Abstract
The transcription factor Nkx6.1 is required for the establishment of functional insulin-producing β-cells in the endocrine pancreas. Overexpression of Nkx6.1 has been shown to inhibit glucagon gene expression while favouring insulin gene activation. Down-regulation resulted in the opposite effect, suggesting that absence of Nkx6.1 favours glucagon gene expression. To understand the mechanism by which Nkx6.1 suppresses glucagon gene expression, we studied its effect on the glucagon gene promoter activity in non-islet cells using transient transfections and gel-shift analyses. In glucagonoma cells transfected with an Nkx6.1-encoding vector, the glucagon promoter activity was reduced by 65%. In BHK21 cells, Nkx6.1 inhibited by 93% Pax6-mediated activation of the glucagon promoter, whereas Cdx2/3 and Maf stimulations were unaltered. Although Nkx6.1 could interact with both the G1 and G3 element, only the former displayed specificity for Nkx6.1. Mutagenesis of the three potential AT-rich motifs within the G1 revealed that only the Pax6-binding site preferentially interacted with Nkx6.1. Chromatin immunoprecipitation confirmed interaction of Nkx6.1 with the glucagon promoter and revealed a direct competition for binding between Pax6 and Nkx6.1. A weak physical interaction between Pax6 and Nkx6.1 was detected in vitro and in vivo suggesting that Nkx6.1 predominantly inhibits glucagon gene transcription through G1-binding competition. We suggest that cell-specific expression of the glucagon gene may only proceed when Nkx6.1, in combination with Pdx1 and Pax4, are silenced in early α-cell precursors.
Keywords: gene regulation, glucagon, Nkx6.1
Abbreviations: CAT, chloramphenicol acetyltransferase; ChIP, chromatin immunoprecipitation; CMV, cytomegalovirus; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; GST, glutathione S-transferase; HD, homeodomain; HDC, HD plus the C-terminal end of Nkx6.1; PD, paired domain; TBP, TATA-binding protein
INTRODUCTION
Pancreas development is orchestrated by the sequential activation of key transcription factors that will dictate the fate of progenitor cells towards exocrine tissue or endocrine entities comprising insulin (β)-, glucagon (α)-, somatostatin (δ)- or PP (pancreatic polypeptide)-producing cells [1]. Members of the HD (homeodomain) family of transcription factors play a crucial role in this process. Gene inactivation in mouse models has shown that induction of Pdx1 and Hb9 [2–4] are pre-requisites for ventral and/or dorsal pancreatic bud formation, while the transient expression of Ngn3 during organogenesis is indispensable for the generation of endocrine precursors [5,6]. The programme of endocrine cell differentiation must then be activated by as yet unknown soluble factors that will stimulate or repress a variety of transcription factors resulting in the generation of the different cell types. Pax4 and Nkx6.1 expression [7,8] are obligatory for the maturation of β-cells whereas Pax6 and Arx are critical for the emergence of α-cells [9,10]. Some of these transcription factors are also important for the subsequent maintenance of islet cell specific hormone gene expression. Thus the β/δ-enriched transcription factor Pdx1, together with E47/Beta2 and MafA, is required for sustaining the β-cell phenotype by positively regulating insulin gene expression [11]. Pax4 and Nkx6.1 β-cell specific expression are mandatory for cell survival as well as robust glucose-induced insulin secretion [12,13].
In contrast to insulin, α-cell-restricted expression of the glucagon gene is predominantly mediated by the interaction of Pax6 with either Cdx2/3 or Maf on the G1 cis-acting element located in the proximal promoter region [14–16]. The latter was previously shown to convey α-cell specificity [17]. Additional transcription factors such as Foxa2, Isl1, E47/Beta2 and Pbx/Prep1, through binding to three islet-specifying promoter elements (G2, G3 and G4), also contribute to expression of the glucagon gene [18–21]. Surprisingly, none of these trans-activating factors are α-cell specific, suggesting that expression of the glucagon gene in mature α-cells could be dependent on the absence of β- and/or δ-cell specific transcription factors such as Pdx1, Pax4 and Nkx6.1. Consistent with this premise, mice carrying one or two ablated alleles of either the pdx1 or pax4 genes harbour pancreatic islets with a reduced number or an absence of insulin-producing cells respectively and increased α-cells [2,7,22]. Repression of Pdx1 function in either mouse islets using antisense RNA, or in the insulinoma INS-1 cell line by overexpression of a dominant-negative form of the transcription factor, resulted in decreased insulin gene expression with a concomitant increase in glucagon gene transcription [23,24]. Furthermore, we have previously demonstrated that ectopic expression of either Pdx1 or Pax4 in the glucagonoma InR1G9 cell line repressed glucagon gene expression. This inhibition was predominantly mediated through direct protein–protein interaction of Pdx1 with Pax6 and Cdx2/3, or by competitive binding between Pax4 and Pax6 to the G1 element [25,26]. Taken together these studies demonstrate that Pax4 and Pdx1 are important in β-cell fate determination as well as in promoting insulin gene expression to the detriment of glucagon gene transcription.
More recently, adenoviral-mediated expression of Nkx6.1 was shown to repress glucagon gene expression in INS-1-derived cell lines expressing glucagon as well as in the αTC1.6 α-cell line [13]. Conversely, repression of Nkx6.1 by RNA interference prompted a 2-fold increase in glucagon mRNA levels in INS-1 cells. EMSAs (electrophoretic mobility-shift assays) revealed a potential binding of Nkx6.1 to the G1 element of the glucagon promoter only in the presence of an Nkx6.1-specific antibody. More convincingly, ChIP (chromatin immunoprecipitation) assays performed in INS-1 cells expressing high levels of Nkx6.1 indicated a direct interaction of the transcription factor with the glucagon promoter [13]. These results substantiate the dogma that β-cell-specific transcription factors repress glucagon gene expression while stimulating insulin gene transcription. However, the mechanism by which Nkx6.1 inhibits glucagon expression remains elusive. The present study was designed to define the direct contribution of Nkx6.1 in glucagon gene transcription repression. To this end, we have used the heterologous BHK-21 cell line to delineate critical promoter regions involved in inhibition as well as potential transcription factors that may be targets of Nkx6.1 repression. We found that Nkx6.1 impaired transcriptional activation of the glucagon gene promoter by Pax6, an effect predominantly occurring through competition for the G1 element rather than protein–protein interaction. Neither Cdx2/3 nor c-Maf trans-activation potentials were altered by Nkx6.1. Finally, overexpression of Nkx6.1 in the InR1G9 glucagonoma cell line repressed endogenous glucagon mRNA levels. We thus propose that Nkx6.1, along with Pax4 and Pdx1, are critical for the establishment of the β-cell phenotype during development and that their absence in α-cells is mandatory for full activation of glucagon gene expression and α-cell differentiation.
MATERIALS AND METHODS
Cell culture and DNA transfection
The glucagon-producing hamster cell line InRIG9 [27] and the non-islet Syrian baby hamster kidney (BHK-21) cell line were grown in RPMI 1640 (Seromed) supplemented with 5% fetal calf serum and 5% newborn calf serum, 2–mM glutamine, 100–units/ml of penicillin and 100–μg/ml of streptomycin. BHK-21 cells were transiently transfected using the calcium phosphate precipitation technique [28] using 10–15–μg of total plasmid DNA per 10-cm Petri dish. The plasmid pSV2A PAP (human alkaline phosphatase cDNA) was added to monitor transfection efficiency. Alternatively, InR1G9 cells were transfected using the DEAE-dextran method as described previously [29]. cDNAs for rat Nkx6.1, hamster Cdx2/3 (both provided by M. S. German, Department of Medicine, University of California, San Fransisco, CA., U.S.A.), mouse Pax4 and quail Pax6 (S. Saule, Institut Curie Section de Recherche, Centre Universitaire, Orsay, France), were subcloned in the expression vector pSG5 (Stratagene). cDNAs for mouse c-Maf (provided by R. Stein, Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, TN, U.S.A.) was cloned in the expression vector pcDNA3 (Invitrogen). In experiments using variable quantities of expression vectors encoding Nkx6.1, Pax6, Cdx2/3 or c-Maf, total amount of DNA was kept constant by adding appropriate amounts of the empty pSG5 vector. Reporter plasmids comprised the CAT (chloramphenicol acetyltransferase) reporter gene under the transcriptional control of different fragments of the rat glucagon gene promoter (−292GluCAT, −138GluCAT, G3–31GluCAT, −31GluCAT) [17] or the CMV (cytomegalovirus) promoter. Empty vector (poCAT) and the prolactin promoter (pFoxCAT-36prl, a gift from M. S. German) were used as negative controls.
In some instances, InR1G9 cells or stable clones thereof expressing either a dominant-negative variant of Pax6 (DN-Pax6) or an empty vector were transiently transfected with the Nkx6.1-expressing vector using TransFectin™ reagent (BioRad). The latter allowed for approximately 75% transfection efficiency as assessed by FACS analysis.
Semi-quantitative PCR
Total RNA was isolated using Trizol reagent (Invitrogen). First-strand cDNA synthesis was performed in the presence of 10–ng/μl random hexamer primers (Promega) and the SuperScript II Reverse Transcriptase (Invitrogen). Semi-quantitative PCR for detection of Nkx6.1 was achieved using the Goldstar DNA Polymerase (Eurogenetech) in the presence of the following primers: sense: 5′-ATCAACGACATCCTGAGC-3′, antisense: 5′-GCTTCTTTCTCCACTTGG-3′. Quantitative PCR was performed using the Quantitect SYBR® Green PCR kit (Qiagen) in a Light-Cycler (Roche Diagnostics). The TBP (TATA-binding protein) transcript was used as an internal control. Primers were as follows: glucagon sense: 5′-GATCATTCCCAGCTTCCCAG-3′, glucagon antisense: 5′-CTGGTAAAGGTCCCTTCAGC-3′, TBP sense: 5′-GCCGAATATAATCCCAAGCG-3′, TBP antisense: 5′-GAAACTTCACATCACAGCTC-3′.
CAT and protein assays
Cell extracts were prepared 48–h after transfection and analysed for CAT and alkaline phosphatase activities as described previously [17]. Quantification of acetylated and non-acetylated forms of CAT was performed using a PhosphorImager (Molecular Dynamics). A minimum of three independent transfections were performed, each of them carried out in duplicate.
EMSAs
Nuclear extracts from BHK-21 cells transfected with either an expression vector encoding the HDC domain of hamster Nkx6.1 (provided by R. Mirmira, Department of Medicine and Pharmacology, University of Virginia, Diabetes Center, Charlottesville, VA, U.S.A.) or an empty vector pBAT12 were prepared according to the method of Schreiber et al. [30]. EMSAs were performed as described previously [31] adding initially 8–μg of nuclear extracts and subsequently labelled oligonucleotides corresponding to the rat glucagon gene promoter elements G1 or G3 sequences [14,15,25]. Nkx6.1 antiserum (provided by O. D. Madsen, Hagedorn Research Institute, Gentofte, Denmark) was added to the binding reaction halfway through the incubation time to confirm identity of the protein. Non-radiolabelled mutant oligonucleotides were used to identify the Nkx6.1-binding site on the G1 element (Figure 4A). Additionally an oligomer corresponding to the A4/A3 sites of the mouse insulin promoter was used as a positive control for Nkx6.1 binding affinity (5′-CATCAGGCCATCTGGTCCCT TATTAAGACT ATAATAACCCTAAGACTAAGTAGAT-3′).
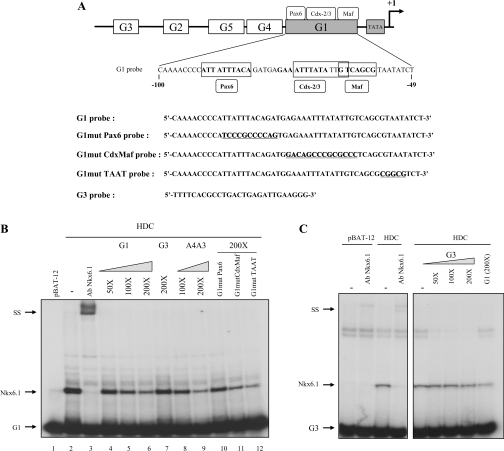
Figure 4. The HD of Nkx6.1 interacts with the G1 element of the glucagon gene promoter.
(A) Schematic representation of the 5′-flanking region of the rat glucagon gene promoter depicting the control elements (G1–G5). The sequence of G1 is shown as well as the binding sites for Pax6, Cdx2/3 and Maf (bold nucleotides). The various probes used in EMSA are depicted and underlined nucleotides represent mutated sequences. Studies of Nkx-6.1 binding to the G1 (−100/−49) and G3 element (−274/−234) are represented in (B) and (C) respectively. Specific interactions of Nkx6.1 on glucagon probes was determined using an anti-Nkx6.1 antibody. pBAT-12 represent nuclear extracts derived from cells transfected with control empty vector. Competition assays were performed with increasing amounts of non-labelled probes ranging from 50- to 200-fold excess for native oligonucleotides and 200-fold excess for mutated oligonucleotides. SS, supershift.
ChIP
ChIP assays were performed as described by Orlando et al. [31a]. Briefly, formaldehyde-cross-linked chromatin extracts were prepared from InR1G9 and fragmented by sonication. Chromatin extract (50–μg) were first pre-cleared with Protein A-sepharose beads (Pharmacia Biotech AB) for 1–h. After centrifugation (10–g for 1–min at 4 °C), supernatants were incubated overnight at 4 °C with 10–μg of an anti-Pax6, anti-Nkx6.1, anti-c-Maf (#BL662; Bethyl), anti-acetyl-histone H4 antibodies (Upstate) and a rabbit IgG (#sc-2027; Santa Cruz Biotechnology). DNA–protein–antibody complexes were then incubated for 3–h with Protein A-sepharose beads and subsequently washed sequentially in a low-salt buffer, high-salt buffer, LiCl buffer and finally a Tris/EDTA buffer as described by Duong et al. [32]. Proteins were eliminated using 200–μg of proteinase K (Applichem) by overnight incubation at 45 °C followed by an additional incubation at 65 °C for 3–h. After phenol extraction, DNA was precipitated, resuspended in water and used as a template for PCR. The PCR primers used for analysis of binding on the glucagon proximal promoter in InR1G9 cells were 5′-GACTAGGCTCATTTGACGTC-3′ and 5′-ATGGAAAGGGCAGTTTGGAG-3′. PCR products were verified on 3% ethidium bromide-stained agarose gels and analysed by real-time PCR using a Light-Cycler.
GST (glutathione S-transferase)-fusion proteins and GST-precipitation
Pax6 GST-fusion proteins have been described previously [14]. GST-fusion proteins were expressed in Escherichia coli and purified according to the manufacturer's protocol (Pharmacia). L-[35S]Methionine-labelled Nkx6.1 was generated in vitro using the TNT system (Promega) and GST-precipitations were performed as described previously [14].
Co-immunoprecipitation and Western blot analysis
Nkx6.1-overexpressing InR1G9 cells were trypsinized, washed and resuspended in a buffer containing 10–mM Hepes (pH–7.9), 10–mM KCl, 0.1–mM EDTA, 0.1–mM EGTA, 2.5–mM DTT (dithiothreitol) and 1.2–mM PMSF. Subsequent to a 15–min incubation on ice, cells were lysed by the addition of 10% Nonidet P40 and intact nuclei were sedimented by centrifugation. Buffer C containing 20–mM Hepes (pH–7.9), 25% glycerol, 250–mM NaCl, 1–mM EDTA, 1–mM EGTA, 1–mM DTT, 1.2–mM PMSF and complemented with a cocktail of protease inhibitors (Roche) was added to the pellet and incubated for 1–h. Nuclear proteins were purified from membrane debris by centrifugation (13–g for 10–min at 4 °C). Approximately 500–μg of nuclear extracts were incubated overnight at 4 °C with a mouse anti-Pax6 antibody (MAB1260; R&D system) coupled to sheep anti-mouse IgG magnetic Dynabeads® (Invitrogen). Beads were then washed extensively with Buffer C and resuspended in 50–μl Lämmli buffer. Next, 10–50–μg of total nuclear extracts (input) and 20–μl out of the 50–μl of immunoprecipitate were separated on SDS/PAGE and transferred on to PVDF membrane (Amersham Biosciences). Membranes were incubated with either an anti-rabbit Pax6 antibody or an anti-rabbit Nkx6.1 antibody and subsequently with a sheep anti-rabbit IgG coupled to horseradish peroxidase. Signals were detected using the Super Signal West Pico Trial Kit (Pierce Chemical Co.).
Statistical analysis
Results are expressed as means±S.E.M. Where indicated, the statistical significance of the differences between groups was estimated using the Student's unpaired t test: * and ** indicate statistical significance with P<0.05 and P<0.02 respectively.
RESULTS
Ectopic expression of Nkx6.1 in InR1G9 cells inhibits the transcriptional activity of the glucagon gene promoter
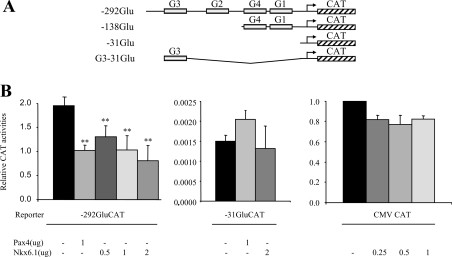
As an initial step to assess the potential inhibitory function of Nkx6.1 on glucagon gene expression, the glucagonoma InR1G9 cell line was co-transfected with a CAT reporter construct driven by a fragment of the glucagon gene promoter conferring full gene activity as well as α-cell specificity (−292GluCAT; Figure 1A) along with increasing amounts of an expression vector harbouring the Nkx6.1 cDNA. Quantitatively similar to Pax4-mediated repression [26], ectopic expression of Nkx6.1 resulted in a dose-dependent inhibition (reaching 65%) of the basal transcriptional activity of the −292GluCAT construct (Figure 1B). In contrast, expression of the minimal glucagon gene promoter construct −31GluCAT (Figures 1A and 1B) or a CAT construct harbouring the viral promoter CMV (Figure 1B) were not significantly modulated by Nkx6.1, indicating that repression was specific to the glucagon gene and conferred by sequences located within the boundaries of −292 and −31 of the promoter. This region harbours five cis-acting elements [33], one of which, the G1, contains three A/T-rich regions resembling putative Nkx6.1-binding sites [13,31,34].
Figure 1. Nkx6.1 inhibits trans-activation of the glucagon gene promoter in InR1G9 cells, but has no effect on a heterologous promoter.
(A) Schematic representation of reporter gene constructs used in the present study. (B) Increasing amounts (0.5–2–μg) of expression vector containing the Nkx6.1 cDNA or 1–μg of expression vector containing the Pax4 cDNA were co-transfected into InR1G9 cells with 3–μg of the indicated reporter constructs. In addition, a reporter construct under the transcriptional control of the ubiquitously active CMV promoter was co-transfected into InR1G9 cells along with increasing amounts of the Nkx6.1 expression vector. Data are presented as relative basal CAT activity. **P<0.02.
Nkx6.1 inhibits glucagon gene transcription by repressing Pax6-mediated activation
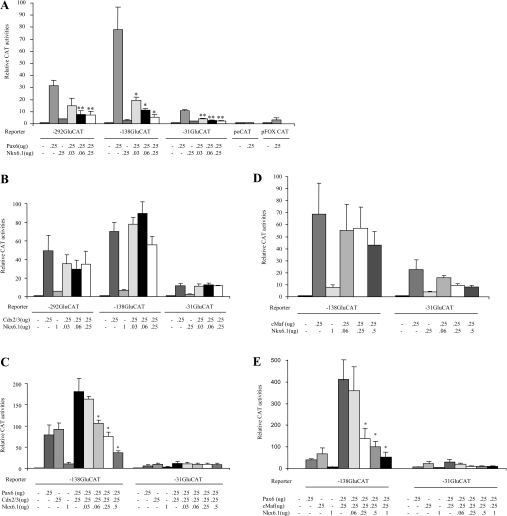
Interaction of the transcription factor Pax6 with either Cdx2/3 or c-Maf on the G1 element of the glucagon gene promoter results in the synergistic activation of transcription [14,16]. We thus investigated whether Nkx6.1 could interfere with activation of the glucagon gene promoter by Pax6, Cdx2/3 and c-Maf, in the non-islet cell line BHK-21 lacking endogenous expression of these transcription factors. As Pax6 interacts with both the G1 and G3 elements of the glucagon gene promoter, we used reporter constructs containing both elements (−292GluCAT) or constructs bearing only the G1 (−138GluCAT) element. As expected, ectopic expression of Pax6 (0.25–μg) resulted in a 33- and 79-fold increase in CAT activity for the −292 and −138GluCAT constructs respectively as compared to control transfections which exhibited low transcriptional activity in the absence of Pax6 (Figure 2A). Of note, Pax6 conveyed a stronger CAT activity to the −138GluCAT reporter containing a single binding site as compared to the −292GluCAT construct, substantiating previous findings that the latter harbours inhibitory regulatory cis-acting elements [35]. Interestingly, Pax6 also induced transcription of the −31GluCAT construct by approximately 10-fold, but not of another construct containing the rat prolactin gene promoter TATA box (pFOXCAT) or of the control poCAT vector indicating a potential weak interaction of the transcription factor with the glucagon basal promoter region (Figure 2A). In contrast to results obtained in InR1G9 cells, overexpression of Nkx6.1 in BHK-21 cells had no significant impact on the basal expression of any of the constructs. However, co-transfection of 0.25–μg of Pax6 along with increasing amounts of Nkx6.1 resulted in a dose-dependent inhibition of Pax6-mediated activation, reaching 75% and 93% suppression for −292GluCAT and −138GluCAT respectively at 0.25–μg of Nkx6.1 (Figure 2A). Interestingly, Nkx6.1 also inhibited the Pax6-mediated activation of the −31GluCAT construct by approximately 50% reinforcing a potential interaction of Pax6 with the glucagon basal promoter (Figure 2A).
Figure 2. Nkx6.1 interferes with Pax6-mediated transcriptional activation of the glucagon gene promoter.
(A) BHK-21 cells were co-transfected with 10–μg of the indicated reporter plasmids and 0.25–μg of expression vectors encoding Pax6 either alone or with 0.03–0.25–μg of Nkx6.1 expression vector. Nkx6.1 (0.25–μg) was transfected alone as a control. (B) BHK-21 cells were co-transfected with 10–μg of the indicated reporter plasmids and 0.25–μg of expression vectors encoding Cdx2/3 either alone or with 0.03–0.25–μg of Nkx6.1 expression vector. Nkx6.1 (1–μg) was transfected alone as a control. (C) BHK-21 cells were co-transfected with 10–μg of the indicated reporter plasmids and 0.25–μg of expression vectors encoding Pax6 and 0.25–μg of Cdx2/3 either alone or with 0.03–0.5–μg of Nkx6.1 expression vector. Nkx6.1 (1–μg) was transfected alone as a control. (D) BHK-21 cells were co-transfected with 10–μg of either −138GluCAT or −31GluCAT along with 0.25–μg of expression vectors encoding c-Maf alone or in combination with 0.06–0.5–μg of Nkx6.1 expression vector. Nkx6.1 (1–μg) was transfected alone as a control. (E) BHK-21 cells were co-transfected with 10–μg of the indicated reporter plasmids and 0.25–μg of expression vectors encoding Pax6 and c-Maf either alone or with 0.06–1–μg of Nkx6.1 expression vector. Nkx6.1 (1–μg) was transfected alone as a control. Data are presented as relative CAT activity. * and ** indicate statistical significance with P<0.05 and P<0.02 respectively.
Nkx6.1 did not significantly inhibit activation of the −292, −138 and −31GluCAT constructs by Cdx2/3, suggesting that binding of the latter to G1 is not subject to regulation by Nkx6.1 (Figure 2B). However, in co-transfection experiments performed in the presence of both Pax6 and Cdx2/3, increasing amounts of Nkx6.1 gradually abrogated the synergistic effect of the two transcription factors on the −138GluCAT construct (180-fold increase) reaching 77% repression using 0.5–μg of Nkx6.1 (Figure 2C). Nkx6.1 did not significantly inhibit the 70-fold increase in trans-activation of the −138GluCAT by c-Maf (Figure 2D). Yet, in the combined presence of Pax6 and c-Maf, the robust 400-fold induction of −138GluCAT was drastically suppressed by Nkx6.1 to levels attained by c-Maf alone (Figure 2E). Although both Cdx2/3 and c-Maf trans-activated the −31GluCAT, addition of Pax6 did not increase transcription of this construct further. Furthermore, increasing amounts of Nkx6.1 did not suppress either Cdx2/3 or c-Maf-mediated increases in −31GluCAT activity. Taken together, these results strongly suggest that Nkx6.1 inhibits glucagon gene expression by interfering with Pax6 mediated-activation via the G1 promoter element, independent of c-Maf and Cdx2/3.
The HD-C-terminal region of Nkx6.1 binds to the G1 element of the glucagon gene promoter
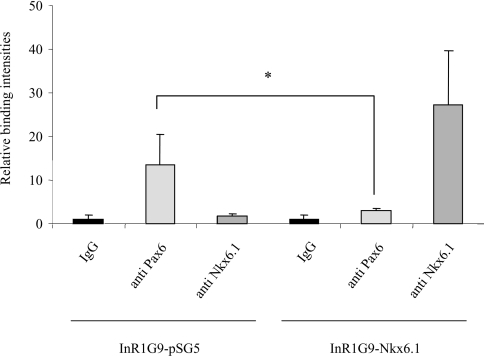
To establish whether Nkx6.1 binds directly to the glucagon gene promoter in vivo, ChIP analysis was performed using InR1G9 cells transfected with either the control vector pSG5 or with the Nkx6.1 expression construct. As expected, the promoter region was co-immunoprecipitated by anti-Pax6 but not by either anti-IgG or anti-Nkx6.1 sera in pSG5-transfected cells (Figure 3, lefthand side). In contrast, cells expressing Nkx6.1 revealed a decreased occupancy of Pax6 to the glucagon promoter with a concomitant increase in Nkx6.1 binding (Figure 3, right-hand side). These results indicate that, when conditionally expressed in α-cells, Nkx6.1 is able to interact with the glucagon gene promoter and repress its expression by potentially interfering with Pax6 binding. To further explore this putative mechanism, we performed EMSAs using the G1 promoter element. This element harbours three A/T-rich sequences reminiscent of Nkx6.1-binding sites: the first sequence, (−90)/5′-ATTATTTA-3′/(−84), was shown to interact with Pax6 and Brain4, while the second, (−72)/5′-ATTTATA-3′/(−64) and the third, (−56)/5′-TAATATCT-3′/(−49) sequences flank the Maf and Cdx2/3 core binding site (Figure 4A). Nuclear extracts derived from BHK-21 cells transfected with a full-length Nkx6.1 cDNA did not produce a retarded complex with the G1 element unless an antibody raised against the transcription factor was added to the reaction mixture (results not shown). The binding was specific to Nkx6.1 as the antibody alone did not generate a complex. These results suggest that a potential protein conformational change may be required in order for Nkx6.1 to interact with the G1. As the N-terminal region of the protein contains a transcriptional repression domain that was shown to interfere with binding of Nkx6.1 to the insulin promoter in vitro [31,36], we repeated the EMSA using a truncated protein containing the HD and the HD containing the C-terminal end of Nkx6.1 (HDC) [37]. A single predominant retarded complex was detected using recombinant HDC (Figure 4B, lanes 1 and 2). The specificity of the complex was confirmed by the addition of anti-Nkx6.1 serum that produced a supershift (Figure 4B, lane 3). The Nkx6.1 complex formed on the labelled G1 was effectively and similarly competed by increasing amounts of either non-radiolabelled G1 or the A4/A3 insulin promoter element bearing an Nkx6.1-binding site (Figure 4B, lanes 4–6 and 8–9). Interestingly, the G3 element, which also interacts with Pax6 but does not contain a conserved A/T-rich region, was unable to compete for binding of Nkx6.1 to the G1 element (Figure 4B, lane 7). To establish the potential contribution of the three G1 putative Nkx6.1binding sites to the formation of the retarded complex, we independently mutated each site (see Figure 4A) and performed competition assays against a bona fide labelled G1 element. Both mutated proximal sequences (−72/−64 and −56/−49) competed as efficiently as the wild-type sequence (compare Figure 4B, lane 6 with lanes 11 and 12) while competition with the most mutated distal sequence corresponding to the Pax6-binding site was completely abrogated (Figure 4B, lane 10). Interestingly, EMSAs performed with labelled G3 and recombinant HDC revealed the formation of a complex that was supershifted in the presence of Nkx6.1 antibodies (Figure 4C). However, in contrast to G1, non-radiolabelled G3 was unable to compete for the complex, indicating low-affinity or non-specific interaction of the HDC with G3. Taken together, these results suggest that Nkx6.1 interacts specifically with the core sequence of the Pax6-binding site within the G1 element of the glucagon gene promoter.
Figure 3. Nkx6.1 interacts with the glucagon promoter and disrupts Pax6 binding in the InR1G9 glucagon-producing cell line.
Histograms representing the relative binding of Pax6 and Nkx6.1 proteins to the glucagon promoter in InR1G9 cells transfected with either an empty vector (pSG5) or the expression vector containing the Nkx6.1 cDNA. After cross-linking, immunoprecipitations were performed with anti-Pax6 or anti-Nkx6.1 as indicated in the Materials and methods section. An anti-histone H4 immunoprecipitation was also performed as a positive control in each experiment (results not shown). Binding capacity was analysed by real-time RT-PCR using a light-cycler (Roche Diagnostics). Binding intensity data are expressed in terms of the IgG immunoprecipitation (non-specific binding) and are presented as the mean±S.E.M. for at least three independent experiments.
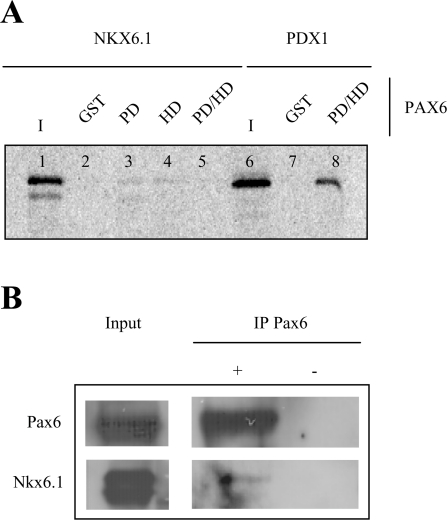
Nkx6.1 interacts with both the PD (paired domain) and HD of Pax6
We next sought to determine whether Nkx6.1 could physically interact with Pax6. To this end, we performed GST precipitation assays using GST-fusion proteins containing the Pax6 PD, HD or a fusion of the PD/HD and 35S-labelled Nkx6.1. A weak protein–protein interaction with Nkx6.1 was observed with all fusion proteins, whereas no precipitation was detected with GST. As previously demonstrated [25], Pdx1 strongly interacted with a Pax6 PD/HD GST-fusion protein (Figure 5). In parallel, co-immunoprecipitation assays using an anti-Pax6 serum were performed on nuclear protein extracts derived from InR1G9 cells transfected with an Nkx6.1 expression vector. Consistent with GST precipitation assays, Nkx6.1 was co-immunoprecipitated along with Pax6, albeit to low levels. These data suggest that Nkx6.1 may potentially interact with Pax6 in the absence of DNA and may contribute to the transcriptional repression of the glucagon gene.
Figure 5. A weak interaction between Nkx6.1 and Pax6 is observed both in vitro and in vivo.
(A) GST-precipitation assay using 10–μg of GST alone, GST-Pax6 PD, HD or PDHD (paired-linker-HD) fusion proteins immobilized on sepharose beads and in vitro synthesized, 35S-labelled Nkx6.1 or Pdx1. Lane input, 10% of the respective in vitro translation reaction used for protein–protein interactions. (B) To confirm in vitro interactions, InR1G9 cells were transfected with Nkx6.1 and co-immunoprecipitation was performed using an anti-mouse Pax6 serum in the presence of sheep anti-mouse IgG magnetic Dynabeads (+). Experiments were also performed in the absence of the Pax6 serum (−). Precipitates were subjected to Western blotting for Pax6 and Nkx6.1. The input for Pax6 was 50–μg while 10–μg was used for Nkx6.1.
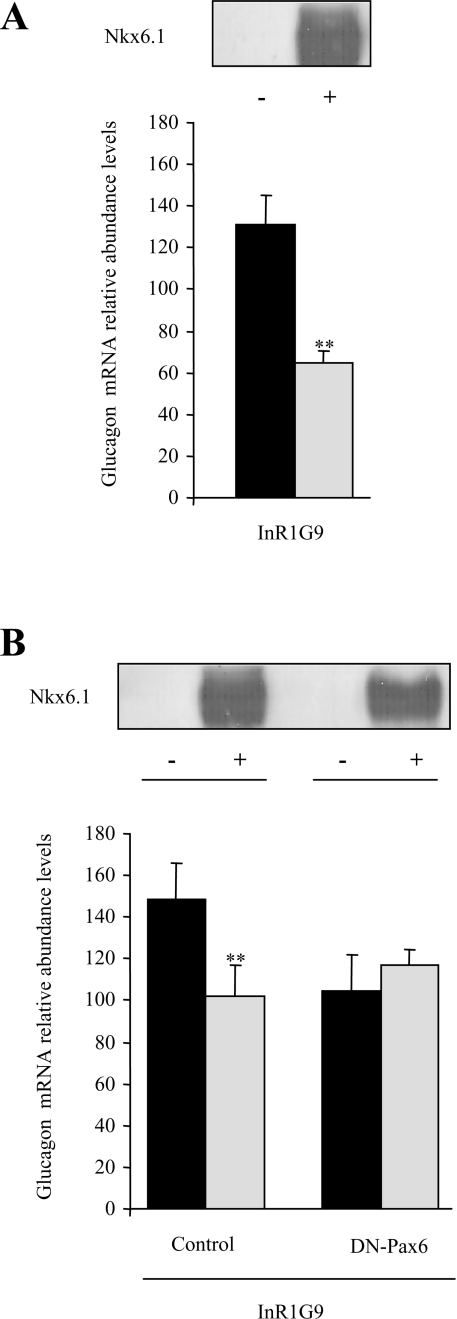
Endogenous levels of glucagon mRNA are repressed in InR1G9 cells expressing Nkx6.1
In order to validate our in vitro data suggesting that repression of glucagon gene expression by Nkx6.1 is mediated through competition with Pax6, InR1G9 cells were transfected with an Nkx6.1 expression vector and endogenous levels of glucagon were evaluated by quantitative PCR. A 46% inhibition of glucagon mRNA relative abundance levels was detected 48–h post-transfection (Figure 6A). To correlate Nkx6.1-mediated repression of glucagon gene expression to decreased Pax6 trans-activation potential, we transfected Nkx6.1 into a clonal cell line expressing a dominant-negative variant of Pax6 (DN-Pax6) at high efficiency (75% of the cells were transfected). The latter binds to its target genes but is no longer able to trans-activate them, as reflected by a 33% decrease in glucagon mRNA levels (Figure 6B, black bars). More importantly, expression of Nkx6.1 in this clonal line did not further decrease glucagon mRNA levels clearly establishing that Nkx6.1 inhibits transcription by directly interfering with Pax6-mediated trans-activation of the hormone gene (Figure 6B).
Figure 6. Overexpression of Nkx6.1 inhibits endogenous levels of glucagon mRNA in InR1G9 cells by competing with Pax6 trans-activation.
(A) InR1G9 cells were transiently transfected with an expression vector encoding Nkx6.1 using the TransFectin™ reagent. This approach permitted 75% transfection efficiency as assessed by FACS analysis. Expression of the Nkx6.1 transgene in transfected cells (+) and its absence in untransfected cells (−) was determined by semi-quantitative PCR. Relative abundance levels of endogenous glucagon mRNA were then evaluated by quantitative PCR and normalized to the housekeeping transcript TBP. (B) Two stable clonal derivatives of the InR1G9 cell line expressing either an empty expression vector (control) or a dominant-negative variant of Pax6 (DN-Pax6) were transiently transfected with Nkx6.1. Expression of Nkx6.1 in non-transfected (−) and transfected (+) cells was evaluated by semi-quantitative PCR. Relative abundance levels of endogenous glucagon mRNA were then evaluated by quantitative PCR and normalized to the housekeeping transcript TBP. ** indicates statistical significance with P<0.02.
DISCUSSION
In contrast to insulin, the α-cell-restricted glucagon gene expression pattern has eluded our understanding as no α-cell-specific transcription factors that are clearly involved in glucagon gene expression have been identified in mature islets. In view of the mutually exclusive expression pattern of both hormones we have previously proposed that, during development, the failure to activate β-cell-specific transcription factors promotes the α-cell phenotype and thus glucagon gene expression. Suggestions of this default pathway have been provided by demonstrating that Pdx1 as well as Pax4 repressed glucagon gene expression, a phenomenon mediated by suppression of Pax6-mediated activation [25,26]. The present study, combined with the work of Schisler et al. [13], further substantiates this model by showing that another β-cell-specific transcription factor, Nkx6.1, also inhibits glucagon gene expression when expressed in α-cells.
We found that the inhibitory effect of Nkx6.1 on glucagon gene expression is predominantly mediated by impairing Pax6-driven activation through the α-cell-specific G1 element of the glucagon gene promoter, whereas the Pax6–G3 interaction does not appear to be a target of repression. Intriguingly, nuclear extracts derived from Nkx6.1-transfected BHK-21 cells revealed no clear identifiable complex corresponding to Nkx6.1 binding to the G1 element unless antiserum was added to the reaction mixture. Similar results have been observed for the insulin and Nkx6.1 gene promoters whereas ChIP assays revealed in vivo binding of the transcription factor to both 5′-flanking regions as well as to the glucagon gene promoter ([13,34] and the present study). These results suggest that interaction with other proteins in the nucleus may be mandatory for Nkx6.1 to bind its consensus sequence. Consistent with the latter, the N-terminal end of Nkx6.1 is composed of a stretch of proline and alanine residues as well as a NK decapeptide that interacts with Groucho/TLE (transducin-like enhancer) producing a potent repressor complex [31,36]. Deletion of this domain from Nkx6.1 revealed the capacity of the HD and C-terminal end to interact with the G1 element. Groucho/TLE may be required to induce a conformational change in the Nkx6.1 protein allowing the complex to interact with its target site. Interestingly, the repressor activity of Nkx6.1 through Groucho/TLE interaction prevails when the conserved consensus ATTA sequence is detected within the binding site. In contrast, when a non-canonical sequence such ATTT is detected near a G/C rich region, Nkx6.1, through its C-terminal end, becomes a transcriptional activator of target genes [37]. Both ATTA and ATTT sequences are found within the G1 element of the glucagon gene promoter whereas no GC-rich regions are detected in the vicinity of the element. Consistent with the latter, no induction of glucagon gene promoter reporter constructs by Nkx6.1 was observed in the heterelogous BHK-21 cell line. Further analysis by mutagenesis of the three A/T-rich regions of G1 revealed that Nkx6.1 preferentially interacted with the core ATTA sequence also recognized by Pax6. The latter finding corroborates our transient transfection data, showing an inhibitory effect of Nkx6.1 on Pax6-mediated glucagon gene activation in both BHK-21 and InR1G9 cells. Thus our results clearly indicate that Nkx6.1 represses glucagon gene expression by directly competing with Pax6 binding at G1. In addition, Nkx6.1, through a weak protein–protein interaction, may also destabilize the Pax6 interaction with G1.
Interestingly, the inhibitory effect of Nkx6.1 was specific to Pax6, as Nkx6.1 did not suppress either Cdx2/3 or c-Maf-stimulated glucagon gene transcription [14–16]. However, the synergistic induction of the glucagon gene promoter by Pax6 and either Cdx2/3 or c-Maf was greatly hampered but not completely abolished, substantiating a selective repression of Pax6 by Nkx6.1. This discriminatory effect was confirmed using a ChIP assay demonstrating decreased occupancy of Pax6 to the glucagon gene promoter when Nkx6.1 was expressed in InR1G9 cells. Interestingly, previous studies have shown that enhancement of glucagon gene transcription by both Cdx2/3 and c-Maf correlated with the ability of these proteins to interact with Pax6 directly and did not necessarily require DNA binding [14,16]. A similar discriminatory inhibition of the Pax6–Cdx2/3 synergistic activation of the glucagon gene was observed with Foxa2 and Pax4 [18,26]. Thus although Cdx2/3 and c-Maf are most likely important in sustaining glucagon gene expression, Pax6 appears to be the key transcription factor conveying regulatory functions imposed by endogenous α-cell activators such as Cdx2/3 and c-Maf, or by suppressors such as Foxa2, on glucagon gene expression. Furthermore, during development, absence and/or silencing of Pax4, Pdx1 and Nkx6.1 expression may permit Pax6-mediated activation of the glucagon gene and thus establishment of the α-cell phenotype. Consistent with the latter, we have previously demonstrated that overexpression of Pdx1 in the INS-1-derived INSrαβ subclone (expressing both insulin and glucagon) eliminated glucagon mRNA as well as protein, and promoted the expression of β-cell-specific genes. In contrast, loss of Pdx1 function in the same cell line suppressed genes restricted to β-cells and prompted α-cell differentiation [24].
Sander et al. [8] have previously demonstrated that ablation of Nkx6.1 in mice resulted in the loss of approximately 94% of β-cells without a concomitant increase in the number of α-cells. The absence of Nkx6.1 may thus not lead to changes in α-cell proliferation but only in glucagon gene transcription. Furthermore, Nkx6.1 is not the sole factor implicated in this process. The latter raises the question as to which factor during development may act as a master switch for cell lineage determination. Recently, the HD-containing transcription factor Arx was shown to be essential for α-cell fate acquisition, a process that involved exclusion of Pax4 expression in precursor cells [10]. Thus it is tempting to speculate that inhibition of Pax4, and potentially Pdx1 and Nkx6.1, by Arx may alleviate their repressive effect on Pax6 which may then stimulate glucagon gene transcription, a pre-requisite for the establishment and maintenance of the α-cell phenotype.
Acknowledgments
This work was supported by the Swiss National Science Foundation (grant no. 32-66907.01 to J. P. and 3100A-107682/1 to B. R. G.) and the Institute for Human Genetics and Biochemistry. We acknowledge the excellent technical skills of Natacha Klages-Jemelin.
References
- 1.Habener J. F., Kemp D. M., Thomas M. K. Minireview: transcriptional regulation in pancreatic development. Endocrinology. 2005;146:1025–1034. doi: 10.1210/en.2004-1576. [DOI] [PubMed] [Google Scholar]
- 2.Johnson J., Carisson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 3.Li H., Arber S., Jessel T. M., Edlund H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat. Genet. 1999;23:67–70. doi: 10.1038/12669. [DOI] [PubMed] [Google Scholar]
- 4.Harrison K. A., Thaler J., Pfaff S. L., Gu H., Kehrl J. H. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat. Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- 5.Gradwohl G., Dierich A., LeMeur M., Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwitzgebel V. M., Scheel D. W., Conners J. R., Kalamaras J., Lee J. E., Anderson D. J., Sussel L., Johnson J. D., German M. S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- 7.Sosa-Pineda B., Chowdhury K., Torres M., Oliver G., Gruss P. The Pax4 gene is essential for differentiation of insulin-producing β-cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 8.Sander M., Sussel L., Conners J., Scheel D., Kalamaras J., Cruz F. D., Schwitzgebel V., Hayes-Jordan A., German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of β-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 9.St-Onge L., Sosa-Pineda B., Chowdhury K., Mansouri A., Gruss P. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 10.Collombat P., Mansouri A., Hecksher-Sorensen J., Serup P., Krull J., Gradwohl G., Gruss P. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aramata S., Han S. I., Yasuda K., Kataoka K. Synergistic activation of the insulin gene promoter by the β-cell enriched transcription factors MafA, Beta2, and Pdx1. Biochim. Biophys. Acta. 2005;1730:41–46. doi: 10.1016/j.bbaexp.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Brun T., Franklin I., St-Onge L., Biason-Lauber A., Schoenle E., Wollheim C. B., Gauthier B. R. The diabetes-linked transcription factor Pax4 promotes β-cell proliferation and survival in rat and human islets. J. Cell Biol. 2004;167:1123–1135. doi: 10.1083/jcb.200405148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schisler J. C., Jensen P. B., Taylor D. G., Becker T. C., Knop F. K., Takekawa S., German M., Weir G. C., Lu D., Mirmira R. G., Newgard C. B. The Nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet β-cells. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7297–7302. doi: 10.1073/pnas.0502168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritz-Laser B., Estreicher A., Klages N., Saule S., Philippe J. Pax-6 and Cdx-2/3 interact to activate glucagon gene expression on the G1 control element. J. Biol. Chem. 1999;274:4124–4132. doi: 10.1074/jbc.274.7.4124. [DOI] [PubMed] [Google Scholar]
- 15.Laser B., Meda P., Constant I., Philippe J. The caudal-related homeodomain protein Cdx-2/3 regulates glucagon gene expression in islet cells. J. Biol. Chem. 1996;271:28984–28994. doi: 10.1074/jbc.271.46.28984. [DOI] [PubMed] [Google Scholar]
- 16.Planque N., Leconte L., Coquelle F. M., Benkhelifa S., Martin P., Felder-Schmittbuhl M. P., Saule S. Interaction of Maf transcription factors with Pax-6 results in synergistic activation of the glucagon promoter. J. Biol. Chem. 2001;276:35751–35760. doi: 10.1074/jbc.M104523200. [DOI] [PubMed] [Google Scholar]
- 17.Morel C., Cordier-Bussat M., Philippe J. The upstream promoter element of the glucagon gene, G1, confers pancreatic α-cell-specific expression. J. Biol. Chem. 1995;270:3046–3055. doi: 10.1074/jbc.270.7.3046. [DOI] [PubMed] [Google Scholar]
- 18.Gauthier B. R., Schwitzgebel V. M., Zaiko M., Mamin A., Ritz-Laser B., Philippe J. Hepatic nuclear factor-3 (HNF-3 or Foxa2) regulates glucagon gene transcription by binding to the G1 and G2 promoter elements. Mol. Endocrinol. 2002;16:170–183. doi: 10.1210/mend.16.1.0752. [DOI] [PubMed] [Google Scholar]
- 19.Ahlgren U., Plaff S. L., Jessell T. M., Edlund T., Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 20.Dumonteil E., Laser B., Constant I., Philippe J. Differential regulation of the glucagon and insulin I gene promoters by the basic helix-loop-helix transcription factors E47 and BETA2. J. Biol. Chem. 1998;273:19945–19954. doi: 10.1074/jbc.273.32.19945. [DOI] [PubMed] [Google Scholar]
- 21.Herzig S., Fuzesi L., Knepel W. Heterodimeric pbx-prep1 homeodomain protein binding to the glucagon gene restricting transcription in a cell type-dependent manner. J. Biol. Chem. 2000;275:27989–27999. doi: 10.1074/jbc.M003345200. [DOI] [PubMed] [Google Scholar]
- 22.Offield M. F., Jetton T. L., Labosky P. A., Ray M., Stein R. W., Magnuson M. A., Hogan B. L., Wright C. V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 23.Lottmann H., Vanselow J., Hessabi B., Walther R. The Tet-On system in transgenic mice: inhibition of the mouse pdx-1 gene activity by antisense RNA expression in pancreatic β-cells. J. Mol. Med. 2001;79:321–328. doi: 10.1007/s001090100229. [DOI] [PubMed] [Google Scholar]
- 24.Wang H., Maechler P., Ritz-Laser B., Hagenfeldt K. A., Ishihara H., Philippe J., Wollheim C. B. Pdx1 level defines pancreatic gene expression pattern and cell lineage differentiation. J. Biol. Chem. 2001;276:25279–25286. doi: 10.1074/jbc.M101233200. [DOI] [PubMed] [Google Scholar]
- 25.Ritz-Laser B., Gauthier B. R., Estreicher A., Mamin A., Brun T., Ris F., Salmon P., Halban P. A., Trono D., Philippe J. Ectopic expression of the β-cell specific transcription factor Pdx1 inhibits glucagon gene transcription. Diabetologia. 2003;46:810–821. doi: 10.1007/s00125-003-1115-7. [DOI] [PubMed] [Google Scholar]
- 26.Ritz-Laser B., Estreicher A., Gauthier B., Mamin A., Edlund H., Philippe J. The pancreatic β-cell-specific transcription factor Pax-4 inhibits glucagon gene expression through Pax-6. Diabetologia. 2002;45:97–107. doi: 10.1007/s125-002-8249-9. [DOI] [PubMed] [Google Scholar]
- 27.Takaki R., Ono J., Nakamura M., Yokogawa Y., Kumae S., Hiraoka T., Yamaguchi K., Hamaguchi K., Uchida S. Isolation of glucagon-secreting cell lines by cloning insulinoma cells. In Vitro Cell. Dev. Biol. 1986;22:120–126. doi: 10.1007/BF02623498. [DOI] [PubMed] [Google Scholar]
- 28.Graham F., Van der Eb A. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 29.Drucker D., Philippe J., Jepeal L., Habener J. Glucagon gene 5′-flanking sequences promote islet cell-specific gene transcription. J. Biol. Chem. 1987;15:15659–15665. [PubMed] [Google Scholar]
- 30.Schreiber E., Matthias P., Muller M., Schaffner W. Identification of a novel lymphoid specific octamer binding protein (OTF-2B) by proteolytic clipping bandshift assay (PCBA) EMBO J. 1988;7:4221–4229. doi: 10.1002/j.1460-2075.1988.tb03319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirmira R. G., Watada H., German M. S. β-cell differentiation factor Nkx6.1 contains distinct DNA binding interference and transcriptional repression domains. J. Biol. Chem. 2000;275:14743–14751. doi: 10.1074/jbc.275.19.14743. [DOI] [PubMed] [Google Scholar]
- 31a.Orlando V., Strutt H., Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 32.Duong D. T., Waltner-Law M. E., Sears R., Sealy L., Granner D. K. Insulin inhibits hepatocellular glucose production by utilizing liver-enriched transcriptional inhibitory protein to disrupt the association of CREB-binding protein and RNA polymerase II with the phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 2002;277:32234–32242. doi: 10.1074/jbc.M204873200. [DOI] [PubMed] [Google Scholar]
- 33.Ritz-Laser B., Mamin A., Brun T., Avril I., Schwitzgebel V. M., Philippe J. The zinc finger-containing transcription factor Gata-4 is expressed in the developing endocrine pancreas and activates glucagon gene expression. Mol. Endocrinol. 2005;19:759–770. doi: 10.1210/me.2004-0051. [DOI] [PubMed] [Google Scholar]
- 34.Iype T., Taylor D. G., Ziesmann S. M., Garmey J. C., Watada H., Mirmira R. G. The transcriptional repressor Nkx6.1 also functions as a deoxyribonucleic acid context-dependent transcriptional activator during pancreatic β-cell differentiation: evidence for feedback activation of the nkx6.1 gene by Nkx6.1. Mol. Endocrinol. 2004;18:1363–1375. doi: 10.1210/me.2004-0006. [DOI] [PubMed] [Google Scholar]
- 35.Philippe J., Morel C., Prezioso V. Glucagon gene expression is negatively regulated by hepatocyte nuclear factor 3β. Mol. Cell. Biol. 1994;14:3514–3523. doi: 10.1128/mcb.14.5.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muhr J., Andersson E., Persson M., Jessell T. M., Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- 37.Taylor D. G., Babu D., Mirmira R. G. The C-terminal domain of the β-cell homeodomain factor Nkx6.1 enhances sequence-selective DNA binding at the insulin promoter. Biochemistry. 2005;44:11269–11278. doi: 10.1021/bi050821m. [DOI] [PubMed] [Google Scholar]