Abstract
Studies comparing the binding of genuine cobalamin (vitamin B12) to that of its natural or synthetic analogues have long established increasing ligand specificity in the order haptocorrin, transcobalamin and intrinsic factor, the high-affinity binding proteins involved in cobalamin transport in mammals. In the present study, ligand specificity was investigated from a structural point of view, for which comparative models of intrinsic factor and haptocorrin are produced based on the crystal structure of the homologous transcobalamin and validated by results of published binding assays. Many interactions between cobalamin and its binding site in the interface of the two domains are conserved among the transporters. A structural comparison suggests that the determinant of specificity regarding cobalamin ligands with modified nucleotide moiety resides in the β-hairpin motif β3-turn-β4 of the smaller C-terminal domain. In haptocorrin, it provides hydrophobic contacts to the benzimidazole moiety through the apolar regions of Arg357, Trp359 and Tyr362. Together, these large side chains may compensate for the missing nucleotide upon cobinamide binding. Intrinsic factor possesses only the tryptophan residue and transcobalamin only the tyrosine residue, consistent with their low affinity for cobinamide. Relative affinity constants for other analogues are rationalized similarly by analysis of steric and electrostatic interactions with the three transporters. The structures also indicate that the C-terminal domain is the first site of cobalamin-binding since part of the β-hairpin motif is trapped between the nucleotide moiety and the N-terminal domain in the final holo-proteins.
Keywords: cobalamin, comparative modelling, haptocorrin, intrinsic factor, structure comparison, transcobalamin, transport proteins
Abbreviations: Cbi, cobinamide; Cbl, cobalamin; DMB, 5,6-dimethylbenzimidazole; HC, haptocorrin; IF, intrinsic factor; TC, transcobalamin; rms, root mean square
INTRODUCTION
The transport of the essential micronutrient Cbl (cobalamin; vitamin B12) from food to cells relies on three successive proteins in mammals, HC (haptocorrin), gastric IF (intrinsic factor) and TC (transcobalamin) [1–3]. Cbl is required by cells as the basis for two enzyme cofactors, methyl-Cbl for methionine synthase and 5′-deoxyadenosyl-Cbl (Ado-Cbl) for methyl-malonyl-CoA mutase [4]. Upon initial uptake of Cbl from food, Cbl becomes bound in the stomach to salivary HC. After proteolysis of HC in the duodenum, Cbl is passed on to IF. Mucosal cells in the ileum absorb the IF–Cbl complex via endocytosis mediated by a specific receptor. In the enterocyte, the IF–Cbl complex is degraded and Cbl is transferred to TC which delivers Cbl to cells via the blood [5]. Only the fraction of Cbl bound to TC is taken up via endocytosis by a specific receptor on most cell types. HC that is also present in plasma cannot facilitate cellular uptake of Cbl except in hepatocytes. TC–Cbl is degraded in lysosomes to release Cbl for further conversion into the important cofactors. Cbl deficiency as a result of impaired intestinal absorption is more common among the elderly population and may lead to disorders including haematologic abnormalities and defects in the nervous system and metabolism [6].
Table 1 summarizes some properties of the human Cbl-transporters. Investigation of their gene structure suggested that the three proteins have a common evolutionary origin and that TC has diverged first from the ancestral gene [7]. The homologous Cbl transporters show about 25% overall sequence identity but several shorter stretches possess considerably higher similarity. Glycosylation is observed for IF and to an even higher extent for HC, but not for TC [2]. The proteins have extraordinary affinity for the vitamin, with similar Kd values <1 pM [8]. However, they show differences in their specificity for Cbl analogues, which are also produced by microorganisms [9].
Table 1. Properties of Cbl transporters in humans.
| Property | IF | HC | TC |
|---|---|---|---|
| Chromosome | 11 | 11 | 22 |
| Source | Mainly gastric mucosa | Gastric mucosa, white blood cells, glands, etc. | Vascular endothelium |
| Occurrence | Stomach, small intestine | Body fluids (mainly saliva, plasma) | Body fluids (mainly plasma) |
| Function | Cbl-transport from proximal intestine to | Salivary HC: binding of dietary HC in the | Cbl-transport from epithelial cells |
| epithelial cells of the ileum | stomach (protection of Cbl from acid | to tissue and organ cells | |
| Filtering out Cbl-analogues (?) | hydrolysis and from uptake by intestinal fauna ?) | Facilitation of Cbl-uptake into cells | |
| Plasma HC: Cbl recruitment from stores, scavenging of Cbl-analogues (?) | |||
| Amino acid content (molecular mass) | 399 (43.4 kDa) | 410 (45.6 kDa) | 409 (45.5 kDa) |
| Carbohydrate content (w/w) | 9–15% | 30–40% | 0 |
| Kd (H2O–Cbl)* | 1 pM | 0.01 pM | 0.005 pM |
*Equilibrium dissociation constants at pH 7.5 and 20 °C taken from [8].
Recently, the structures of human and bovine TC in complex with Cbl have been solved by X-ray crystallography [10]. These allowed the detailed description of the Cbl-binding mode and represent the prototype for the family of mammalian Cbl transporters since no structures have been reported yet for IF or HC. Their structure determination by X-ray crystallography is likely to remain challenging due to difficulties with crystallization of these highly glycosylated proteins. Detailed structural information for IF and HC is of importance for understanding their way of functioning in greater detail, especially the interactions with the ligand Cbl, the role of glycosylation and the recognition by specific cell surface receptors.
In particular, knowledge of the Cbl-binding mode for all three proteins is essential to rationalize the observed diversity in ligand specificity. IF shows the highest specificity for Cbl and is suggested to select in the first place genuine Cbl among Cbl analogues in the diet, while HC as the least-specific binder may remove other Cbl analogues from the circulation that can harmfully compete for the binding site of Cbl in methionine synthase and methyl-malonyl-CoA mutase [9]. The precise physiological role of HC within the complex process of Cbl internalization is not yet well understood. Furthermore, the structural basis for ligand specificity is of interest in view of the design of Cbl-based bioconjugates. Such bioconjugates show potential as cytotoxic drugs or imaging agents in cancer treatment [11] or may function as Cbl competitors to selectively block cell growth in the treatment of AIDS-related lymphoma [12] or cancer [13]. These applications are based on the observation that proliferating cells express more surface receptors for holo-TC, and thus take up proportionally more Cbl, than cells in the stationary phase [11,14].
In the present study we analyse all three Cbl transporters with regard to structural determinants of ligand specificity. For this purpose, we produced comparative models of Cbl-complexed human IF and HC guided by our recently determined X-ray structures of human and bovine TC [10]. The models are validated with the help of available experimental data for a variety of structural features and are compared with the human TC structure.
METHODS
Modelling
The comparative modelling of IF and HC is based on the structures of human and bovine TC determined by X-ray crystallography (PDB accession codes 2BB5 and 2BB6 respectively [10]) and on the multiple sequence alignment of IF and HC with these two TC forms shown in Figure 1. This alignment was extracted from a larger multiple sequence alignment to increase its accuracy, employing the following 16 mammalian Cbl-transporting proteins: TC from man (SwissProt entry P20062), chimpanzee (XP_525562), orangutan (Q5REL7), dog (XP_543481), rat (Q9R0D6), mouse (O88968) and cow (Q9XSC9); IF from man (P27352), dog (Q5XWD5), rat (P17267), mouse (P52787) and cow (XP_873322); HC from man (P20061), dog (XP_855361), cow (XP_873306) and hog (P17630). The alignment was performed with CLUSTALW v.1.82 [15] with the Gonnet 250 scoring matrix. Using MODELLER 8v1, 10 models were calculated both for IF and HC [16] using default parameters. The Cbl ligand of the TC template structures were included as rigid body (‘block residue’ definition in MODELLER) whereas water molecules were excluded from the templates. Conserved disulfide bridges were automatically used as geometric restraints for IF and HC. The best model was chosen according to MODELLER's lowest objective function criterion. The respective best model was subsequently refined iteratively by geometry idealization with REFMAC5 [17] and model rebuilding with the graphics program O [18]. The energy profile (MODELLER's DOPE score) of the models was compared with those of the experimentally determined TC structures. The model geometry was analysed with MOLPROBITY [19] regarding Ramachandran plot, van der Waals contacts, hydrogen bonds and steric clashes and with PROCHECK [20] regarding amino acid stereochemistry. Cavities in the models accessible to a probe of 1.4 Å (1 Å=0.1 nm) radius (corresponding to a water molecule) were found with the program CAVENV [21] and visualized with O. rms (root mean square) deviations of Cα-atoms in a pair of models were calculated by DALI [22]. The electrostatic potential on the protein surface was calculated with APBS [23] assuming neutral pH. Protein structures were visualized using PYMOL (DeLano Scientific). Atomic accessibility areas were calculated with NACCESS (S. L. Hubbard and J. M. Thornton, University College London, London, U.K.).
Figure 1. Sequence alignment of human and bovine TC with human IF and HC.
This alignment together with the known three-dimensional structures of the two TC forms were used for comparative modelling of IF and HC. One letter amino acid codes in italics represent secondary structure elements in the α-domain and β-domain respectively. α-helices (α) and β-strands (β) are numbered consecutively from the N- to the C-terminus. Residues participating in hydrophobic interactions with the ligand Cbl are highlighted in bold. Residues participating in hydrogen-bonds to Cbl are marked by the letter for the involved corrin side chain above the residue (those residues employing their side chain are additionally highlighted in grey). The histidine residue which co-ordinates to the Co ion of Cbl in human and bovine TC (after displacement of H2O at the upper axial position of H2O–Cbl) and cysteine residues of disulfide bridges are marked by rectangular boxes. Potential asparagine-linked glycosylation sites in IF and HC (based on the Asn-X-Thr/Ser consensus sequence) are underlined. See Supplementary Figure 1 (at http://www.BiochemJ.org/bj/403/bj4030431add.htm) for a colour version of this figure.
RESULTS AND DISCUSSION
Description of the IF and HC models and comparison with TC
Comparative models were produced for the human forms of IF and HC on the basis of the sequence alignment with human and bovine TC (Figure 1) and guided by the recently determined crystal structures of human and bovine TC [10]. The overall correctness of the modelling is indicated by the absence of bad overlaps and large holes. The models show compact globular domains, well-defined hydrogen-bonds for buried polar residues and secondary structure elements, as well as extended hydrophobic domain cores. Small cavities were found which in size and location were similar to the water-occupied cavities observed in the crystal structures of TC. The energy profiles of the models were similar to those of the TC template structures. IF and HC show reasonable stereochemistry with regard to rms deviations from ideal geometry and Ramachandran plot statistics (see Supplementary Table 1 at http://www.BiochemJ.org/bj/403/bj4030431add.htm).
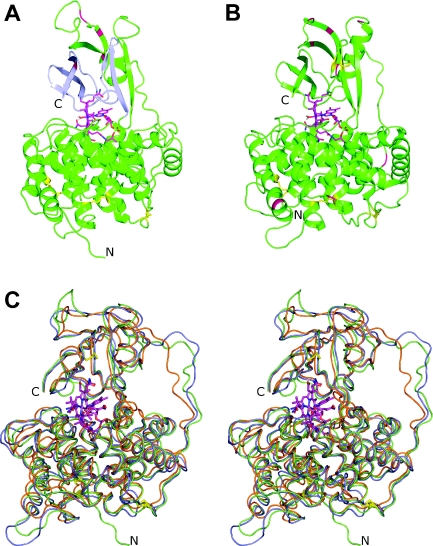
As the template protein TC, the comparative models of IF and HC possessed a two-domain architecture in which a larger N-terminal domain (‘α-domain’), composed of 12 α-helices in an α6-α6 barrel and a short 3/10 helix, is connected by a flexible linker region to a smaller C-terminal domain (‘β-domain’) consisting mainly of two β-sheets. The Cbl ligand, with its DMB (5,6-dimethylbenzimidazole) moiety co-ordinated to the Co ion (‘base-on’ form), was tightly enclosed in the domain interface (Figure 2). The sequence identity of human IF and HC as well as the rms deviations of superimposed Cα-atoms of their models are given in Table 2, including also the two TC forms. The regions of elevated similarity between the Cbl transporters in the sequence alignment of Figure 1 were mapped on the experimentally determined human TC structure. Regarding the α-domain, these regions coincided with the inner six (even-numbered) helices, i.e. the core of the domain, while similarity was spread over the entire β-domain.
Figure 2. Comparative models of IF and HC.
(A) Ribbon representation of the IF model showing secondary structure elements, the orientation of the ligand Cbl (magenta) with respect to the N- and C-terminal domain, the disulfide bridges (yellow sticks) and potential asparagine-linked glycosylation sites (red). The part of the β-domain coloured in blue corresponds to the 50 C-terminal residues which were removed from the protein in the Cbl-binding studies of Tang et al. [26], see text. (B) Ribbon representation of HC using the colour scheme of (A). (C) Stereo view of superimposed backbone traces of human Cbl-transporters. The comparative models of IF and HC are shown in green and blue respectively, and the human TC structure as determined by X-ray crystallography is coloured orange.
Table 2. Comparison of primary and tertiary structures of Cbl-transporters.
The upper diagonal gives sequence identities (relative to the longer sequence in each pair) and, in parenthesis, sequence similarity (‘positives’ in the substitution matrix of Henikoff and Henikoff [36]). The lower diagonal reports rms deviations of Cα-atoms for equivalenced residues in the models superimposed with DALI [22]. The number of these residues is given in parenthesis.
| Human IF | Human HC | Human TC | Bovine TC | |
|---|---|---|---|---|
| Human IF | – | 30% (48%) | 23% (38%) | 22% (38%) |
| Human HC | 1.3 Å (387) | – | 27% (45%) | 25% (43%) |
| Human TC | 1.6 Å (379) | 1.5 Å (389) | – | 73% (81%) |
| Bovine TC | 1.2 Å (381) | 1.1 Å (395) | 1.2 Å (409) | – |
Experimental proof of the presence of a two-domain architecture in human IF has been given previously [24,25]. Two fragments, approx. 30 kDa and 20 kDa in size, were obtained from natural proteolysis of recombinant IF (50 kDa) and closely corresponded to the α- and β-domain of TC according to their N-terminal sequencing. Each fragment alone was able to bind Cbl with high affinity, and a mixture of both fragments, provided Cbl was present, could be assembled into a ternary complex which behaved in a very similar way to uncleaved IF–Cbl with respect to Cbl-binding and receptor recognition.
The finding [26] that truncation of 50 amino acids from the C-terminal end of human IF abolished its Cbl-binding ability is consistent with our IF model, since this truncation leads to the loss of all hydrogen-bond interactions and most of the hydrophobic contacts between Cbl and the β-domain of IF.
All secondary structure elements of TC were also present in the models of IF and HC (Figure 1). Helices α3, α5 and α13 were shorter in IF than in the other proteins, whereas particularly extended loops are found between helices α4 and α5 in TC, between α8 and α9 in HC and between strands β1 and β2 in IF. The flexible linker is three and four residues longer in IF and HC respectively. Swapping of amino acids was observed between IF and HC in the hydrophobic core of the β-domain, involving the hydrophobic contact Leu325–Phe370 in IF and Phe335–Leu381 in HC. This concerted mutation is located at the same position as that between human and bovine TC which involves a leucine residue and a valine residue.
Disulfide bridges
Human and bovine TC show three disulfide bridges in the N-terminal α-domain. In human TC, these are Cys3–Cys249, Cys147–Cys187 and Cys98–Cys291. Results from initial multiple sequence alignments indicated that in IF all three disulfide bridges are conserved, whereas for HC the conservation of only the first two bridges is obtained. The presence of the third bridge, however, appears possible since one of the involved cysteine residues (Cys82) is already conserved and the second (Cys285) is shifted away from the conserved position by only one amino acid towards the C-terminus. An alignment featuring the conservation of all three disulfide bridges also in HC can thus be obtained by moving, both for TC and IF, one insertion gap from the nearby linker region to a position N-terminal of the involved cysteine residue. A model of HC calculated with the third bridge shows excellent stereochemistry in the loops hosting the bridging cysteine residues, i.e. the introduction of a restraint for this third bridge does not lead to distorted backbone geometry.
Apart from the three conserved disulfide bridges, an additional fourth bridge was found in the model of HC, where Cys365 and Cys370 connect the β-strands β4 and β5 in the C-terminal domain. No restraint for this bridge was present during modelling and initial refinement. The possibility of the presence of such a bridge was raised earlier based on a primary sequence analysis [27]. A multiple alignment of four known HC sequences (from man, dog, cow and hog) shows that the fourth disulfide bridge is a feature unique to the human form. In conclusion, the following disulfide bridges were present in our models: Cys8–Cys228, Cys125–Cys164, Cys85–Cys270 in IF and Cys3–Cys242, Cys132–Cys174, Cys82–Cys285, Cys365–Cys370 in HC.
Glycosylation sites
All glycosylation sites that were predicted on the basis of the Asn-X-Thr/Ser consensus sequence were found to be solvent-exposed in our comparative models (Figure 2). The IF model showed glycosylation sites exclusively on the β-domain, as was confirmed experimentally [24]. Asn293 was suggested to be glycosylated due to the inability to identify this particular residue during sequencing. HC also possessed two potential glycosylation sites on the α-domain which fall on the surface region recently proposed as a receptor-recognition site of TC [10]. The presence of carbohydrates on the α-domain of HC, but not on TC, may therefore explain how cell-surface receptors for TC discriminate between the two plasma Cbl transporters TC and HC. Moreover, assuming that the receptor-recognition site is located on the α-domain alone [26], the lack of carbohydrates on the α-domain of IF may account for the observation that glycosylation does not effect receptor-binding of IF [28]. Recently, however, the presence of both domains was found to be required for binding of IF to its specific receptor cubilin [25].
Cbl environment in IF and HC
The lower side of Cbl (termed the α-side) is occupied by the nucleotide moiety and is embedded in the protein (see discussion below). The upper axial side (termed the β-side) contains the ligand H2O in our models. This ligand can interact via hydrogen-bonds with the main-chain oxygen of Pro149 in IF or Gln159 in HC. It possesses solvent accessibility which is, however, restricted by the C-terminal part of the loop between helices α7 and α8. The accessibility of the β-side to solvent is in agreement with experimental data on kinetics for binding of Cbl with different upper ligands. The fact that very similar binding constants are obtained for Cbl with small ligands, such as CN or H2O, and larger ligands such as 5′-deoxyadenosyl, has long been considered as evidence that the β-side is at least partly solvent-exposed and thus does not play a role in determining ligand specificity [29]. As concluded in [1], the transporters show comparable affinities for the various Cbl forms and do not generally discriminate between Cbl with different upper axial ligands (see Supplementary Table 2 at http://www.BiochemJ.org/bj/403/bj4030431add.htm). A special feature in this respect (with unknown physiological function) may be attributed to TC which treats H2O–Cbl differently to CNCbl and N3Cbl in that it replaces the H2O ligand with a histidine side chain. Spectroscopic studies on the kinetics of ligand exchange at the upper axial position of H2O–Cbl upon binding to any of the three transporters [8], revealed reactivity of Co with externally supplied histidine for IF- and HC-bound Cbl, but not for TC-bound Cbl. In fact, the recent X-ray structure revealed that upon binding of H2O–Cbl to TC, the side chain of a histidine residue forms a co-ordination bond to the Co ion [10]. The Co-co-ordinating histidine residue in TC is not conserved in IF or HC, as is visible from the sequence alignment in Figure 1, and consequently, H2O remains co-ordinated.
Most of the polar and hydrophobic interactions between Cbl and TC are also observed in our comparative models of IF and HC (Table 3). Water-mediated polar interactions are not included in this comparison. IF lacks the hydrogen-bonds to the oxygen atom of the corrin side chains a and f which are present in both TC and HC. Regarding hydrophobic contacts, IF and HC show identical interactions, with an exception being the smaller Val351 in IF at the position of Tyr362 in HC and TC close to the DMB, and an additional contact of a threonine Cγ atom in IF. TC differs from those hydrophobic interactions in three regions. First, the tryptophan residue close to DMB (Trp348 in IF, Trp359 in HC) is occupied by a Ser359 and three water molecules. Secondly, an additional contact exists at the pyrrole ring A of the corrin macrocycle and involves Met270 in human TC. Thirdly, the phenylalanine side chain is absent at the upper side of the corrin macrocycle between rings C and D. All three transporters have the hydrophobic nature of the protein environment in van der Waals contact with the DMB moiety in common, as has been concluded previously from the observation that CN− is unable to displace DMB from the Co ion in IF [30].
Table 3. Interactions between Cbl and its human transport proteins.
| Cbl atom | IF | HC | TC |
|---|---|---|---|
| Co (co-ordination bond) | – | – | His173 Nϵ |
| Hydrogen-bond interactions (entries in parentheses show weak hydrogen-bonding, i.e. N-O distance>3.4 Å) | |||
| Side chain a: | |||
| O28 | – | Asn217 Nδ | Asn224 Nδ |
| N29 | (Asp153 Oδ1/Oδ2) | (Asp163 Oδ2) | Asp176 Oδ2 |
| Asp204 Oδ1/Oδ2 | Asn217 Oδ | Asn224 Oδ | |
| Side chain b: | |||
| O33 | – | – | – |
| N34 | Asn378 O | Ser389 O | (Leu388 O) |
| Side chain c: | |||
| O39 | Phe370 N | Leu381 N | Leu379 N |
| N40 | Phe370 O | Leu381 O | Leu379 O |
| (Leu377 O) | Leu388 O | Leu387 O | |
| Side chain d: | |||
| O44 | (Val352 N) | (Ile363 N) | (Leu363 N) |
| N45 | Val352 O | Ile363 O | Leu363 O |
| (Trp368 O) | (Trp379 O) | (Trp377 O) | |
| Side chain e: | |||
| O51 | Ser112 Oγ | Asn120 Nδ | – |
| N52 | Ser112 O | Thr119 O | Thr134 O |
| Side chain f: | |||
| O58 | – | (Gln123 Nϵ) | Gln138 Nϵ |
| N59 | – | – | – |
| Side chain g: | |||
| O62 | (Gln252 Nϵ) | (Gln266 Nϵ) | (Gln273 Nϵ) |
| Tyr115 Oη | (Tyr122 Oη) | (Tyr137 Oη) | |
| N63 | (Gln252 Oϵ) | Gln266 Oϵ | Gln273 Oϵ |
| Asp153 Oδ1 | Asp163 Oδ1 | Asp176 Oδ2 | |
| Nucleotide arm: | |||
| O4 | His73 Nϵ | Arg357 Nη1 | Gln86 Nϵ |
| O5 | (Ser347 N) | Ser358 N | Leu358 N |
| Hydrophobic interactions | |||
| DMB | Thr346 Cγ | Arg357 Cβ | – |
| Trp348 | Trp359 | Ser359 | |
| Val351 | Tyr362 | Tyr362 | |
| Gly380 | Gly391 | Gly390 | |
| Nucleotide arm | Gly72 | Gly70 | Gly85 |
| Leu76 | Leu74 | Leu89 | |
| Leu119 | Leu126 | Leu141 | |
| Methyl groups: | |||
| Ring A | – | – | Met270 |
| Tyr206 | Phe219 | Tyr226 | |
| Ring C | Tyr367 | Tyr378 | Phe376 |
| Tyr399 | Tyr410 | Trp409 | |
| Ring D | Tyr115 | Tyr122 | Tyr137 |
| Phe150 | Phe160 | – |
Compared to the solvent-accessible surface of Cbl bound to TC (≈80 Å2 or 6.6%), the Cbl ligand shows an accessibility twice as high (≈163 Å2 or 13.3%) when bound to IF and half the accessibility when bound to HC (≈39 Å2 or 3.2%). In IF, the 5′-hydroxy group of the nucleotide moiety appears easily accessible to solvent water molecules. The corrin side chains d, e and f as well as an oxygen of the phosphate group possess restricted accessibility while side chain a–c are inaccessible. The same holds for HC where, in addition, side chain f is inaccessible.
Protein surface properties
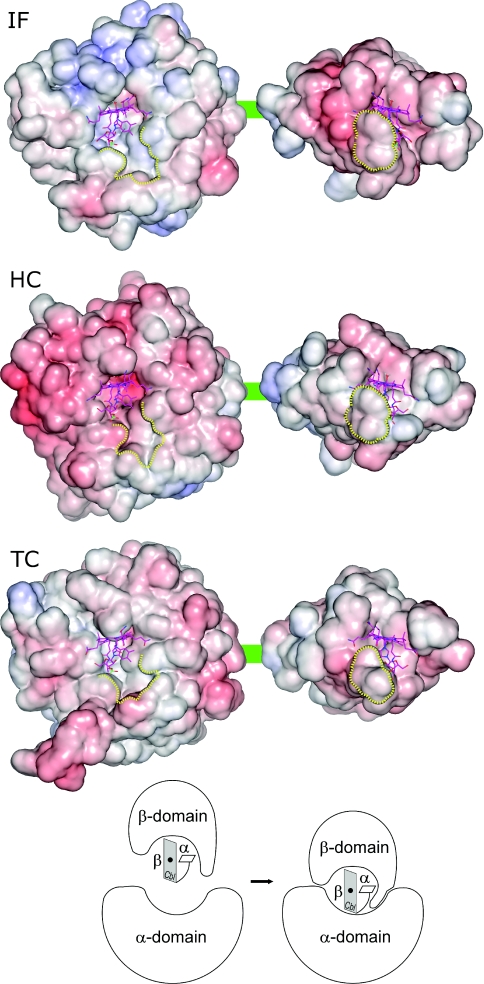
As already found for human TC, no strong electrostatic potential is calculated from the models at the interface of Cbl and either domain of IF or HC, as can be expected for binding of a neutral ligand (Figure 3). Regarding the domain contact area, a moderate degree of complementarity in the potential appears to exist only for IF which shows the unique feature of a slightly positively charged interface on the α-domain. Instead, the interface surfaces of HC and TC are neutral to slightly negative.
Figure 3. Surface properties of Cbl transport proteins.
The electrostatic potential (blue, positive potential; red, negative potential) is mapped on the surface of each protein. The proteins are presented here in an artificial ‘open’ conformation in which the α-domain (left) is separated from the β-domain (right) to allow the view on their Cbl-hosting interface. The green line represents the linker region (Figure 1). A neutral or slightly negatively potential dominates on all interface regions with the exception of the α-domain of IF, thus possessing moderate charge complementarity. Shape complementarity with the ligand Cbl is much more significant; about half of the surface of Cbl is in contact with the α-domain (most of the upper β-side of the corrin ring and side chains a and g) and with the β-domain (most of the lower α-side of the corrin ring and side chains c and d). A region of the β-domain (bordered by a yellow line) wraps around part of the nucleotide moiety on the α-side of Cbl and fits into a depression on the α-domain (also bordered yellow). This region corresponds to the turn between β-strands β3 and β4 (see Figure 4A); in the holo-form structures, it is trapped between the α-domain and Cbl (bottom scheme). The scheme illustrates the wrapping of the turn around the α-side of Cbl at the step of complex formation with the α-domain, but this conformational change may be induced already upon binding of Cbl to the β-domain.
Figure 3 also illustrates the shape complementarity between the ligand and the two domains of the transporters. A depression in the otherwise flat interface of the α-domain to the β-domain accommodates about half of the ligand and a small portion of the β-domain, which we identified as the turn between strands β3 and β4. In the holo-form of the transporters, this turn is in part wrapped around the nucleotide moiety of Cbl and appears trapped between the α-domain and Cbl. This spatial arrangement could hardly be achieved if Cbl was already bound to the α-domain. As a consequence, it seems necessary for a correct formation of the holo-form that the protein binds Cbl first with its β-domain. Subsequently, the α-domain can attach to the Cbl–β-domain complex and induce (if not already accomplished) the wrapping of the mentioned turn in the β-domain around the nucleotide moiety of Cbl. In this latter step, a large amino acid residue is involved in IF (Trp348) and HC (Trp359), but only a small amino acid residue in TC (Ser359). An additional indication for the sequence of Cbl-binding ‘β-domain before α-domain’ is the observation for IF that Cbl binds to the isolated β-domain with higher affinity than to the isolated α-domain [25].
Ligand specificity of the three Cbl transporters
An early study [9] demonstrated that the affinity of both IF and TC for 14 different Cbl analogues decreases upon modification of the corrin side chains or the nucleotide moiety. In contrast, HC tolerates major structural differences and even the total lack of the nucleotide moiety. A later study focused on the influence of modifications at the nucleotide moiety on ligand recognition [31]. These assays allowed the Cbl-binding specificities to be ranked in the order HC ≪ TC < IF. The likely physiological role thereof implies that the most specific protein IF filters out various Cbl analogues before they pass to the plasma, whereas HC in the blood plasma acts as a scavenger of potentially toxic Cbl analogues, thus exploiting its lowest level of discrimination among the three transporters.
Attempts to explain the observed differences in specificity have so far been unsuccessful. Earlier, a correlation between His–Co co-ordination on the upper side of the corrin ring (as is observed for TC) and high ligand selectivity was ruled out by comparative kinetic analysis [8]. The lack of correlation is now corroborated by the present finding that the His–Co co-ordination is present neither in IF nor in HC. Analysis of conservation among the primary structure of the transporters, even if supplemented by the detailed knowledge of Cbl interactions with TC [10], did not suffice to identify crucial residues for ligand specificity. This prompted us to produce and examine comparative models for IF and HC in order to address this issue at the level of tertiary structures.
In the following, our structural analysis of ligand specificity is based on observations from affinity measurements that employ a variety of Cbl analogues [9,31]. The affinity of a transport protein for a given Cbl analogue is expressed as the ratio of the apparent affinity constant to that for CN–Cbl and is determined by the ability of an analogue to competitively inhibit the binding of [57Co]CN–Cbl [9]. Supplementary Table 2 (at http://www.BiochemJ.org/bj/403/bj4030431add.htm) presents a compilation of investigated Cbl analogues and their affinity relative to CN–Cbl. Modifications can be grouped into those concerning the corrin side chains (a–g) and those concerning the nucleotide arm, in particular the DMB moiety. The first three analogues in Supplementary Table 2 involve the replacement of a neutral amide group with a negatively charged carboxy group at one of the corrin side chains b, d or e (see Figure 4 and the on-line multimedia adjuncts at http://www.BiochemJ.org/bj/403/bj4030431add.htm). In all three protein models, each of these replacements leads to the loss of hydrogen-bond interactions to main chain oxygen atoms (Table 3). A notable effect of the replacement is observed for chains b and d but not for chain e. A difference between the modification at chain b or d on one hand and chain e on the other hand is the spatial freedom for the latter chain to adapt its conformation according to the introduced electrostatic repulsion while the b- and d-chains are tightly packed against the protein. Regarding the modification at chain b, the negative charge of the introduced O− suffers from electrostatic repulsion to the backbone oxygens and additionally to two residues in IF (Glu379 and Asp383), and to only one residue in TC (Asp393) but no other residue in HC. Likewise, the electrostatic surface potential in the contact region of chain d is more negative for IF than for the other two transporters (Figure 3). This explains the trend of increasing specificity in the order HC < TC < IF regarding these modifications.
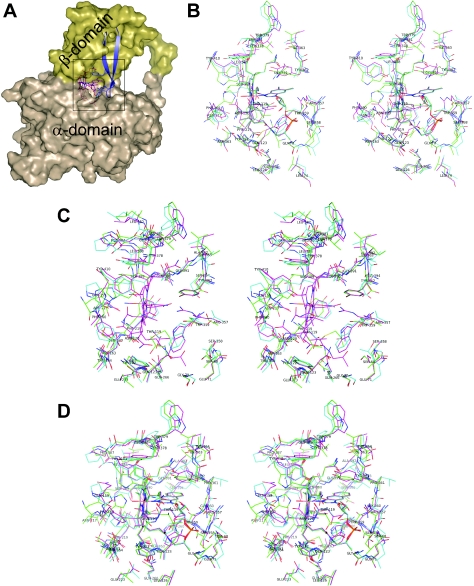
Figure 4. Structural analysis of protein–ligand interactions.
(A) Overview of the ligand position in the transporter with an emphasis on the β-hairpin motif in the β-domain. The ligand (here H2O–Cbl) is shown as magenta sticks, the β-strands 3 and 4 as blue ribbons and the rest of the protein with its surface in brown (α-domain) or yellow (β-domain). (B–D) Stereo views of superimposed structures of the transporters in a 5 Å neighbourhood of the ligand (carbon atoms of TC in green, of IF in blue and of HC in magenta). All labels refer to HC. (B) Interactions of H2O–Cbl with the proteins. (C) Interactions of the nucleotide-lacking dicyanocobinamide (Cbl-analogue number 16 in Supplementary Table 2 at http://www.BiochemJ.org/bj/403/bj4030431add.htm). Only HC can fill the space occupied by the nucleotide moiety of Cbl after appropriate changes in the side chain conformations of Arg357, Trp359 and Tyr362. (D) Environment of a cobamide (here NZA–Cba, Cbl-analogue number 7) as an example of analogues with small modifications at the DMB moiety. For each panel of this Figure, an on-line multimedia adjunct is provided at http://www.BiochemJ.org/bj/403/bj4030431add.htm to facilitate the visualization of the structural features.
Chain d (as well as chain c) is fully surrounded by the β-domain alone, i.e. the domain proposed to first accommodate the ligand. Electrostatic repulsion of a modified Cbl will thus disturb the binding process at the earliest step. In contrast, chains b and e are located in the domain interface, and their repulsion will be fully effective only at the subsequent step of formation of the domain interface. This difference is expected to account for the lower affinity of the d-monocarboxylic Cbl-analogue with respect to the b- and e-forms. Analogue 4 combines the three modifications and confirms the mentioned trend in specificity. The swapping of chain e from below the corrin plane to above (analogue 5) is neither connected to significant loss of affinity nor to differences in affinity among the three proteins and, as judged from our models, does not result in steric clash.
The second group of analogues in Supplementary Table 2 (at http://www.BiochemJ.org/bj/403/bj4030431add.htm) involves modifications at the DMB moiety on the lower side (α-side) of Cbl. This group can be divided into analogues which preserve the co-ordination bond between the Co ion and the nitrogen atom N3B of DMB (analogues 6–14 in ‘base-on’ form) and those which do not (analogues 15–19 in ‘base-off’ form). Some analogues were examined in both studies (analogues 7, 10, 12, 16). In the study by Kolhouse and Allen [9], affinity ratios are systematically lower than those obtained by Stupperich and Nexø [31]. These lower affinities might be due to contamination of protein samples with trace amounts of CN–Cbl or other CN–Cbl analogues, as stated by the authors [9]. Stupperich and Nexø concluded that modifications on the DMB moiety are tolerated by all binding proteins as long as the Co–DMB co-ordination is left intact [31]. Upon disruption of this co-ordination bond, both IF and, to a slightly smaller extent, TC show strongly decreased affinity, in contrast with HC which maintains most of its affinity. Generally, the size of the nucleotide's base had only minor effects on the affinity constants. These conclusions are in full agreement with structural features of our protein models. Smaller modifications at the DMB can be accommodated by the environment of the proteins around the nucleotide base (Figure 4 and the on-line multimedia adjuncts at http://www.BiochemJ.org/bj/403/bj4030431add.htm). For base-off analogues, the models suggest that in the final holo-form of the transporter, the nucleotide moiety should approach the Co ion in a similar way to a base-on form in order to avoid major steric clashes.
A striking difference among the Cbl-transporters is the ability of HC to bind with high affinity even those analogues that lack the nucleotide moiety, such as Cbi (cobinamide). In all transporter models, the lower side of Cbl, which hosts the nucleotide moiety, is surrounded by a region built of the β-hairpin motif β3-turn-β4. It resembles an ‘arm’ at one side of the β-domain as ‘body’ (Figure 4A). HC shows the unique feature of three large residues in van der Waals contact with the nucleotide. Two aromatic side chains, Trp359 and Tyr362, flank the DMB group and the apolar part of the Arg357 side chain packs against the ribose group (Figure 4B and the on-line multimedia adjuncts at http://www.BiochemJ.org/bj/403/bj4030431add.htm). Trp359 is present in IF (Trp348) but absent in TC (Ser359 is at this position). In contrast, Tyr362 is also present in TC but absent in IF (Val351 is found instead). At the position of Arg357 there is a small polar residue both in IF (Thr346) and in TC (Ser357). These structural differences suggest that the space of the nucleotide moiety of Cbl can be filled upon binding of the nucleotide-lacking Cbi to HC by the side chains of Arg357, Trp359 and Tyr362 without major main chain rearrangements (Figure 4C and the on-line multimedia adjuncts at http://www.BiochemJ.org/bj/403/bj4030431add.htm). Appropriate side chain conformations of these three residues may enable HC to provide hydrophobic interactions towards the apolar lower side of Cbi and polar contacts towards the solvent. In contrast, IF and TC provide only one aromatic side chain and thus leave a large part of the apolar lower side of Cbi accessible to solvent molecules. Major conformational rearrangements in the turn and adjacent amino acids of β-strands 3 and 4 might improve the interaction between Cbi and the hairpin, but remain in any case less optimal than in HC, as is evident from the binding assays. In addition, the positively charged arginine residue can balance the negative charge at the α-side of Cbi, stabilizing this Cbl analogue in HC.
We predict that mutations in HC at one or several positions of the three residues mentioned significantly change the binding of Cbl analogues. Replacing these large residues with smaller residues, such as serine or threonine as present in TC or IF, is expected to cause deterioration in the packing of the apolar lower side of Cbi to the hairpin motif. Likewise, mutations to render TC and IF equal to HC at the three residues should produce less selective forms of these two Cbl transporters. In addition, the results of such a study may answer the question of whether or not binding of Cbl-analogues without the nucleotide moiety also involves structural rearrangements outside the interface between the analogue and the protein, for instance in regions determining the flexibility of the hairpin motif.
Binding of Cbl bioconjugates to TC
In addition to the Cbl analogues compiled in Supplementary Table 2 (at http://www.BiochemJ.org/bj/403/bj4030431add.htm), binding of Cbl bioconjugates has been studied in great detail for TC [12,32–34]. The effect that the attachment of a molecule to Cbl had on the interaction with TC was systematically investigated in competitive binding assays using [57Co]CN–Cbl, varying the type and location of the attached conjugate. Attachments at the corrin side chain e showed the smallest decrease in binding (about a factor of 2). Increasingly strong impairment of binding was observed for attachment on side chains in the rows b, d and c. Biotin conjugates attached either to the Co ion or the 5′-hydroxy group of the ribose moiety had little effect on binding [12]. These results are rationalized by the three-dimensional structure of human holo-TC [10], showing that sites of conjugation with minor impact on binding are indeed those which suffer less from steric problems with the surrounding protein, i.e. the β-axial position of Co, the 5′-hydroxy group and the side chain e. These locations can accommodate conjugates without restriction on size if an appropriate spacer moiety is inserted between Cbl and the conjugate [12,32,34].
For the use of Cbl derivatives as therapeutic or diagnostic agents, the correct binding of the derivative to TC is not the only requirement. It is of importance that a complex of TC with a Cbl-derivative shows cellular uptake and intracellular trafficking similar to that of the complex of TC with natural Cbl. McLean et al. [13] investigated the ability of monomeric and dimeric Cbl derivatives to block the growth of leukaemia cells. They found that all those bioconjugates with modifications at the side chain e were good inhibitors of cell proliferation in that they act as competitors with natural Cbl for binding to TC and, at the same time, do not allow coenzyme activity. The constraint that Cbl conjugation should not alter the binding affinity to the transport protein too much holds additionally for IF and HC if the resulting derivative is intended for oral administration. Our comparative models suggest that the functional groups of Cbl can accommodate conjugates in a similar way when bound to IF or TC. In the case of HC, however, available space at the corrin side chains appears more restricted.
In conclusion, the present study has investigated the first structural comparison of the three human Cbl transport proteins, in particular their interaction with Cbl and Cbl analogues. This has permitted a rationalization of biochemical data in the literature about the difference in ligand specificity and the binding of bioconjugates. The protein models indicate the importance of the β3-turn-β4 hairpin motif in the smaller β-domain with regard to the high affinity of HC for Cbi. Three large amino acid side chains are proposed to supply hydrophobic contacts to the apolar side of Cbi, allowing HC, in contrast with IF and TC, to compensate for the lack of the nucleotide. The position of the hairpin's turn, trapped partially between the α-domain and the nucleotide moiety of Cbl, is suggested to require Cbl-binding first to the β-domain before the compact sandwich complex α-domain–Cbl–β-domain can be achieved. Polar interactions at the corrin side chains of Cbl are very similar in the three proteins; a finding that is consistent with their similar equilibrium dissociation constants. Many interactions of corrin side chains involve the protein backbone and their modification to carboxylic acids causes in general electrostatic repulsion from the tightly packed protein environment. Subtle differences in the structure of the protein may manifest themselves in the kinetics of binding to the different analogues but resolution of such features requires more detailed assays in addition to those assays relating to Cbl available to date. New experimental approaches have recently been presented in the literature [35].
Online data
Acknowledgments
This work was supported by the Ministero dell'Istruzione dell'Università e della Ricerca, Rome (FIRB2003-RBNE03PX83).
References
- 1.Nexø E. Cobalamin binding proteins. In: Kräutler B., Arigoni D., Golding T., editors. Vitamin B12 and B12-proteins. Weinheim: Wiley-VCH; 1998. pp. 461–475. [Google Scholar]
- 2.Alpers D. H., Russell-Jones G. J. Intrinsic Factor, Haptocorrin, and their Receptors. In: Banerjee R., editor. Chemistry and Biochemistry of Vitamin B12. New York: John Wiley & Sons; 1999. pp. 411–439. [Google Scholar]
- 3.Rothenberg S. P., Quadros E. V., Regec A. Transcobalamin II. In: Chemistry and Biochemistry of Vitamin B12. In: Banerjee R., editor. New York: John Wiley & Sons; 1999. pp. 441–473. [Google Scholar]
- 4.Banerjee R., Ragsdale S. W. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 5.Quadros E. V., Regec A. L., Kahn K. M., Quadros E., Rothenberg S. P. Transcobalamin II synthesized in the intestinal villi facilitates transfer of cobalamin to the portal blood. Am. J. Physiol. 1999;277:G161–G166. doi: 10.1152/ajpgi.1999.277.1.G161. [DOI] [PubMed] [Google Scholar]
- 6.Wolters M., Strohle A., Hahn A. Cobalamin: a critical vitamin in the elderly. Prev. Med. 2004;39:1256–1266. doi: 10.1016/j.ypmed.2004.04.047. [DOI] [PubMed] [Google Scholar]
- 7.Li N., Seetharam S., Seetharam B. Genomic structure of human transcobalamin II: comparison to human intrinsic factor and transcobalamin I. Biochem. Biophys. Res. Commun. 1995;208:756–764. doi: 10.1006/bbrc.1995.1402. [DOI] [PubMed] [Google Scholar]
- 8.Fedosov S. N., Berglund L., Fedosova N. U., Nexø E., Petersen T. E. Comparative analysis of cobalamin binding kinetics and ligand protection for intrinsic factor, transcobalamin, and haptocorrin. J. Biol. Chem. 2002;277:9989–9996. doi: 10.1074/jbc.M111399200. [DOI] [PubMed] [Google Scholar]
- 9.Kolhouse J. F., Allen R. H. Absorption, plasma transport, and cellular retention of cobalamin analogues in the rabbit. Evidence for the existence of multiple mechanisms that prevent the absorption and tissue dissemination of naturally occurring cobalamin analogues. J. Clin. Invest. 1977;60:1381–1392. doi: 10.1172/JCI108899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wuerges J., Garau G., Geremia S., Fedosov S. N., Petersen T. E., Randaccio L. Structural basis for mammalian vitamin B12 transport by transcobalamin. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4386–4391. doi: 10.1073/pnas.0509099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell-Jones G., McTavish K., McEwan J., Rice J., Nowotnik D. Vitamin-mediated targeting as a potential mechanism to increase drug uptake by tumours. J. Inorg. Biochem. 2004;98:1625–1633. doi: 10.1016/j.jinorgbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Pathare P. M., Wilbur D. S., Heusser S., Quadros E. V., McLoughlin P., Morgan A. C. Synthesis of cobalamin-biotin conjugates that vary in the position of cobalamin coupling. Evaluation of cobalamin derivative binding to transcobalamin II. Bioconjugate Chem. 1996;7:217–232. doi: 10.1021/bc9600022. [DOI] [PubMed] [Google Scholar]
- 13.McLean G. R., Pathare P. M., Wilbur D. S., Morgan A. C., Woodhouse S. V., Schrader J. W., Ziltener H. J. Cobalamin analogues modulate the growth of leukemia cells in vitro. Cancer Res. 1997;57:4015–4022. [PubMed] [Google Scholar]
- 14.Amagasaki T., Green R., Jacobsen D. W. Expression of transcobalamin II receptors by human leukemia K562 and HL-60 Cells. Blood. 1990;76:1380–1386. [PubMed] [Google Scholar]
- 15.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sali A., Blundell T. L. Comparative modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 17.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 18.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models to electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 19.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 21.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Holm L., Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 23.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedosov S. N., Fedosova N. U., Berglund L., Moestrup S. K., Nexø E., Petersen T. E. Assembly of the intrinsic factor domains and oligomerization of the protein in the presence of cobalamin. Biochemistry. 2004;43:15095–15102. doi: 10.1021/bi048924c. [DOI] [PubMed] [Google Scholar]
- 25.Fedosov S. N., Fedosova N. U., Berglund L., Moestrup S. K., Nexø E., Petersen T. E. Composite organization of the cobalamin binding and cubilin recognition sites of intrinsic factor. Biochemistry. 2005;44:3604–3614. doi: 10.1021/bi047936v. [DOI] [PubMed] [Google Scholar]
- 26.Tang L. H., Chokshi H., Hu C. B., Gordon M. M., Alpers D. H. The intrinsic factor (IF)-cobalamin receptor binding site is located in the amino-terminal portion of IF. J. Biol. Chem. 1992;267:22982–22986. [PubMed] [Google Scholar]
- 27.Fedosov S. N., Berglund L., Nexø E., Petersen T. E. Sequence, S-S bridges, and spectra of bovine transcobalamin expressed in Pichia pastoris. J. Biol. Chem. 1999;37:26015–26020. doi: 10.1074/jbc.274.37.26015. [DOI] [PubMed] [Google Scholar]
- 28.Gordon M. M., Hu C., Chokshi H., Hewitt J. E., Alpers D. H. Glycosylation is not required for ligand or receptor binding by expressed rat intrinsic factor. Am. J. Physiol. 1991;260:G736–G742. doi: 10.1152/ajpgi.1991.260.5.G736. [DOI] [PubMed] [Google Scholar]
- 29.Gräsbeck R. Intrinsic factor and the transcobalamins with reflections on the general function and evolution of soluble transport proteins. Scand. J. Clin. Lab. Invest. Suppl. 1967;95:7–18. [PubMed] [Google Scholar]
- 30.Lien E. L., Ellenbogen L., Law P. Y., Wood J. M. Studies on the mechanism of cobalamin binding to hog intrinsic factor. J. Biol. Chem. 1974;249:890–894. [PubMed] [Google Scholar]
- 31.Stupperich E., Nexø E. Effect of the cobalt-N co-ordination on the cobamide recognition by the human vitamin B12 binding proteins intrinsic factor, transcobalamin and haptocorrin. Eur. J. Biochem. 1991;199:299–303. doi: 10.1111/j.1432-1033.1991.tb16124.x. [DOI] [PubMed] [Google Scholar]
- 32.Pathare P. M., Wilbur D. S., Hamlin D. K., Heusser S., Quadros E. V., McLoughlin P., Morgan A. C. Synthesis of cobalamin dimmers using isophthalate cross-linking of corrin ring carboxylates and evaluation of their binding to transcobalamin II. Bioconjugate Chem. 1997;8:161–172. doi: 10.1021/bc970003+. [DOI] [PubMed] [Google Scholar]
- 33.Wilbur D. S., Hamlin D. K., Pathare P. M., Heusser S., Vessella R. L., Buhler K. R., Stray J. E., Daniel J., Quadros E. V., McLoughlin P., Morgan A. C. Synthesis and nca-radioiodination of arylstannyl-cobalamin conjugates. Evaluation of aryliodo-cobalamin conjugate binding to transcobalamin II and biodistribution in mice. Bioconjugate Chem. 1996;7:461–474. doi: 10.1021/bc960033x. [DOI] [PubMed] [Google Scholar]
- 34.Wilbur D. S., Pathare P. M., Hamlin D. K., Rothenberg S. P., Quadros E. V. Radioiodination of cyanocobalamin conjugates containing hydrophilic linkers: preparation of a radioiodinated cyanocobalamin monomer and two dimmers, and assessment of their binding with transcobalamin II. Bioconjugate Chem. 1999;10:912–920. doi: 10.1021/bc9900340. [DOI] [PubMed] [Google Scholar]
- 35.Fedosov S. N., Grissom C. B., Fedosova N. U., Moestrup S. K., Nexø E., Petersen T. E. Application of a fluorescent cobalamin analogue for analysis of the binding kinetics. A study employing recombinant human transcobalamin and intrinsic factor. FEBS J. 2006;273:4742–4753. doi: 10.1111/j.1742-4658.2006.05478.x. [DOI] [PubMed] [Google Scholar]
- 36.Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anton D. L., Hogenkamp H. P. C., Walker T. E., Matwiyoff N. A. Carbon-13 nuclear magnetic resonance studies of the monocarboxylic acids of cyanocobalamin. Assignments of the b-, d-, and e-monocarboxylic acids. J. Am. Chem. Soc. 1980;102:2215–2219. [Google Scholar]
- 38.Nexø E., Olesen H., Hansen M. R. Strength of binding of methyl-, 5′-deoxyadenosyl-, cyano- and hydroxocobalamin to human transcobalamin I and II and intrinsic factor. In: Zagalak B., Friedrich W., editors. Vitamin B12. Berlin, New York: Walter de Gruyter; 1979. pp. 851–854. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.