Abstract
Previously, we have demonstrated that co-expression of ChSy-1 (chondroitin synthase-1), with ChPF (chondroitin-polymerizing factor) resulted in a marked augmentation of glycosyltransferase activities and the expression of the chondroitin polymerase activity of ChSy-1. These results prompted us to evaluate the effects of co-expression of the recently cloned CSS3 (chondroitin sulfate synthase-3) with ChPF, because ChSy-1 and CSS3 have similar properties, i.e. they possess GalNAcT-II (N-acetylgalactosaminyltransferase-II) and GlcAT-II (glucuronyltransferase-II) activities responsible for the elongation of CS (chondroitin sulfate) chains but cannot polymerize chondroitin chains by themselves. Co-expressed CSS3 and ChPF showed not only substantial GalNAcT-II and GlcAT-II activities but also chondroitin polymerase activity. Interestingly, co-expressed ChSy-1 and CSS3 also exhibited polymerase activity. The chain length of chondroitin formed by the co-expressed proteins in various combinations was different. In addition, interactions between any two of ChSy-1, CSS3 and ChPF were demonstrated by pull-down assays. Moreover, overexpression of CSS3 increased the amount of CS in HeLa cells, while the RNA interference of CSS3 resulted in a reduction in the amount of CS in the cells. Altogether these results suggest that chondroitin polymerization is achieved by multiple combinations of ChSy-1, CSS3 and ChPF. Based on these characteristics, we have renamed CSS3 ChSy-2 (chondroitin synthase-2).
Keywords: chondroitin sulfate, chondroitin polymerization, glycosyltransferase, glycosaminoglycan, protein interaction, proteoglycan
Abbreviations: ChPF, chondroitin-polymerizing factor; ChSy, chondroitin synthase; ChGn, chondroitin β1,4-N-acetylgalactosaminyltransferase; CS, chondroitin sulfate; CSS3, chondroitin sulfate synthase-3; GAG, glycosaminoglycan; GalNAcT, β1,4-N-acetylgalactosaminyltransferase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GlcAT, β1,3-glucuronyltransferase; HAS, hyaluronan synthase; HS, heparan sulfate; PG, proteoglycan; RT, reverse transcriptase; siRNA, small interfering RNA; TM, thrombomodulin
INTRODUCTION
CS (chondroitin sulfate) is a class of GAG (glycosaminoglycan) that is distributed on the surface of virtually all cells and in the extracellular matrix. CS is covalently linked to a specific serine residue in the core protein and occurs as CS-PGs (CS-proteoglycans). Many of the physiological roles of CS-PGs are performed by CS side chains, with core proteins largely playing the role of a scaffold to make CS functionally available for binding to a variety of ligands. A growing body of evidence indicates biological roles for GAGs including CS and HS (heparan sulfate) in signalling, cell differentiation, cell–cell or cell–matrix interactions, and morphogenesis [1–3]. Previously, genetic manipulations of proteins responsible for the biosynthesis of GAGs have been performed in various model organisms of vertebrates and invertebrates [4–6]. In mice, a deficiency of an enzyme which synthesizes or modifies GAG backbones, leads to neonatal lethality not only with abnormal organogenesis but also with the aberration of signalling pathways of morphogens and growth factors [4]. In sqv mutants of Caenorhabditis elegans, defects in proteins involved in the biosynthesis of GAGs, particularly chondroitin, lead to a perturbation of vulval invagination in addition to abnormal cytokinesis in the early embryonic stages [6–8].
The biosynthesis of CS is performed by various glycosyltransferases [9]. First, the synthesis of the linkage tetrasaccharide sequence, the so-called GAG–protein linkage region (GlcAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser), is initiated by the addition of a Xyl residue to a specific serine residue of the core protein, followed by sequential transfer of two Gal residues, and is completed by the addition of the GlcA residue [9]. Each transfer reaction is catalysed by the corresponding glycosyltransferase. Following completion of the synthesis of the linkage tetrasaccharide sequence, polymerization of disaccharide-repeating units occurs by the alternate transfer of GlcA and GalNAc [9]. Subsequently, a number of sulfotransferases modify the chondroitin backbone with sulfate at specific positions, resulting in the structural diversity of CS [10].
Previously, a series of glycosyltransferases and sulfotransferases responsible for the biosynthesis of CS have been cloned. We and others have revealed three chondroitin-synthesizing enzymes, ChSy-1 (chondroitin synthase-1) and ChGn-1 and -2 (chondroitin GalNAc transferases 1 and 2) [11–15]. Whereas ChSy-1 shows dual enzymatic [GalNAcT-II (N-acetylgalactosaminyltransferase-II) and GlcAT-II (glucuronyltransferase-II)] activities [11] responsible for chain elongation, ChGn-1 and -2 catalyse chain initiation and elongation, exhibiting activities of GalNAcT-I (N-acetylgalactosaminyltransferase-I) and GalNAcT-II [12–15]. Also, chondroitin GlcAT, involved in chain elongation, has been identified by others [16]. However, none of these enzymes have shown the chondroitin polymerase activities required for the synthesis of a long stretch of the disaccharide-repeating region. We recently identified ChPF (chondroitin-polymerizing factor), which shows weak yet significant homology to ChSy-1 [17,18]. Although ChPF lacks a DXD motif characteristic of Golgi-residing glycosyltransferases, co-expression of ChPF and ChSy-1 resulted in a marked augmentation of not only the glycosyltransferase activities but also the polymerase activites of ChSy-1 [17], which may indicate that ChPF acts as a chaperone, conferring much stronger glycosyltransferase activities on ChSy-1 or stabilizing ChSy-1 as in the case of EXT2, which appears to function as a chaperone to express the polymerase activity of EXT1 for HS biosynthesis [19,20].
After the discovery of ChPF, an additional ChSy family member, called CSS3 (chondroitin sulfate synthase-3), was identified by Yada et al. [21]. Although this new member, like ChSy-1, possesses dual enzymatic (GalNAcT-II and GlcAT-II) activities, the glycosyltransferase activities were much weaker than those of ChSy-1. In addition, no functional analysis of CSS3 in terms of chondroitin-polymerizing activities was performed. Thus in the present study, we re-evaluated the properties of CSS3 and found unexpected characteristics. Based on these characteristics, we have renamed CSS3 ChSy-2 (chondroitin synthase-2).
EXPERIMENTAL
Materials
UDP-[U-14C]GlcA (285.2 mCi/mmol) and UDP-[3H]GalNAc (10 Ci/mmol) were purchased from NEN Life Science Products. Unlabelled UDP-GlcA and UDP-GalNAc were obtained from Sigma. Chondroitin (a chemically desulfated derivative of whale cartilage chondroitin sulfate A) and Arthrobacter aurescens chondroitinase ABC (EC 4.2.2.4) were purchased from Seikagaku Corporation. Purified α-TM (α-thrombomodulin) [22] was provided by the research institute, Dai-ichi Pharmaceutical Co., and contained a linkage-region tetrasaccharide, GlcAβ1-3Galβ1-3Galβ1-4Xyl [23]. Superdex™ Peptide HR10/30 and Superdex™ 200 10/300 GL columns were obtained from Amersham Biosciences.
Construction of a soluble form of CSS3 (ChSy-2)
The cDNA fragment of a truncated form of CSS3 (ChSy-2), lacking the first 129 N-terminal amino acids of CSS3, was amplified by RT (reverse transcriptase)-PCR with the Marathon-Ready™ cDNA library derived from human brain (Clontech) as a template using a 5′-primer (5′-CCCTCGAGGGCCGAGGGGGAGCCCGA-3′) containing an in-frame XhoI site and a 3′-primer (5′-CCCTCGAGCTGTCAGGAGAGAGTTCGATT-3′) containing a XhoI site located 3 bp downstream of the stop codon. The PCR was carried out using KOD-Plus DNA polymerase (Toyobo Biochemicals) for 30 cycles of 94 °C for 30 s, 58 °C for 30 s and 68 °C for 150 s in 5% (v/v) DMSO. The PCR fragments were digested with XhoI, and both ends of the fragments were partially filled by a Klenow fragment (New England Biolabs) with dCTP and dTTP. The pGIR201protA [24] vector digested with BamHI was also partially filled with dATP and dGTP. The resultant fragments were subcloned into pGIR201protA, resulting in the fusion of CSS3 (ChSy-2) with the insulin signal sequence and the Protein A sequence present in the vector. An NheI fragment containing the above fusion protein sequence was inserted into the XbaI site of the expression vector pEF-BOS [25]. The nucleotide sequence of the amplified cDNA was determined in a 377 DNA sequencer (PerkinElmer Applied Biosystems). Soluble forms of ChSy-1 and ChPF were constructed previously [11,17].
Expression of a soluble form of CSS3 (ChSy-2) and enzyme assays
The expression plasmid (6 μg) was transfected into COS-1 cells on 100-mm plates using FuGENE™ 6 (Roche Molecular Biochemicals) according to the manufacturer's instructions. For co-transfection experiments, the CSS3 (ChSy-2) and ChSy-1 or ChPF expression plasmids (3 μg each) were co-transfected into COS-1 cells on 100-mm plates using FuGENE™ 6 as above. At 2 days after transfection, 1 ml of the culture medium was collected and incubated with 10 μl of IgG–Sepharose (Amersham Pharmacia Biotech) for 1 h at 4 °C. The beads recovered by centrifugation were washed with and then resuspended in the assay buffer, and tested for GalNAcT and GlcA transferase activities as described below. Assays for GalNAcT-II and GlcAT-II were performed using chondroitin as an acceptor and UDP-GalNAc or UDP-GlcA as a sugar donor respectively, as described previously [26,27]. Polymerization reactions using α-TM as an acceptor were co-incubated in reaction mixtures containing the following constituents in a total volume of 20 μl, i.e. 1 nmol of α-TM, 0.25 mM UDP-[3H]GalNAc (5.28×105 d.p.m.), 0.25 mM UDP-GlcA, 100 mM Mes (pH 6.5), 10 mM MnCl2 and 10 μl of the resuspended beads. The mixtures were incubated at 37 °C overnight, and the 3H-labelled products were then separated by gel filtration chromatography on a Superdex peptide column equilibrated and eluted with 0.2 M NH4HCO3. Fractions (0.4 ml each) were collected at a rate of 0.4 ml/min, and the measurement of radioactivity was carried out by liquid scintillation spectrophotometry.
Characterization of the enzyme reaction products
Products of polymerization reactions on α-TM were isolated by gel filtration on a Superdex peptide column with 0.2 M NH4HCO3 as the eluent. The [3H]GalNAc-labelled oligosaccharide chains were released from α-TM by alkaline reduction treatment using 1.0 M NaBH4/0.05 M NaOH and then exhaustively digested with chondroitinase ABC using 50 m-units of the enzyme for 1 h as described previously [28]. An aliquot of the enzyme digest was subjected to gel filtration on Superdex peptide as described above. To determine the size of the reaction products, the remaining aliquot was subjected to gel filtration on a Superdex 200 column with 0.2 M NH4HCO3 as the eluent. The calibration of the Superdex 200 column was performed using a series of size-defined commercial polysaccharides.
Pull-down assays
The cDNA fragment of a truncated form of ChPF, lacking the first 61 N-terminal amino acids of the putative ChPF, was amplified using a 5′-primer (5′-CGGAATTCAACTCGGTGCAGCCCGGAGC-3′) containing an in-frame EcoRI site and a 3′-primer (5′-CGGGATCCGCTCTGGTTTTGGGGGAGAAG-3′) containing a BamHI site. The cDNA fragment of a truncated form of ChSy-1, lacking the first 45 N-terminal amino acids of ChSy-1, was also amplified using a 5′-primer (5′-GCTCTAGAGGCTGCCGGTCCGGGCAG-3′) containing an in-frame XbaI site and a 3′-primer (5′-GCTCTAGACAATCTTAAAGGAGTCCTATGTA3′) containing a XbaI site. Each DNA fragment was inserted into the pcDNA3Ins-His expression vector, resulting in the fusion of the protein with the insulin signal sequence and His6 sequence present in the vector. Combinations of these constructs and the Protein A-tagged expression vectors were transfected into COS-1 cells on 100-mm plates using FuGENE™ 6 (Roche Molecular Biochemicals) according to the manufacturer's instructions. At 2 days after transfection, 1 ml of the culture medium was collected and incubated with 10 μl of Ni2+-NTA (Ni2+-nitrilotriacetate) agarose (Qiagen) overnight at 4 °C. The beads recovered by centrifugation were washed three times with TBS [Tris-buffered saline; 50 mM Tris/HCl (pH 7.5) and 0.15 NaCl] containing Tween 20 and subjected to SDS/PAGE (7% gel), and proteins were transferred to a PVDF membrane. The membrane, after blocking in PBS containing 2% non-fat skimmed milk powder and 0.1% Tween 20, was incubated with IgG antibody, and then treated with an anti-mouse IgG conjugated with horseradish peroxidase (Amersham Bioscience). Proteins bound to the antibody were visualized with an ECL® advance kit (Amersham Bioscience).
Northern blot analysis
A commercial human 12-lane multiple tissue Northern blot (Clontech) membrane was used for the analysis. The membrane was probed with a gel-purified, radiolabelled (>1×109 c.p.m./μg), 1.25-kb CSS3 (ChSy-2)-specific fragment corresponding to nucleotides 1419–2659 of the CSS3 (ChSy-2) cDNA (Genbank™ accession No. AB175496).
Establishment of an expression vector for CSS3 (ChSy-2) and preparation of cells that stably overexpress CSS3 (ChSy-2)
The cDNA fragment encoding CSS3 (ChSy-2) was amplified from a human brain Marathon-Ready cDNA library as a template using a 5′-primer (5′-CGGAATTCACAGCCGCGATGGCTGTGCGCT-3′) containing a EcoRI site and a 3′-primer (5′-CCCTCGAGCTGTCAGGAGAGAGTTCGATT-3′) containing a XhoI site. PCR was carried out with KOD-Plus DNA polymerase (Toyobo Biochemicals) for 30 cycles of 94 °C for 30 s, 53 °C for 42 s and 68 °C for 180 s in 5% (v/v) DMSO. The PCR fragments were subcloned into the EcoRI-XhoI site of the pCMV expression vector (Invitrogen). The nucleotide sequence of the amplified cDNA was determined in a 377 DNA sequencer (PerkinElmer Applied Biosystems). The expression plasmid (6.7 μg) was transfected into HeLa cells on 100-mm plates using FuGENE™ 6 (Roche Applied Science) according to the manufacturer's instructions. Transfectants were cultured in the presence of 1000 μg/ml of G418 (Geneticin). Resultant colonies were removed and propagated for experiments.
RNA interference of the CSS3 (ChSy-2) gene
A 25-mer double-stranded RNA composed of sense 5′-AUCCAAGACCUUCACAAUAGCAAAAAG-3′ and antisense 5′-UUUUGCUAUUGUGAAGGUCUUGGAUAU-3′ sequences and of sense 5′-GUCUUAUCCCAAAGCAGAAUGUAAAAG-3′ and antisense 5′-UUUACAUUCUGCUUUGGGAUAAGACAU-3′ sequences for CSS3 (ChSy-2) was designed and purchased from iGENE (Tsukuba). A silencing scrambled RNA composed of sequences with no homology to known human sequences (iGENE) was used as a control. HeLa cells were transfected with 10 nM siRNA (small interfering RNA) by using TransIT-TKO transfection reagent (Takara).
Quantitative real-time RT-PCR
Total RNA was extracted from HeLa cells using a QuickPrep total RNA extraction kit (Amersham Bioscience). The cDNA was synthesized from approx. 1 μg of total RNA using Moloney murine leukemia virus-reverse transcriptase (Promega) and an oligo(dT)20-M4 adaptor primer (Takara). The primer sequences used were as follows: CSS3 (ChSy-2), a forward primer 5′-GGAAATTCAGTATGGCTACCG-3′ and a reverse primer 5′-CTCTGAAGAAAGGCTTGC-3′; and GAPDH (glyceraldehyde-3-phosphate dehydrogenase), a forward primer 5′-ATGGGTGTGAACCATGAGAAGTA-3′ and a reverse primer 5′-GGCAGTGATGGCATGGAC-3′. Quantitative real-time RT-PCR was performed using a FastStart DNA Master plus SYBR Green I (Roche Diagnostics) in a LightCycler ST300 (Roche Diagnostics). The expression level of CSS3 (ChSy-2) mRNA was normalized to that of the GAPDH transcript.
Derivatization of GAGs from HeLa cells using a fluorophore, 2-aminobenzamide
Cells were homogenized in acetone and air-dried. The dried materials were digested with heat-pretreated (60 °C for 30 min) actinase E (0.1 mg) in 200 μl of 0.1 M sodium borate (pH 8.0), containing 10 mM calcium acetate at 60 °C for 24 h. Following incubation, each sample was treated with trichloroacetic acid and the resultant precipitate was removed by centrifugation (800 g for 5 min). The soluble fraction was extracted with ether. The aqueous phase was neutralized with 1.0 M sodium carbonate and adjusted to contain 80% ethanol. The resultant precipitate was dissolved in 50 mM pyridine acetate and subjected to gel filtration on a PD-10 column using 50 mM pyridine acetate as an eluent. The flow-through fractions were collected and evaporated dry. The dried sample was dissolved in water. Digestion with 5 m-units of chondroitinase ABC was conducted as described previously [29] at 37 °C for 1 h in a total volume of 10 μl. Reactions were terminated by boiling for 1 min. Each digest was derivatized with 2-aminobenzamide, then analysed by HPLC as reported previously [30].
RESULTS
Glycosyltransferase activity of CSS3 (ChSy-2)
Previous experiments have revealed that co-expression with ChPF resulted in a dramatic augmentation of the glycosyltransferase activities of ChSy-1 [17], suggesting that ChPF serves as a molecular chaperone, stabilizing ChSy-1. In addition, the properties of CSS3 (ChSy-2) [21] were reminiscent of those of ChSy-1 [11], which possesses dual glycosyltransferase activities of GalNAcT-II and GlcAT-II, required for the biosynthesis of disaccharide repeating units of CS. Hence, co-expression of CSS3 (ChSy-2) with ChPF was performed. To facilitate the functional analysis of CSS3 (ChSy-2), a soluble form of CSS3 (ChSy-2) was generated by replacing the first 129 amino acids of the protein with a cleavable insulin signal sequence and a Protein A IgG-binding domain as described in the Experimental section, and the soluble protein was expressed in COS-1 cells as a recombinant protein fused with the Protein A IgG-binding domain. The fused protein expressed in the medium was adsorbed on to IgG–Sepharose beads to eliminate endogenous glycosyltransferases, and then the protein-bound beads were used as an enzyme source. When CSS3 (ChSy-2) bound to beads was evaluated for glycosyltransferase activities using chondroitin as an acceptor and either UDP-GalNAc or UDP-GlcA as a donor substrate, only weak activities were detected compared with those of ChSy-1 (Table 1). These results were similar to the findings of Yada et al. [21], who reported weak activities of GalNAcT-II and GlcAT-II for CSS3 (ChSy-2), which were approximately one-tenth of those of ChSy-1. However, when soluble CSS3 (ChSy-2) was co-expressed with soluble ChPF, significant glycosyltransferase activities were detected in the culture medium (Table 1). Notably, co-expression of soluble CSS3 (ChSy-2) with soluble ChSy-1 markedly augmented both the GalNAcT-II and GlcAT-II activities (Table 1).
Table 1. Comparison of GalNAcT-II and GlcAT-II activities of fusion proteins secreted into the culture medium by transfected COS-1 cells.
Polymer chondroitin was used as an acceptor substrate, and the values are the means±S.E. of three determinations. ChSy-1/ChPF, CSS3 (ChSy-2)/ChPF and ChSy-1/CSS3 (ChSy-2) represent coexpressed ChSy-1 and ChPF, CSS3 (ChSy-2) and ChPF, and ChSy-1 and CSS3 (ChSy-2), respectively. ND, not detected (<0.01 nmol/mg/h).
| Protein | GalNAcT-II activity (nmol/mg/h) | GlcAT-II activity (nmol/mg/h) |
|---|---|---|
| ChSy-1 | 1.0±0.2 | 3.3±0.5 |
| CSS3 (ChSy-2) | 0.1±0.01 | 0.1±0.01 |
| ChPF | ND | ND |
| ChSy-1/ChPF | 14.2±2.3 | 54.8±3.5 |
| CSS3 (ChSy-2)/ChPF | 5.2±0.1 | 2.6±0.2 |
| ChSy-1/CSS3 (ChSy-2) | 38.1±2.9 | 34.4±3.2 |
Polymerase activity of CSS3 (ChSy-2)
Previously, co-expressed soluble ChSy-1 and soluble ChPF showed not only a dramatic augmentation in glycosyltransferase activities of ChSy-1 but also polymerizing activities for disaccharide-repeating units of CS onto α-TM [17] (a part-time proteoglycan and bears a truncated linkage region tetrasaccharide [23]). Hence, we measured the polymerizing activity upon the co-expression of soluble CSS3 (ChSy-2) with soluble ChPF or soluble ChSy-1. When α-TM was used as an acceptor substrate for the polymerization assay in the presence of UDP-[3H]GalNAc and UDP-GlcA, incubation of both the co-expressed putative enzyme complexes yielded radiolabelled saccharide chains on α-TM (results not shown).
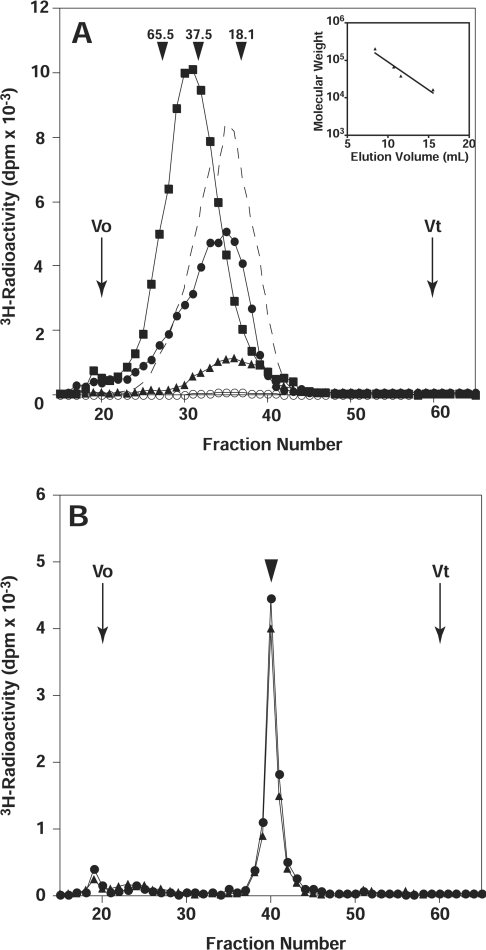
We then examined whether the length of the chondroitin chains formed by the co-expressed proteins in various combinations was different. For this analysis, glycosyltransferase activities detected for various combinations of the proteins were normalized against the GalNAcT-II activity, where all of the enzyme sources exhibited the same amount of GalNAcT-II activity. Each reaction product obtained with α-TM was subjected to reductive β-elimination using NaBH4/NaOH, and the released radiolabelled saccharides were analysed by gel filtration chromatography using a Superdex 200 column as shown in Figure 1(A). The lengths of chondroitin chains formed by co-expressed CSS3 (ChSy-2) and ChPF or CSS3 (ChSy-2) and ChSy-1 were comparable with those of commercial chondroitin chains (a chemically desulfated derivative of whale cartilage chondroitin sulfate A), although the co-expressed CSS3 (ChSy-2) and ChPF polymerized chondroitin chains slightly longer than the co-expressed CSS3 (ChSy-2) and ChSy-1. In contrast, co-expressed ChSy-1 and ChPF formed relatively longer chondroitin chains compared with commercial chondroitin chains (Figure 1A). In addition, these enzyme sources exhibited chondroitin-polymerizing activities on an authentic synthetic substrate, GlcAβ1-3Galβ1-O-C2H4NHCbz (where Cbz is benzyloxycarbonyl), which shares the disaccharide sequence with the GAG-protein linkage region tetrasaccharide, although the efficacy of polymerization appeared to be much lower than that with α-TM as an acceptor substrate (results not shown). It should be noted that no polymerization was induced on any substrate tested either by using one of the soluble forms of ChSy-1, CSS3 (ChSy-2) or ChPF or by using a mixture of separately expressed soluble forms of any two of ChSy-1, CSS3 (ChSy-2), and ChPF (Figure 1A). These results clearly suggest the critical requirement of the co-expressed proteins in combinations of any two of ChSy-1, CSS3 (ChSy-2), and ChPF for the chondroitin polymerization.
Figure 1. Comparison of the chain length of chondroitin formed using various combinations of CSS3 (ChSy-2), ChSy-1 and ChPF.
(A) α-TM was tested as an acceptor for the polymerization reaction, where co-expressed ChSy-1 and ChPF, CSS3 (ChSy-2) and ChPF, or CSS3 (ChSy-2) and ChSy-1 were used as an enzyme source as described in the Experimental section. [3H]GalNAc-labelled products were first isolated by gel-filtration, subjected to reductive β-elimination using NaBH4/NaOH, and then rechromatographed on a Superdex 200 column (1.0 cm×30 cm) as shown in this Figure. Effluent fractions (0.4 ml each) were analysed for radioactivity. The broken line indicates commercial chondroitin chains (a chemically desulfated derivative of whale cartilage chondroitin sulfate A). ■, products obtained using co-expressed ChSy-1 and ChPF; ●, products obtained using co-expressed CSS3 (ChSy-2) and ChPF; ▲, products obtained using co-expressed CSS3 (ChSy-2) and ChSy-1; ○, products obtained using a mixture of separately expressed soluble forms of CSS3 (ChSy-2) and ChPF. The inset shows the calibration curve, showing a linear relation between the log Mr and the elution volume, which was generated using the data obtained with size-defined commercial polysaccharides; dextran (average Mr: 200000, 65500, 37500, and 18100; all from Sigma). (B) The [3H]GalNAc-labelled products were first isolated by gel-filtration and subjected to reductive β-elimination using NaBH4/NaOH. The 3H-labelled products were digested with chondroitinase ABC and chromatographed on a Superdex peptide column (1.0 cm×30 cm). Effluent fractions (0.4 ml each) were analysed for radioactivity. The arrowhead indicates the elution position of the authentic disaccharide, GlcAβ1–3GalNAc. ●, products obtained using co-expressed CSS3 (ChSy-2) and ChPF; ▲, products obtained using co-expressed CSS3 (ChSy-2) and ChSy-1.
To identify radiolabelled oligosaccharides synthesized on α-TM as chondroitin chains, the reaction products obtained with α-TM in both cases using CSS3 (ChSy-2) were subjected to digestion by chondroitinase ABC. The digests were separated by gel-filtration chromatography using a Superdex Peptide column, resulting in a small peak of the linkage hexasaccharide attached to the core protein around the void volume and a large peak, which should contain monosaccharide, GalNAc, and disaccharide, ΔHexAα1-3GalNAc (Figure 1B). These findings indicated that chondroitin chains were polymerized on the truncated linkage tetrasaccharide sequence of α-TM.
Interactions among ChSy-1, CSS3 (ChSy-2) and ChPF
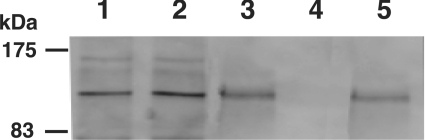
As shown above, co-expression of CS-synthesizing enzymes resulted in a marked augmentation of not only glycosyltransferase activities but also polymerase activity. In view of these results, interactions between these molecules were expected. Thus the interactions of these molecules were evaluated by conducting pull-down assays. For this analysis, soluble forms of ChSy-1 and CSS3 (ChSy-2) fused with Protein A at their N-termini (ChSy-1–ProA and ChSy-2–ProA), and soluble forms of ChSy-1 and ChPF tagged with the 6 x His epitope at their N-termini (ChSy-1–His and ChPF–His) were generated as described in the Experimental section. To evaluate the interactions among ChSy-1, CSS3 (ChSy-2) and ChPF, co-expression of ChSy-2–ProA with ChPF–His or ChSy-1–His was performed. First, to confirm that the co-expression of any two proteins in different combinations yields active enzymes, the culture medium from each transfection experiment was purified with IgG–Sepharose and evaluated for enzyme activities. Glycosyltransferase activities were detected in the medium from all transfectants (results not shown). Next, to evaluate the association among these proteins, pull-down assays were performed. In addition, to ensure specificity, we tried these assays with human EXT-1 (a HS co-polymerase) which is expected to interact with human EXT-2 (a HS co-polymerase) [20,31] but not with ChSy-1, CSS3 (ChSy-2) or ChPF. Ni2+-NTA agarose was added to the culture medium to pull down His-tagged proteins, then the proteins were subjected to SDS/PAGE followed by Western blotting using a primary antibody to Protein A-tagged proteins, and detection used an ECL® advance kit. No band was detected for the transfectant of either ChSy-2–ProA alone (results not shown) or for the co-transfectant of ChSy-2–ProA and EXT-1–His (Figure 2, lane 4). In contrast, proteins with a molecular mass of approximately 120 kDa, corresponding to ChSy-2–ProA, were detected for the co-transfectant of ChSy-2–ProA and ChSy-1–His (Figure 2, lane 1) or ChSy-2–ProA and ChPF–His (Figure 2, lane 2). As expected, co-expressed ChSy-1–ProA and ChPF–His (Figure 2, lane 3) or EXT-2–ProA and EXT-1–His (Figure 2, lane 5) showed similar results. These results indicated that ChSy-1, CSS3 (ChSy-2) and ChPF interact with each other.
Figure 2. The pull-down assays of co-expressed CSS3 (ChSy-2), ChSy-1, ChPF, EXT-1 and EXT-2 in different combinations.
Culture medium from cells co-expressing ChSy-2–ProA and ChPF–His, ChSy-1–His, or EXT-1–His, co-expressing ChSy-1–ProA and ChPF–His, or co-expressing EXT-2–ProA and EXT-1–His was purified with Ni2+-NTA agarose and subjected to SDS/PAGE. Separated proteins were transferred to PVDF membrane by Western blotting, allowed to react with IgG as a primary antibody, and visualized using the ECL® advance kit. Lane 1, ChSy-2–ProA and ChSy-1–His; lane 2, ChSy-2–ProA and ChPF–His; lane 3, ChSy-1–ProA and ChPF–His; lane 4, ChSy-2–ProA and EXT-1–His; lane 5, EXT-2–ProA and EXT-1–His.
Expression pattern of CSS3 (ChSy-2)
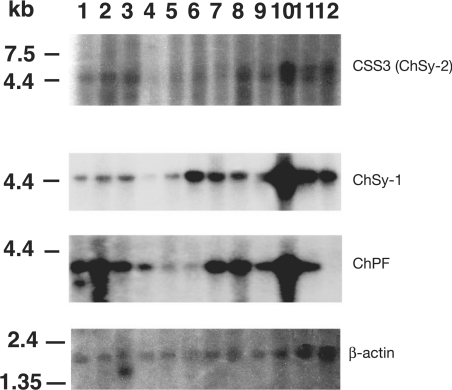
Northern blot analysis of CSS3 (ChSy-2) mRNA showed a single band of approx. 4.2 kb for all human tissues examined (Figure 3). The expression level of CSS3 (ChSy-2) mRNA was low compared with the levels of ChSy-1 and ChPF. In addition, whereas ChSy-1 and ChPF exhibited similar expression patterns except for the marked absence of ChPF expression in the peripheral blood leukocytes, the expression pattern of CSS3 (ChSy-2) was distinct in that the highest expression was observed in skeletal muscle, small intestine, lung and peripheral blood leucocytes.
Figure 3. Northern blot analysis of the ChSy family members in human tissues.
Northern blots with RNA from various human tissues were hybridized with a probe for CSS3 (ChSy-2), ChSy-1, ChPF or β-actin as described in the Experimental section. Lane 1, brain; lane 2, heart; lane 3, skeletal muscle; lane 4, colon; lane 5, thymus; lane 6, spleen; lane 7, kidney; lane 8, liver; lane 9, small intestine; lane 10, placenta; lane 11, lung; lane 12, peripheral blood leucocytes.
Involvement of CSS3 (ChSy-2) in CS biosynthesis in cultured cells
Because the activity and expression of CSS3 (ChSy-2) were weaker than those of ChSy-1, we finally determined the physiological relevance of CSS3 (ChSy-2) in HeLa cells that express ChSy-1, CSS3 (ChSy-2) and ChPF endogenously. When the expression levels of ChSy-1, CSS3 (ChSy-2) and ChPF mRNAs were normalized to those of the GAPDH transcript, the values of ChSy-1, CSS3 (ChSy-2) and ChPF/GAPDH (×10−3 copies) were 13.3, 0.8 and 3.7 respectively. To examine the physiological relevance of CSS3 (ChSy-2) in HeLa cells, we first asked whether overexpression of CSS3 (ChSy-2) increased the amount of CS. A pCMV-Script expression vector, which possesses the cytomegalovirus promoter and the neomycin-resistance gene, and harbors the open reading frame of human CSS3 (ChSy-2), was transfected into HeLa cells, and positive selection was then carried out in the presence of a neomycin analogue, G418. Each of the resultant colonies was removed and propagated for experiments, and the expression of CSS3 (ChSy-2) was measured by quantitative real-time RT-PCR. As shown in Table 2, the disaccharide composition and the amount of CS isolated from the two stable clones with the different expression levels of CSS3 (ChSy-2) introduced (ChSy-2-1 and ChSy-2-2 cells) were analysed by HPLC as described in the Experimental section. The results showed that while the disaccharide composition in ChSy-2-1 and ChSy-2-2 cells was similar to that in the control HeLa cells, the amount of CS was increased in the two stable clones, corresponding to the expression level of CSS3 (ChSy-2) (Table 2). These results indicated that overexpression of CSS3 (ChSy-2) increased the amount of CS in HeLa cells.
Table 2. Disaccharide composition of CS in control and transfected HeLa cells.
HeLa cells were transfected with vector alone (mock), plasmids containing CSS3 (ChSy-2), control siRNA or CSS3 (ChSy-2) siRNA. For experimental details, see the Experimental section. The values are expressed as pmol of disaccharide per mg of dried homogenate of these cells and the means from two to three independent experiments. ΔDi-0S, ΔHexAα1-3GalNAc; ΔDi-6S, ΔHexAα1-3GalNAc(6S); ΔDi-4S, ΔHexAα1-3GalNAc(4S); ΔDi-diSD, ΔHexA(2S)α1-3GalNAc(6S); ΔDi-diSE, ΔHexAα1-3GalNAc(4S,6S). Relative expression of the CSS3 (ChSy-2) transcript were quantified by quantitative real-time RT-PCR. Normalization of the data was performed using the GAPDH mRNA levels.
| Disaccharide composition [pmol/mg (mol%)] | |||||||
|---|---|---|---|---|---|---|---|
| Cell type | ΔDi-0S | ΔDi-6S | ΔDi-4S | ΔDi-diSD | ΔDi-diSE | Total disaccharide (pmol/mg) | Relative expression |
| HeLa (n=3) | 3.1 (6) | 9.8 (19) | 34.7 (69) | 1.8 (4) | 0.8 (2) | 50.3 | 1.0 |
| ChSy-2-1 (n=3) | 6.0 (5) | 16.6 (13) | 97.0 (73) | 7.9 (6) | 4.2 (3) | 131.8 | 7.0 |
| ChSy-2-2 (n=3) | 3.7 (5) | 9.3 (12) | 61.3 (77) | 2.8 (4) | 2.1 (2) | 79.3 | 2.5 |
| Control siRNA (n=2) | 4.0 (8) | 11.3 (22) | 33.7 (65) | 2.2 (4) | 0.4 (1) | 51.6 | 1.0 |
| ChSy-2 siRNA (n=2) | 1.4 (3) | 10.4 (25) | 27.4 (66) | 1.8 (4) | 1.4 (1) | 42.4 | 0.6 |
Prompted by these observations, we next examined whether the knockdown of CSS3 (ChSy-2) expression by RNA interference decreases the amount of CS as described in the Experimental section. The efficiency of gene silencing was determined by quantitative real-time RT-PCR. As shown in Table 2, transfection of the CSS3 (ChSy-2) siRNA (ChSy-2 siRNA cells) resulted in a 40% knockdown of the CSS3 (ChSy-2) mRNA and an 18% reduction in the amount of CS when compared with that of the control siRNA, whereas the disaccharide composition in ChSy-2 siRNA cells was similar to that in the control siRNA cells. These findings altogether suggest that CSS3 (ChSy-2), although its levels of activity and expression are lower than those of ChSy-1, can participate in the biosynthesis of CS through its interaction with ChSy-1 or ChPF.
DISCUSSION
In the present study, we have demonstrated that CSS3 (ChSy-2) can polymerize chondroitin chains onto α-TM bearing the linkage region tetrasaccharide and an authentic synthetic linkage region analogue through co-operation with ChPF or ChSy-1. Thus polymerization of the chondroitin backbone was achieved in vitro by multiple CS-synthesizing glycosyltransferases. Although co-expressed CSS3 (ChSy-2) and ChSy-1 or CSS3 (ChSy-2) and ChPF showed chondroitin polymerase activity with α-TM, the chain of chondroitin formed by these complexes was shorter than that formed by co-expressed ChSy-1 and ChPF. These results may suggest that the enzyme complex consisting of CSS3 (ChSy-2) and ChPF or CSS3 (ChSy-2) and ChSy-1 is responsible for the formation of shorter CS chains and the complex of ChSy-1 and ChPF produces longer CS chains in vivo. The results are reminiscent of the three bifunctional glycosyltransferases, HAS (hyaluronan synthase) 1, HAS2 and HAS3, which are involved in the biosynthesis of hyaluronic acid [32]. Although all of these enzymes can synthesize hyaluronic acid, the chain length of each of the hyaluronic acid products of HAS1, HAS2 and HAS3 is different; the molecular masses range from 2×105 to approx. 1×106, 2×106 Da and 1×105 to 1×106 Da respectively, for the three enzymes [32,33]. In view of these and our findings, the chain length of CS may be dependent on which enzyme complex is formed in a given tissue expressing CSS3 (ChSy-2), ChSy-1 and ChPF.
CSS3 (ChSy-2) showed weaker GalNAcT-II and GlcAT-II activities than did ChSy-1. Similarly, Yada et al. [21] reported weak GalNAcT-II and GlcAT-II activities of CSS3 (ChSy-2), being approximately one-tenth to one-twentieth those of ChSy-1. However, the present study revealed that co-expressed CSS3 (ChSy-2) and ChPF or CSS3 (ChSy-2) and ChSy-1 showed a substantial increase in GalNAcT-II and GlcAT-II activities as in the case of ChSy-1 with ChPF. Although the underlying mechanism by which glycosyltransferase activities of CSS3 (ChSy-2) could be augmented by co-expression with ChPF remains unclear, ChPF may function as a molecular chaperone, through stabilization and promotion of the proper folding of CSS3 (ChSy-2) or enhancement of the affinity of CSS3 (ChSy-2) for the acceptor substrates. Surprisingly, co-expressed ChSy-1 and CSS3 (ChSy-2) exhibited chondroitin polymerase activity as well. Although it is unknown which enzyme, ChSy-1 or CSS3 (ChSy-2), exhibited the polymerizing activities, ChSy-1 and CSS3 (ChSy-2) may be stabilized by forming a heterodimeric complex, as in the case of co-expression of EXT1 and EXT2, which form an enzyme complex for the polymerization of HS [31].
In the present study, we demonstrated that ChSy-1, CSS3 (ChSy-2) and ChPF interacted with each other. These interactions appear to be critical for the functions of these enzymes, because the enzyme complexes, but not individual components, showed strong glycosyltransferase or polymerase activities (see Table 1 and Figure 1A). Recent experiments have demonstrated that interactions of glycosyltransferases with chaperone-like proteins are important for exerting relevant activities of glycosyltransferases. For instance, such interactions have been shown in HS biosynthesis. Although EXT1 or EXT2 shows glycosyltransferase activities required for elongation of HS chains, polymerization of HS chains had not been achieved by individual enzyme components [20]. Other studies, however, revealed that co-expression of EXT1 with EXT2 resulted not only in an augmentation of glycosyltransferase activities of EXT1 but also in the expression of HS polymerase activity [19,20,31]. Another such example is O-mannosylation of α-dystroglycan, the abnormal glycosylation of which is the primary cause of some forms of congenital muscular dystrophy [34]. POMT1 and POMT2, which are homologous to yeast protein O-mannosyltransferases, are responsible for O-mannosylation of α-dystroglycan. Although expression of either POMT1 or POMT2 alone is insufficient for O-mannosylation of α-dystroglycan, co-expressed POMT1 and POMT2 could transfer a Man residue to a specific serine residue of α-dystroglycan [35]. Thus, in some cases, glycan synthesis is conducted by an enzyme complex, which contains a chaperone-like protein in addition to a glycosyltransferase. Yada et al. [21] reported that the genomic organization of CSS3 (ChSy-2) was quite similar to that of ChSy-1. The ChSy-1 and ChSy-2 genes consist of three exons each quite similar in length. In C. elegans and Drosophila melanogaster, there is only a single gene for each orthologue corresponding to ChSy-1 or CSS3 (ChSy-2) [8,11], suggesting that CSS3 (ChSy-2) resulted from a gene duplication of ChSy-1 or vice versa.
Although CSS3 (ChSy-2) exhibited a widespread expression, its expression pattern was different from that of ChSy-1 or ChPF, which in contrast showed similar expression patterns to each other (see Figure 3). As CSS3 (ChSy-2) requires ChPF or ChSy-1 to exert CS-synthesizing activity, the difference in expression pattern among these proteins suggests that CSS3 (ChSy-2) participates in the biosynthesis of CS indeed in the peripheral blood leucocytes, because its expression level is relatively high in these cells, where no ChPF mRNA is apparently expressed (see Figure 3) despite the marked production of CS.
Although it is unclear why multiple CS-synthesizing enzymes exist, such redundancy may explain why no cell lines in which the biosynthesis of CS is completely disrupted have been reported so far. As demonstrated here, polymerization of CS could be achieved by combinations of any two of ChSy-1, CSS3 (ChSy-2) and ChPF, and therefore a deficiency of any one of them may be compensated by the other proteins to some extent. Thus it may be difficult to establish cell lines defective completely in CS biosynthesis. In C. elegans, there is a single orthogue for each of ChSy and ChPF. In view of the fact that a defect of ChSy or ChPF leads to a dramatic decrease in the amount of chondroitin in C. elegans [7,8,36], ChSy and ChPF may be absolutely required for CS biosynthesis. However, in mammals, six chondroitin-synthesizing proteins exist. Thus it may be difficult to investigate specific roles of each protein involved in the biosynthesis of CS in mammalian cells where functional redundancy exists.
Acknowledgments
We thank Mariko Ueda for technical assistance. This work was supported in part by CREST (Core Research for Evolutional Science and Technology) of the JST (Japan Science and Technology) Corporation (H. K. and K. S.), the Science Research Promotion Fund of the Japan Private School Promotion Foundation, and Grants-in-aid for Scientific Research #16590075 (H. K.) and for Scientific Research on Priority Areas #14082207 (K. S.) from MEXT, Japan
References
- 1.Kjellén L., Lindahl U. Proteoglycans: structures and interactions. Annu. Rev. Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 2.Salmivirta M., Lidholt K., Lindahl U. Heparan sulfate: a piece of information. FASEB J. 1996;10:1270–1279. doi: 10.1096/fasebj.10.11.8836040. [DOI] [PubMed] [Google Scholar]
- 3.Perrimon N., Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 4.Lowe J. B., Marth J. D. A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 5.Princivalle M., Agostini A. D. Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis. Int. J. Dev. Biol. 2002;46:267–278. [Google Scholar]
- 6.Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., Kitagawa H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 2003;13:612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Hwang H.-Y., Olson S. K., Esko J. D., Horvitz R. H. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature. 2003;423:439–443. doi: 10.1038/nature01634. [DOI] [PubMed] [Google Scholar]
- 8.Mizuguchi S., Uyama T., Kitagawa H., Nomura K., Dejima K., Gengyo-Ando K., Mitani S., Sugahara K., Nomura K. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature. 2003;423:443–448. doi: 10.1038/nature01635. [DOI] [PubMed] [Google Scholar]
- 9.Silbert J. E., Sugumaran G. Biosynthesis of chondroitin/dermatan sulfate. IUBMB Life. 2002;54:177–186. doi: 10.1080/15216540214923. [DOI] [PubMed] [Google Scholar]
- 10.Kusche-Gullberg M., Kjellén L. Sulfotransferases in glycosaminoglycan biosynthesis. Curr. Opin. Struct. Biol. 2003;13:605–611. doi: 10.1016/j.sbi.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Kitagawa H., Uyama T., Sugahara K. Molecular cloning and expression of a human chondroitin synthase. J. Biol. Chem. 2001;276:38721–38726. doi: 10.1074/jbc.M106871200. [DOI] [PubMed] [Google Scholar]
- 12.Uyama T., Kitagawa H., Tamura J., Sugahara K. Molecular cloning and expression of human chondroitin N-acetylgalactosaminyltransferase: the key enzyme for chain initiation and elongation of chondroitin/dermatan sulfate on the protein linkage region tetrasaccharide shared by heparin/heparan sulfate. J. Biol. Chem. 2002;277:8841–8846. doi: 10.1074/jbc.M111434200. [DOI] [PubMed] [Google Scholar]
- 13.Gotoh M., Sato T., Akashima T., Iwasaki H., Kameyama A., Mochizuki H., Yada T., Inaba N., Zhang Y., Kikuchi N., et al. Enzymatic synthesis of chondroitin with a novel chondroitin sulfate N-acetylgalactosaminyltransferase that transfers N-acetylgalactosamine to glucuronic acid in initiation and elongation of chondroitin sulfate synthesis. J. Biol. Chem. 2002;277:38189–38196. doi: 10.1074/jbc.M203619200. [DOI] [PubMed] [Google Scholar]
- 14.Sato T., Gotoh M., Kiyohara K., Akashima T., Iwasaki H., Kameyama A., Mochizuki H., Yada T., Inaba N., Togayachi A., et al. Differential roles of two N-acetylgalactosaminyltransferases, CSGalNAcT-1, and a novel enzyme, CSGalNAcT-2. Initiation and elongation in synthesis of chondroitin sulfate. J. Biol. Chem. 2003;278:3063–3071. doi: 10.1074/jbc.M208886200. [DOI] [PubMed] [Google Scholar]
- 15.Uyama T., Kitagawa H., Tanaka J., Tamura J., Ogawa T., Sugahara K. Molecular cloning and expression of a second chondroitin N-acetylgalactosaminyltransferase involved in the initiation and elongation of chondroitin/dermatan sulfate. J. Biol. Chem. 2003;278:3072–3078. doi: 10.1074/jbc.M209446200. [DOI] [PubMed] [Google Scholar]
- 16.Gotoh M., Yada T., Sato T., Akashima T., Iwasaki H., Mochizuki H., Inaba N., Togayachi A., Kudo T., Watanabe H., et al. Molecular cloning and characterization of a novel chondroitin sulfate glucuronyltransferase that transfers glucuronic acid to N-acetylgalactosamine. J. Biol. Chem. 2002;277:38179–38188. doi: 10.1074/jbc.M202601200. [DOI] [PubMed] [Google Scholar]
- 17.Kitagawa H., Izumikawa T., Uyama T., Sugahara K. Molecular cloning of a chondroitin polymerizing factor that cooperates with chondroitin synthase for chondroitin polymerization. J. Biol. Chem. 2003;278:23666–23671. doi: 10.1074/jbc.M302493200. [DOI] [PubMed] [Google Scholar]
- 18.Yada T., Gotoh M., Sato T., Shionyu M., Go M., Kaseyama H., Iwasaki H., Kikuchi N., Kwon Y.-D., Togayachi A., et al. Chondroitin sulfate synthase-2. Molecular cloning and characterization of a novel human glycosyltransferase homologous to chondroitin sulfate glucuronyltransferase, which has dual enzymatic activities. J. Biol. Chem. 2003;278:30235–30247. doi: 10.1074/jbc.M303657200. [DOI] [PubMed] [Google Scholar]
- 19.Busse M., Kusche-Gullberg M. In vitro polymerization of heparan sulfate backbone by the EXT proteins. J. Biol. Chem. 2003;278:41333–41337. doi: 10.1074/jbc.M308314200. [DOI] [PubMed] [Google Scholar]
- 20.Kim B.-T., Kitagawa H., Tanaka J., Tamura J., Sugahara K. In vitro heparan sulfate polymerization: crucial roles of core protein moieties of primer substrates in addition to the EXT1–EXT2 interaction. J. Biol. Chem. 2003;278:41618–41623. doi: 10.1074/jbc.M304831200. [DOI] [PubMed] [Google Scholar]
- 21.Yada T., Sato T., Kaseyama H., Gotoh M., Iwasaki H., Kikuchi N., Kwon Y.-D., Togayachi A., Kudo T., Watanabe H., et al. Chondroitin sulfate synthase-3. Molecular cloning and characterization. J. Biol. Chem. 2003;278:39711–39725. doi: 10.1074/jbc.M304421200. [DOI] [PubMed] [Google Scholar]
- 22.Nawa K., Sakano K., Fujiwara H., Sato Y., Sugiyama N., Teruuchi T., Iwamoto M., Marumoto Y. Presence and function of chondroitin-4-sulfate on recombinant human soluble thrombomodulin. Biochem. Biophys. Res. Commun. 1990;171:729–737. doi: 10.1016/0006-291x(90)91207-9. [DOI] [PubMed] [Google Scholar]
- 23.Nadanaka S., Kitagawa H., Sugahara K. Demonstration of the immature glycosaminoglycan tetrasaccharide sequence GlcAβ1-3Galβ1-3Galβ1-4Xyl on recombinant soluble human α-thrombomodulin. An oligosaccharide structure on a ‘part-time’ proteoglycan. J. Biol. Chem. 1998;273:33728–33734. doi: 10.1074/jbc.273.50.33728. [DOI] [PubMed] [Google Scholar]
- 24.Kitagawa H., Paulson J. C. Cloning of a novel α2,3-sialyltransferase that sialylates glycoprotein and glycolipid carbohydrate groups. J. Biol. Chem. 1994;269:1394–1401. [PubMed] [Google Scholar]
- 25.Mizushima S., Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitagawa H., Tsutsumi K., Ujikawa M., Goto F., Tamura J., Neumann K. W., Ogawa T., Sugahara K. Regulation of chondroitin sulfate biosynthesis by specific sulfation: acceptor specificity of serum β-GalNAc transferase revealed by structurally defined oligosaccharides. Glycobiology. 1997;7:531–537. doi: 10.1093/glycob/7.4.531. [DOI] [PubMed] [Google Scholar]
- 27.Kitagawa H., Ujikawa M., Tsutsumi K., Tamura J., Neumann K. W., Ogawa T., Sugahara K. Characterization of serum β-glucuronyltransferase involved in chondroitin sulfate biosynthesis. Glycobiology. 1997;7:905–911. doi: 10.1093/glycob/7.7.905. [DOI] [PubMed] [Google Scholar]
- 28.Sugahara K., Shigeno K., Masuda M., Fujii N., Kurosaka A., Takeda K. Structural studies on the chondroitinase ABC-resistant sulfated tetrasaccharides isolated from various chondroitin sulfate isomers. Carbohydr. Res. 1994;255:145–163. doi: 10.1016/s0008-6215(00)90976-5. [DOI] [PubMed] [Google Scholar]
- 29.Sugahara K., Ohkita Y., Shibata Y., Yoshida K., Ikegami A. Structural studies on the hexasaccharide alditols isolated from the carbohydrate-protein linkage region of dermatan sulfate proteoglycans of bovine aorta. Demonstration of iduronic acid-containing components. J. Biol. Chem. 1995;270:7204–7212. doi: 10.1074/jbc.270.13.7204. [DOI] [PubMed] [Google Scholar]
- 30.Kinoshita A., Sugahara K. Microanalysis of glycosaminoglycan-derived oligosaccharides labeled with a fluorophore 2-aminobenzamide by high-performance liquid chromatography: application to disaccharide composition analysis and exosequencing of oligosaccharides. Anal. Biochem. 1999;269:367–378. doi: 10.1006/abio.1999.4027. [DOI] [PubMed] [Google Scholar]
- 31.Senay C., Lind T., Muguruma K., Tone Y., Kitagawa H., Sugahara K., Lidholt K., Lindahl U., Kusche-Gullberg M. The EXT1/EXT2 tumor suppressors: catalytic activities and role in heparan sulfate biosynthesis. EMBO Rep. 2000;1:282–286. doi: 10.1093/embo-reports/kvd045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itano N., Kimata K. Mammalian hyaluronan synthases. IUBMB Life. 2002;54:195–199. doi: 10.1080/15216540214929. [DOI] [PubMed] [Google Scholar]
- 33.Itano N., Sawa T., Yoshida M., Lenas P., Yamada Y., Imagawa M., Shinomura T., Hamaguchi M., Yoshida Y., Ohnuki Y., et al. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J. Biol. Chem. 1999;274:25085–25092. doi: 10.1074/jbc.274.35.25085. [DOI] [PubMed] [Google Scholar]
- 34.Michele D. E., Campbell K. P. Dystrophin–glycoprotein complex: post-translational processing and dystroglycan function. J. Biol. Chem. 2003;278:15457–15460. doi: 10.1074/jbc.R200031200. [DOI] [PubMed] [Google Scholar]
- 35.Manya H., Chiba A., Yoshida A., Wang X., Chiba Y., Jigami Y., Margolis R. U., Endo T. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:500–505. doi: 10.1073/pnas.0307228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Izumikawa T., Kitagawa H., Mizuguchi S., Nomura K. H., Nomura K., Tamura J., Gengyo-Angyo K., Mitani S., Sugahara K. Nematode chondroitin polymerizing factor showing cell-/organ-specific expression is indispensable for chondroitin synthesis and embryonic cell division. J. Biol. Chem. 2004;279:53755–53761. doi: 10.1074/jbc.M409615200. [DOI] [PubMed] [Google Scholar]