights into the molecular basis of the catalytic properties of these aldolases. Sac-KDGA crystallized in two space groups, P3121 and P6522, for which diffraction data were obtained to a maximum resolution of 1.8 and 2.5 Å (1 Å=0.1 nm) respectively (Table 5). The structure could be determined by molecular replacement using the co-ordinates of previously elucidated Sso-KDGA [25] (50% amino acid sequence identity) as the starting model. Initial electron-density maps were of good quality and allowed the models in space groups P3121 and P6522 to be refined to acceptable R factors of 15.7 and 17.6% respectively (Table 5). Both space groups have two molecules in the asymmetric unit, with both of them containing all 288 residues of the native amino acid sequence. The total of four independently refined molecules are nearly identical with pairwise rmsds (root mean square deviations) of their Cα atom positions of less than 0.20 Å. Sac-KDGA has a tetrameric structure (Figure 2A) in which each subunit consists of a (β/α)8 barrel with a C-terminal helical extension (Figure 2B). The tetramer forms a ring-like structure with a hollow core of ∼20 Å diameter. The active site of each subunit is located at the C-terminal end of the β-barrel (Figure 2B) and is only accessible from the inside of the tetrameric ring. This structure not only is similar to the structure of Sso-KDGA, as is evident from the rmsd between 288 matching Cα atoms of only 1.0 Å, but also is similar to the structures of other crystallized members of the NAL subfamily, illustrating the structural conservation within this group of pyruvate-dependent aldolases [25].
Figure 2. Overall structure of Sac-KDGA and reaction mechanism.
(A) Overall structure of Sac-KDGA tetramer. (B) Orthogonal views of the Sac-KDGA monomer. The Schiff-base-forming Lys-153 is indicated in stick presentation with pyruvate (yellow) attached. To show the layout of the substrate-binding pocket, the loop of the neighbouring subunit carrying Arg-105 is also shown, together with Arg-105 and Arg-234 (orange). (C) General scheme of the aldol condensation reaction catalysed by the KDGAs.
The active site is among the most highly conserved regions in the protein. It consists of a pyruvate-binding cavity close to the catalytic Lys-153 (Figure 3A), which is at the bottom of a wider hydrophilic pocket (Figure 3B), accessible from within the tetrameric ring (Figure 2B). The entrance to this hydrophilic pocket is somewhat constricted by the flexible side chains of two arginine residues, Arg-234 and Arg-105′, from a neighbouring subunit (Figures 2B and 3B). The Sac-KDGA active site residues adopt somewhat different conformations as a function of pH. Most conspicuously, at pH 7.5, the catalytic Lys-153 side chain displays some conformational variability. In its most populated conformation, its side chain points away from the pyruvate binding cavity and buries its amino group into the interior of the protein in a hydrogen bond with the Ser-96 side chain (results not shown). Since Sac-KDGA has an activity optimum at approx. pH 6.5, data were also collected from a crystal that had been transferred to a cryosolution at pH 6.3 before freezing. At this pH, the lysine residue is in a conformation that would allow Schiff-base formation and catalysis (results not shown).
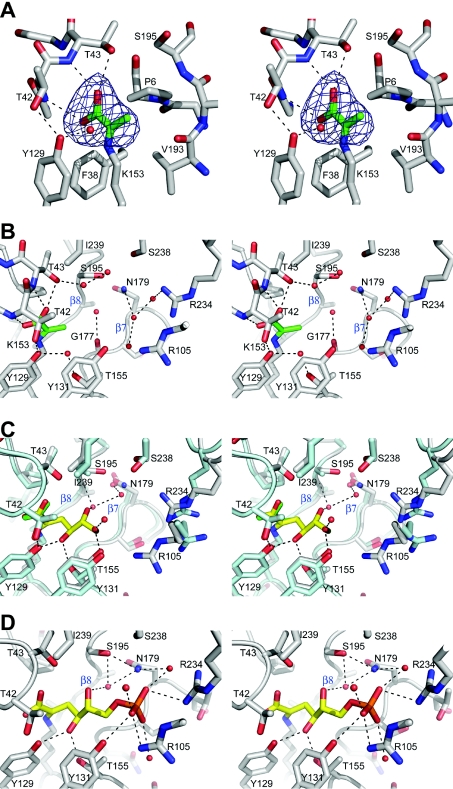
Figure 3. Stereo views of the Sac-KDGA active site.
(A) Weighted difference density map of pyruvate (green) bound to Lys-153 in the pyruvate-binding cavity of Sac-KDGA. Map is contoured at 4 σ. (B) The substrate-binding pocket of Sac-KDGA with pyruvate. Strands β7 and β8 are indicated. (C) Superposition of the Pyr–Sac-KDGA (green/grey) and KDG–Sso-KDGA complex ([25], yellow/cyan). Hydrogen-bonding interactions with the pyruvate moiety have been omitted for clarity, and the orientation is slightly different from that in (B). Water molecules shown are from the KDG–Sso-KDGA complex. (D) KDPG (yellow) modelled in the active site of Sac-KDGA on the basis of the KDG–Sso-KDGA complex [25]. Water molecules from the crystal structure of native Sac-KDGA, which would have favourable interactions with the ligand are also shown. Some hydrogen-bonding interactions have been omitted for clarity and the orientation is slightly different from that of (B) and (C).
To assess the structure of the Schiff-base intermediate with pyruvate, a crystal was soaked in a pyruvate-containing solution at pH 6.3, after which data were collected to 2.5 Å resolution. The well-defined pyruvate moiety is buried inside the active site and makes close interactions with backbone atoms of Thr-42, Thr-43, Val-193 and Gly-177, and the ring of Pro-6, thereby ensuring the high specificity of the enzyme for pyruvate (Figure 3A). This part of the active site and the pyruvate-binding mechanism are highly conserved, with Tyr-129 participating in both substrate binding and in the substrate-assisted catalysis, by relaying a proton between the pyruvate carboxylate and the aldehyde of the acceptor [25,36].
Despite the different conformations of the Lys-153 side chain, the backbone conformations in the active-site region of the Sac-KDGA structure are rigid and remain almost invariant upon pyruvate binding. As this rigidity was also observed for the Sso-KDGA structure [25], it may correlate with thermostability. Interestingly, the active sites of NALs from mesophilic H. influenzae and E. coli are nearly identical with the KDGA active sites [36,37], indicating that the backbone conformation itself is not an adaptation to higher temperatures. The thermostable KDGAs appear to have stabilized this region, and therefore the protein, by rigidifying the loop that carries Tyr-129, which displays a higher flexibility in H. influenzae and E. coli NAL [36,37].
In contrast with the strict requirement for pyruvate, the ability of KDGA to accept both 2-keto-3-deoxy(-6-phospho)gluconate and 2-keto-3-deoxy(-6-phospho)galactonate, in their D- as well as their L-enantiomer conformations, already indicates that the specificity for the acceptor substrate is much more relaxed than for the donor substrate. Previously, the structures of D-KDG and D-KDGal Schiff-base intermediates in Sso-KDGA [25] have revealed the binding mode for these non-phosphorylated substrates during catalysis. The binding modes of the carbon backbone of the GA moieties show considerable variation across the independently determined structures and between D-KDG and D-KDGal. Still, in all cases, the C-4 hydroxy group maintains a hydrogen bond with the Tyr-130 (Tyr-129 in Sac-KDGA) hydroxy group (Figure 3C), which is important for catalysis. The C-5 and C-6 hydroxy groups show more flexibility and have different hydrogen-bonding partners for D-KDG and D-KDGal [25]. These hydrogen-bonding partners are conserved in the Sac-KDGA structure as well as in the sequence of Sto-KDGA (Figures 1 and 3), which is in agreement with their comparable aldolase activity on pyruvate and GA. No structural data are available for the binding of the L-enantiomers of both KDG and KDGal in the KDGA active site, but the hydrophilic nature of the pocket allows their flexible binding in a similar way as the D-enantiomers. This not only provides a structural explanation for the lack of stereocontrol of the KDGAs with their natural non-phosphorylated substrates, but also gives important clues about the applicability of these enzymes with non-physiological substrates.
Sulfolobus KDGAs contain a novel phosphate-binding motif
All KDGAs show higher activity with GAP than with GA (Table 4 and [23]), which may be of physiological relevance in the semi-phosphorylated Entner–Doudoroff metabolic pathway in Sulfolobus spp. It was suggested recently that starvation conditions may favour the slower non-phosphorylated route, whereas otherwise a KDG kinase may phosphorylate KDG to KDPG [24]. The possible binding modes of these larger phosphorylated substrates were investigated by using the Sso-KDGA and Sac-KDGA intermediate structures (Figure 3C) as templates. In Sac-KDGA, the position of a bulky and highly polar phosphate group at the C-6 position is restricted on one side by the apolar side chains of Ile-239 (replaced by leucine in Sso-KDGA and Sto-KDGA) and Thr-42 (Figure 3C). This side of the pocket would not be able to provide favourable interactions to the phosphate. Model building shows that minor rearrangements of the C-6 moiety would bring the phosphate closer to the entrance of the pocket and within hydrogen-bonding distance of the polar side chains of Arg-234 and Tyr-131, and, additionally, Arg-105′ of a neighbouring subunit (Figure 3D). These three residues are absolutely conserved among the KDGAs, with confirmed KDPGA activity from Sulfolobus spp. and T. tenax (Figure 1), and form a novel phosphate-binding motif. In addition, water-mediated contacts would be possible with the nearby side chains of Ser-238, Asn-179 and Ser-195 (Figure 3D). The promiscuity of binding of KDGA, which has already been revealed for the GA moiety [25], is also present in the bound phosphate through these water-mediated contacts and the large conformational flexibility of the Arg-105′ and Arg-234 side chains (Figure 3D). Charge complementarity between the phosphate group and the two conserved arginine residues allows efficient targeting of GAP to the active site and additional enzyme–substrate interactions may also increase substrate affinity. These additional interactions provide a structural explanation for the higher activity observed with GAP and the higher catalytic efficiency with KDPG than KDG. On the other hand, the phosphate group does not influence catalysis directly and does not significantly affect the conformation of the GA moiety (Figure 3D), in line with the observation that the KDGAs do not absolutely require phosphorylated substrates.
The novel phosphate-binding motif is only conserved among the KDGAs from Sulfolobus spp. and T. tenax. In putative KDGAs identified in other thermophilic archaea such as Thermoplasma and Picrophilus torridus, the two arginine residues of the motif are not conserved (Figure 1), neither is the genomic context similar to that of Sulfolobus spp. Significantly, these organisms use a non-phosphorylative Entner–Doudoroff pathway [38,39], in which their KDGAs do not require the dual KDGA/KDPGA functionality of KDGAs active in a semi-phosphorylative pathway. To our knowledge, the KDGAs from Sulfolobus spp. and T. tenax are the only members of the NAL subfamily identified so far that act on phosphorylated substrates and contain this motif. Therefore the motif may be used to identify aldolases from this family with KDPGA activity.
Intriguingly, many of the enzymes of the (β/α)8 fold family that specifically act on phosphorylated substrates contain a conserved phosphate-binding site [40], which differs entirely from the phosphate-binding motif of the KDGAs. In these enzymes, phosphate moieties are often bound between the diverging ends of strands β7 and β8 by their backbone amide groups [40]. Our structural analysis reveals that this structural β7–β8 feature is not conserved in the Sulfolobus KDGAs. In the Sac-KDGA structure, a depression exists between strands β7 and β8, where two water molecules reside, hydrogen-bonding to Gly-177, the side chain of Ser-178 and the main-chain amides and side chains of Asn-179 and Ser-195 (Figure 3B). Manual docking indicates that a phosphate group cannot be accommodated at this position; the backbone amide groups in Sac-KDGA and Sso-KDGA are not oriented properly for an optimal interaction with the phosphoryl oxygen atoms, and the side chain of Asn-179 in Sac-KDGA would give steric hindrance (Figures 3B and 3D). In addition, Asn-179 is replaced by an aspartate residue in both Sso-KDGA and Sto-KDGA (Figure 1), which would give unfavourable interactions with a negatively charged phosphate moiety. These structural differences led us to conclude that this β7–β8 site is not favourable for phosphate binding and support the novel phosphate-binding mechanism that is outlined above.
The phosphorylated KDPG is also a substrate for the KDPG aldolases from E. coli and Pseudomonas putida. These type-I aldolases also have a (β/α)8 fold, but they do not belong to the NAL subfamily and the details of their catalytic mechanisms are different [41,42]. The KDPG aldolase from E. coli has, for example, an rmsd of 2.8 Å over 171 matching Cα atoms with Sac-KDGA. Remarkably, these aldolases do contain a hydrophilic space between the β7 and β8 strands with the structural features of a phosphate-binding site. Significantly, in both the E. coli and P. putida KDPG aldolase crystal structures, a sulfate ion is bound at this β7–β8 site [41,42]. The overall similarity between sulfate and phosphate ions strongly suggests that, in contrast with the Sulfolobus KDGAs, the β7–β8 site functions as a binding site for the phosphate group of their substrate. Interestingly, the KDPG aldolases from E. coli and P. putida do not perform well without the phosphate group on the GA moiety. This indicates that their active site confers considerably more spatial specificity towards their phosphorylated acceptor substrate than the open hydrophilic binding site of the Sulfolobus KDGAs. The strict specificity of the bacterial KDPG aldolases fits their biological function, but makes them less suitable for biotechnological applications. In contrast, the unique phosphate-binding motif of the archaeal KDGAs incorporates the flexibility that is necessary for promiscuous substrate binding. This flexibility may extend to, or be easily adapted to, the conversion of a wider range of acceptor substrates.
Structural requirements for acceptor substrates of KDGAs
KDGAs display substrate promiscuity towards their natural aldehyde substrates [22–24]. It was reported recently that Sso-KDGA can also accept glyceraldehyde acetonide in the synthesis reaction [21]. Furthermore, the structures of Sac-KDGA and Sso-KDGA suggest that the substrate-binding site is able to accept a wide range of acceptor substrates. To identify the structural requirements for aldehydes as acceptor, the activities of Sac-KDGA, Sso-KDGA and Sto-KDGA were assayed with a range of non-physiological aldehyde acceptors (Table 4, and results not shown). Like their other properties, the substrate specificities of the different Sulfolobus KDGAs are quite comparable and will not be discussed separately.
First, the importance of the C-6 moiety was investigated by testing the 2-carbon aldehydes glycolaldehyde and glyoxylate. The available structural information suggests that the binding site should be able to accommodate and orient these smaller substrates through hydrogen bonds with the C-5 hydroxy group. Indeed, activity rates with these substrates were similar to or only slightly lower than that of GA (Table 4), establishing that the KDGAs do accept substrates with only two carbon atoms. The reduced activity with glyoxylate may be the result of its resemblance to pyruvate and weak competitive inhibition of Schiff-base formation. In comparison, KDPG aldolases from P. putida, E. coli and Zymomonas mobilis [43] react quite well with glyoxylate, but react with very low rates only with glycolaldehyde. This provides additional evidence that the KDGAs from Sulfolobus can perform well without a negative charge on the substrate and establishes that they have a catalytic range which is distinctly different from the bacterial KDPG aldolases.
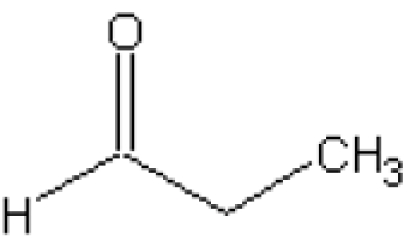
After having established that the C-6 hydroxy group is dispensable, the importance of the hydroxy group at the C-5 position was investigated. In the Sso-KDGA KDG and KDGal complexes, this hydroxy group has hydrogen-bonding interactions with either Tyr-131 or water-mediated contacts with Gly-177 and Ser-195 ([25], see also Figure 3C). Two aldehydes were tested that do not contain such a hydroxy group at C-5, and this showed that acetaldehyde is not accepted and propionaldehyde only at a very low rate (<0.1% of GA) (Table 4). This suggests that the polar hydroxy substituent at the C-5 position is indispensable for proper binding and orientation of the aldehyde in the open hydrophilic binding pocket.
Next, the ability of the KDGAs to accept aldehydes with one more carbon atom than their natural substrates was explored using the simple four-carbon aldotetroses in all possible stereoisomers. The large hydrophilic binding pocket should be able to provide favourable interactions for the additional hydroxy groups of these sugars. Indeed, all aldotetroses tested were readily accepted in synthesis reactions at 5–25% of the GA rate. As all stereoisomers are accepted, albeit with a substantially higher rate for the reaction with L-threose, the KDGAs show a limited enantio-selectivity on these larger substrates. The relative rates are again higher than those of the different KDPG aldolases of P. putida, E. coli and Z. mobilis [43], which accepted these compounds at 0–0.5% of the GAP rate only.
In contrast, the larger aldopentoses that were tested were accepted at low rates (<0.5% of GA) (Table 4). The binding site should, however, be large enough to accommodate these five-carbon acceptors. The low rates with the aldopentoses are likely to be due to a reduced effective concentration of the reactive open-chain conformation of these sugars, which is only ∼0.1% of the total aldopentose concentration at 28 °C [44]. Significantly, the activity of the cleavage of KDO [2-keto-3-deoxyoctonate (2-oxo-3-deoxyoctonate)] (5 mM) to D-arabinose and pyruvate, is much higher than its condensation, reaching 0.146±0.006 unit/mg (S. acidocaldarius), 0.470±0.040 unit/mg (S. solfataricus) and 0.393±0.025 unit/mg (S. tokodaii). Activity appears to be correlated with the effective concentration of the open-chain forms of the substrates, which is probably much higher for the ketone KDO than for the aldehyde D-arabinose, suggesting that the KDGAs do not catalyse the ring-opening step of sugar substrates. Ring opening as the rate-limiting step may also play a role with the aldotetroses [45], and product cyclization may effectively limit the rate of the reverse aldol reaction [24,46].
Taken together, these results define the substrate requirements, or lack thereof, of these KDGAs. The substrate requirements are primarily restricted to the substituents of the carbon atom immediately adjacent to the aldehyde moiety, making glycolaldehyde the smallest acceptor substrate that can still be used by the KDGAs. Despite the absence of enantioselectivity in the condensation reaction, the hydroxy group at the C-5 position of the synthesis product is indispensable for catalysis by providing a significant binding affinity of the aldehyde substrate. Our measurements do not rule out the possibility that additional hydrogen-bonding interactions of other parts of an aldehyde acceptor may partially compensate for the decrease in binding affinity. The structure of the binding sites of Sac-KDGA and Sso-KDGA combined with our activity experiments explain the general lack of specific requirements on the structure of the acceptor beyond the glycolaldehyde moiety. This suggests that these enzymes should be applicable in the biosynthesis of a broad range of compounds which carry this structural motif.
The three different Sulfolobus KDGAs are not the only NAL family members which accept several different (non-phosphorylated) aldehydes in the condensation reaction with pyruvate. Other members of the NAL subfamily also have broad acceptor specificity, indicating the general versatility of their binding sites. Strikingly, although their binding sites are very similar, acceptors are bound very differently in different enzymes. In NAL, for example, substrate analogues may be accommodated in a different part of the active-site pocket [36]. Given the similarity between active sites and substrates, it was previously suggested that the substrates sialic acid and KDG would bind in a similar way and that substrate specificity within the NAL subfamily would be modulated in a subtle way by only a few amino acid mutations [36]. The structural and biochemical evidence for Sac-KDGA and Sso-KDGA [25] now give striking illustrations of how structurally similar binding sites may bind substrates in entirely different ways. This suggests that a limited number of mutations in the active site may already lead to novel substrate specificities. This has been demonstrated by the creation of NAL enzymes with DHDPS activity [47] or with L-KDO aldolase activity [48]. The thermostability, ease of production and well-characterized acceptor specificity of the Sulfolobus KDGAs make these enzymes highly suitable platforms for such an approach.
Acknowledgments
This research was performed as part of the IBOS Programme (Integration of Biosynthesis and Organic Synthesis) of Advanced Chemical Technologies for Sustainability (ACTS). In addition we thank Theo Sonke (DSM, Geleen, The Netherlands) and Maurice Franssen (Wageningen University) for fruitful discussions.
References
- 1.Machajewski T. D., Wong C. H. The catalytic asymmetric aldol reaction. Angew. Chem. Int. Ed. Engl. 2000;39:1352–1375. doi: 10.1002/(sici)1521-3773(20000417)39:8<1352::aid-anie1352>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 2.Samland A. K., Sprenger G. A. Microbial aldolases as C-C bonding enzymes: unknown treasures and new developments. Appl. Microbiol. Biotechnol. 2006;71:253–264. doi: 10.1007/s00253-006-0422-6. [DOI] [PubMed] [Google Scholar]
- 3.Gijsen H. J., Qiao L., Fitz W., Wong C. H. Recent advances in the chemoenzymatic synthesis of carbohydrates and carbohydrate mimetics. Chem. Rev. 1996;96:443–474. doi: 10.1021/cr950031q. [DOI] [PubMed] [Google Scholar]
- 4.Takayama S., McGarvey G. J., Wong C. H. Microbial aldolases and transketolases: new biocatalytic approaches to simple and complex sugars. Annu. Rev. Microbiol. 1997;51:285–310. doi: 10.1146/annurev.micro.51.1.285. [DOI] [PubMed] [Google Scholar]
- 5.Brock T. D., Brock K. M., Belly R. T., Weiss R. L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 6.Grogan D. W. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 1989;171:6710–6719. doi: 10.1128/jb.171.12.6710-6719.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki T., Iwasaki T., Uzawa T., Hara K., Nemoto N., Kon T., Ueki T., Yamagishi A., Oshima T. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles. 2002;6:39–44. doi: 10.1007/s007920100221. [DOI] [PubMed] [Google Scholar]
- 8.DeLong E. F., Pace N. R. Environmental diversity of bacteria and archaea. Syst. Biol. 2001;50:470–478. [PubMed] [Google Scholar]
- 9.Stedman K. M., Schleper C., Rumpf E., Zillig W. Genetic requirements for the function of the archaeal virus SSV1 in Sulfolobus solfataricus: construction and testing of viral shuttle vectors. Genetics. 1999;152:1397–1405. doi: 10.1093/genetics/152.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aravalli R. N., Garrett R. A. Shuttle vectors for hyperthermophilic archaea. Extremophiles. 1997;1:183–191. doi: 10.1007/s007920050032. [DOI] [PubMed] [Google Scholar]
- 11.Cannio R., Contursi P., Rossi M., Bartolucci S. An autonomously replicating transforming vector for Sulfolobus solfataricus. J. Bacteriol. 1998;180:3237–3240. doi: 10.1128/jb.180.12.3237-3240.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.She Q., Singh R. K., Confalonieri F., Zivanovic Y., Allard G., Awayez M. J., Chan-Weiher C. C., Clausen I. G., Curtis B. A., De Moors A., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawarabayasi Y., Hino Y., Horikawa H., Jin-no K., Takahashi M., Sekine M., Baba S., Ankai A., Kosugi H., Hosoyama A., et al. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 2001;8:123–140. doi: 10.1093/dnares/8.4.123. [DOI] [PubMed] [Google Scholar]
- 14.Chen L., Brugger K., Skovgaard M., Redder P., She Q., Torarinsson E., Greve B., Awayez M., Zibat A., Klenk H. P., et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 2005;187:4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klimczak L. J., Grummt F., Burger K. J. Purification and characterization of DNA polymerase from the archaebacterium Sulfolobus acidocaldarius. Nucleic Acids Res. 1985;13:5269–5282. doi: 10.1093/nar/13.14.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi M., Rella R., Pensa M., Bartolucci S., De Rosa M., Gambacorta A., Raia C. A., Dell'Aversano Orabona N. Structure and properties of a thermophilic and thermostable DNA polymerase isolated from Sulfolobus solfataricus. Syst. Appl. Microbiol. 1986;7:337–341. [Google Scholar]
- 17.Dionne I., Robinson N. P., McGeoch A. T., Marsh V. L., Reddish A., Bell S. D. DNA replication in the hyperthermophilic archaeon Sulfolobus solfataricus. Biochem. Soc. Trans. 2003;31:674–676. doi: 10.1042/bst0310674. [DOI] [PubMed] [Google Scholar]
- 18.Zillig W., Stetter K. O., Janekovic D. DNA-dependent RNA polymerase from the archaebacterium Sulfolobus acidocaldarius. Eur. J. Biochem. 1979;96:597–604. doi: 10.1111/j.1432-1033.1979.tb13074.x. [DOI] [PubMed] [Google Scholar]
- 19.Reiter W. D., Palm P., Zillig W. Analysis of transcription in the archaebacterium Sulfolobus indicates that archaebacterial promoters are homologous to eukaryotic pol II promoters. Nucleic Acids Res. 1988;16:1–19. doi: 10.1093/nar/16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell S. D., Jackson S. P. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 1998;6:222–228. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 21.Lamble H. J., Danson M. J., Hough D. W., Bull S. D. Engineering stereocontrol into an aldolase-catalysed reaction. Chem. Commun. 2005:124–126. doi: 10.1039/b413255f. [DOI] [PubMed] [Google Scholar]
- 22.Lamble H. J., Heyer N. I., Bull S. D., Hough D. W., Danson M. J. Metabolic pathway promiscuity in the archaeon Sulfolobus solfataricus revealed by studies on glucose dehydrogenase and 2-keto-3-deoxygluconate aldolase. J. Biol. Chem. 2003;278:34066–34072. doi: 10.1074/jbc.M305818200. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed H., Ettema T. J., Tjaden B., Geerling A. C., van der Oost J., Siebers B. The semi-phosphorylative Entner–Doudoroff pathway in hyperthermophilic archaea: a re-evaluation. Biochem. J. 2005;390:529–540. doi: 10.1042/BJ20041711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamble H. J., Theodossis A., Milburn C. C., Taylor G. L., Bull S. D., Hough D. W., Danson M. J. Promiscuity in the part-phosphorylative Entner–Doudoroff pathway of the archaeon Sulfolobus solfataricus. FEBS Lett. 2005;579:6865–6869. doi: 10.1016/j.febslet.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 25.Theodossis A., Walden H., Westwick E. J., Connaris H., Lamble H. J., Hough D. W., Danson M. J., Taylor G. L. The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner–Doudoroff pathway in Sulfolobus solfataricus. J. Biol. Chem. 2004;279:43886–43892. doi: 10.1074/jbc.M407702200. [DOI] [PubMed] [Google Scholar]
- 26.Buchanan C. L., Connaris H., Danson M. J., Reeve C. D., Hough D. W. An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated substrates. Biochem. J. 1999;343:563–570. [PMC free article] [PubMed] [Google Scholar]
- 27.Horecker B. L., Kornberg A. The extinction coefficients of the reduced band of pyridin nucleotides. J. Biol. Chem. 1948;175:385–390. [PubMed] [Google Scholar]
- 28.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 29.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 31.Terwilliger T. C. Using prime-and-switch phasing to reduce model bias in molecular replacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2144–2149. doi: 10.1107/S0907444904019535. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 33.McRee D. E. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 34.Siebers B., Brinkmann H., Dorr C., Tjaden B., Lilie H., van der Oost J., Verhees C. H. Archaeal fructose-1,6-bisphosphate aldolases constitute a new family of archaeal type class I aldolase. J. Biol. Chem. 2001;276:28710–28718. doi: 10.1074/jbc.M103447200. [DOI] [PubMed] [Google Scholar]
- 35.Dobson R. C., Valegård K., Gerrard J. A. The crystal structure of three site-directed mutants of Escherichia coli dihydrodipicolinate synthase: further evidence for a catalytic triad. J. Mol. Biol. 2004;338:329–339. doi: 10.1016/j.jmb.2004.02.060. [DOI] [PubMed] [Google Scholar]
- 36.Barbosa J. A., Smith B. J., DeGori R., Ooi H. C., Marcuccio S. M., Campi E. M., Jackson W. R., Brossmer R., Sommer M., Lawrence M. C. Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme. J. Mol. Biol. 2000;303:405–421. doi: 10.1006/jmbi.2000.4138. [DOI] [PubMed] [Google Scholar]
- 37.Izard T., Lawrence M. C., Malby R. L., Lilley G. G., Colman P. M. The three-dimensional structure of N-acetylneuraminate lyase from Escherichia coli. Structure. 1994;2:361–369. doi: 10.1016/s0969-2126(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 38.Reher M., Schonheit P. Glyceraldehyde dehydrogenases from the thermoacidophilic euryarchaeota Picrophilus torridus and Thermoplasma acidophilum, key enzymes of the non-phosphorylative Entner–Doudoroff pathway, constitute a novel enzyme family within the aldehyde dehydrogenase superfamily. FEBS Lett. 2006;580:1198–1204. doi: 10.1016/j.febslet.2006.01.029. [DOI] [PubMed] [Google Scholar]
- 39.Budgen N., Danson M. J. Metabolism of sugar via a modified Entner–Doudoroff pathway in the thermoacidophilic archaebacterium Thermoplasma acidophilum. FEBS Lett. 1986;196:207–210. [Google Scholar]
- 40.Nagano N., Orengo C. A., Thornton J. M. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 41.Allard J., Grochulski P., Sygusch J. Covalent intermediate trapped in 2-keto-3-deoxy-6- phosphogluconate (KDPG) aldolase structure at 1.95-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3679–3684. doi: 10.1073/pnas.071380898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bell B. J., Watanabe L., Rios-Steiner J. L., Tulinsky A., Lebioda L., Arni R. K. Structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003;59:1454–1458. doi: 10.1107/s0907444903013192. [DOI] [PubMed] [Google Scholar]
- 43.Shelton M. C., Cotterill I. C., Novak S. T. A., Poonawala R. M., Sudarshan S., Toone E. J. 2-Keto-3-deoxy-6-phosphogluconate aldolases as catalysts for stereocontrolled carbon–carbon bond formation. J. Am. Chem. Soc. 1996;118:2117–2125. [Google Scholar]
- 44.Drew K. N., Zajicek J., Bondo G., Bose B., Serianni A. S. 13C-labeled aldopentoses: detection and quantitation of cyclic and acyclic forms by heteronuclear 1D and 2D NMR spectroscopy. Carbohydr. Res. 1998;307:199–209. [Google Scholar]
- 45.Serianni A. S., Pierce J., Huang S.-G., Barker R. Anomerization of furanose sugars: kinetics of ring-opening reactions by 1H and 13C saturation-transfer NMR spectroscopy. J. Am. Chem. Soc. 1982;104:4037–4044. [Google Scholar]
- 46.Midelfort C. F., Gupta R. K., Meloche H. P. Specificity of 2-keto-3-deoxygluconate-6-P aldolase for open chain form of 2-keto-3-deoxygluconate-6-P. J. Biol. Chem. 1977;252:3486–3492. [PubMed] [Google Scholar]
- 47.Joerger A. C., Mayer S., Fersht A. R. Mimicking natural evolution in vitro: an N-acetylneuraminate lyase mutant with an increased dihydrodipicolinate synthase activity. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5694–5699. doi: 10.1073/pnas.0531477100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu C. C., Hong Z., Wada M., Franke D., Wong C. H. Directed evolution of D-sialic acid aldolase to L-3-deoxy-manno-2-octulosonic acid (L-KDO) aldolase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9122–9126. doi: 10.1073/pnas.0504033102. [DOI] [PMC free article] [PubMed] [Google Scholar]