Abstract
Wheat (Triticum aestivum) contains a previously unknown type of xylanase (EC 3.2.1.8) inhibitor, which is described in the present paper for the first time. Based on its >60% similarity to TLPs (thaumatin-like proteins) and the fact that it contains the Prosite PS00316 thaumatin family signature, it is referred to as TLXI (thaumatin-like xylanase inhibitor). TLXI is a basic (pI≥9.3 in isoelectric focusing) protein with a molecular mass of approx. 18–kDa (determined by SDS/PAGE) and it occurs in wheat with varying extents of glycosylation. The TLXI gene sequence encodes a 26-amino-acid signal sequence followed by a 151-amino-acid mature protein with a calculated molecular mass of 15.6–kDa and pI of 8.38. The mature TLXI protein was expressed successfully in Pichia pastoris, resulting in a 21–kDa (determined by SDS/PAGE) recombinant protein (rTLXI). Polyclonal antibodies raised against TLXI purified from wheat react with epitopes of rTLXI as well as with those of thaumatin, demonstrating high structural similarity between these three proteins. TLXI has a unique inhibition specificity. It is a non-competitive inhibitor of a number of glycoside hydrolase family 11 xylanases, but it is inactive towards glycoside hydrolase family 10 xylanases. Progress curves show that TLXI is a slow tight-binding inhibitor, with a Ki of approx. 60–nM. Except for zeamatin, an α-amylase/trypsin inhibitor from maize (Zea mays), no other enzyme inhibitor is currently known among the TLPs. TLXI thus represents a novel type of inhibitor within this group of proteins.
Keywords: heterologous expression, thaumatin-like protein (TLP), thaumatin-like xylanase inhibitor (TLXI), slow tight binding, wheat (Triticum aestivum), xylanase inhibitor
Abbreviations: CEC, cation-exchange chromatography; EST, expressed sequence tag; GH, glycoside hydrolase family; [I]/[E]50, ratio of inhibitor concentration to enzyme concentration necessary to obtain 50% inhibition; 4-MUX2, 4-methylumbelliferyl-β-D-xylobioside; PNGase F, peptide N-glycosidase F; RACE, rapid amplification of cDNA ends; SPR, surface plasmon resonance; TAXI, Triticum aestivum xylanase inhibitor; TFMSA, trifluoromethanesulfonic acid; TLP, thaumatin-like protein; (r)TLXI, (recombinant) thaumatin-like xylanase inhibitor; XIP, xylanase inhibitor protein
INTRODUCTION
The plant cell is protected from its surrounding environment by the cell wall, which forms a structurally heterogeneous barrier. In the case of plant attack, pathogenic micro-organisms produce a diverse array of enzymes which depolymerize the different polysaccharides in the cell walls [1]. One type of such enzymes are xylanases (also referred to as endo-β-1,4-xylanases or endoxylanases, EC 3.2.1.8). They depolymerize xylan, which, next to cellulose, is one of the most abundant polysaccharides in the cell wall of higher plants. It consists of a main chain of β-1,4-xylopyranosyl residues that, depending on the origin, may be replaced with, e.g., glucuronyl, acetyl or arabinofuranosyl groups to form heteroxylans. Xylanases hydrolyse the β-1,4-xylosidic linkages in the xylan main chain [2]. The majority of the xylanases belong either to glycoside hydrolase family 10 (GH10) or to the structurally unrelated glycoside hydrolase family 11 (GH11) (http://afmb.cnrs-mrs.fr/CAZY/ [3]). In both families, a pair of glutamate residues catalyses the cleavage of the glycosidic bond, one acting as a nucleophile and the other as the acid–base catalyst. Recently, a xylanase was shown to be indispensable in the infection of plants by the pathogen Botrytis cinerea [4]. Xylanases are produced not only by micro-organisms, but also by plants. The latter belong to GH10 and play important physiological roles in several tissues, such as contribution to seed germination and fruit ripening [5].
At the same time, some plants produce proteins which can inhibit xylanases. Over the last decade, studies have revealed the presence of two types of proteinaceous xylanase inhibitors in cereals, i.e. the TAXI (Triticum aestivum xylanase inhibitor)-type [6,7] and the XIP (xylanase inhibitor protein)-type inhibitors [8,9]. These proteins have been purified and characterized biochemically, genetically and structurally.
TAXI-type proteins occur in common wheat (Triticum aestivum), durum wheat (Triticum durum), barley (Hordeum vulgare) and rye (Secale cereale) [10,11] as monomeric (40–kDa) as well as heterodimeric (30+10–kDa) basic (pI≥8.0) proteins which specifically inhibit GH11 xylanases [12]. Crystallographic analysis of a complex between a GH11 xylanase of Aspergillus niger and TAXI-I showed His374 of TAXI-I to be a key residue in xylanase inhibition. This histidine residue interacts in the active site with the two active glutamate residues of the xylanase, clearly indicating a competitive type of inhibition [13]. XIP-type proteins have been isolated from the above-cited cereals as well as from maize (Zea mays) and rice (Oryza sativa) [11,14,15]. These monomeric proteins (30–kDa, pI≥6.7) inhibit GH10 and GH11 xylanases, provided that they are from fungal origin [15]. The crystal structures of XIP-I in complex with GH10 Aspergillus nidulans xylanase on the one hand, and with GH11 Penicillium funiculosum xylanase on the other hand, reveal that XIP-I possesses an independent enzyme-binding site for each family of xylanases. Like TAXI, XIP is a competitive inhibitor, interacting in the active site of the xylanases [16]. For both families, the inhibition mechanism is based on substrate mimicry.
A regulatory role of TAXI and XIP in plant development is disaffirmed by their lack of effectiveness against endogenous xylanases, their distinct specificity towards xylanases of microbial origin, the ability of TAXI to inhibit two GH11 xylanases of the cereal pathogen Fusarium graminearum [17] and the fact that both TAXI and XIP genes are induced by pathogens and wounding [18].
The present study reports on the existence of a third, structurally unrelated, type of xylanase inhibitor in wheat which belongs to the thaumatin family. It is further referred to as TLXI (thaumatin-like xylanase inhibitor). More particularly, the purification of this protein from wheat, the identification, cloning and heterologous expression of its corresponding gene is described. Additionally, the biochemical characteristics and the kinetic parameters of inhibition of both native and recombinant TLXI are discussed.
EXPERIMENTAL
Materials
Wheat (cultivar Soissons) (from Aveve) wholemeal was prepared using a Cyclotec 1093 sample mill. All electrophoresis and chromatography media, and molecular mass and pI markers were from GE Healthcare, unless specified otherwise. The producers of the kits and enzymes used in cloning and heterologous expression of TLXI are mentioned below. Bacillus subtilis GH11 xylanase and an Aspergillus aculeatus GH10 xylanase were supplied by Puratos (by Ir Filip Arnaut). Two GH11 xylanases from Trichoderma longibrachiatum (also known as Trichoderma reesei), i.e. Xyn I and Xyn II, GH11 xylanases from A. niger and Trichoderma viride, and Xylazyme-AX tablets were from Megazyme. GH10 xylanases from Aspergillus oryzae and Penicillium purpurogenum were kindly made available by VTT Biotechnology (from Professor Maija Tenkanen, now at Department of Applied Chemistry and Microbiology, University of Helsinki, Helsinki, Finland) and the Laboratorio de Bioquimíca (Professor Jaime Eyzaguirre, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, Santiago, Chile) respectively. Dr Nathalie Juge (Institute of Food Research, Norwich, U.K.) kindly provided a Pseudomonas fluorescens GH10 and a Penicillium funiculosum GH11 xylanase. Thermophilic Thermobacillus xylanilyticus GH10 and GH11 xylanases were made available by Dr Michael O’Donohue (INRA, Reims, France). A. niger GH10 xylanase was purified from an A. niger CBS 110.42 culture filtrate [19]. Grindamyl H 640 bakery enzyme, containing the wild-type B. subtilis GH11 xylanase, and Biobake 710, containing the above-cited A. niger GH11 xylanase, were from Danisco and Quest International respectively. The Pro-Q Emerald 300 Glycoprotein Stain kit was from Invitrogen. Thaumatin, oat spelt xylan, horseradish-peroxidase-conjugated goat anti-rabbit antibodies, substrate (3,3′,5,5′-tetramethylbenzidine) for the horseradish peroxidase, PNGase F (peptide N-glycosidase F) and all other chemicals were from Sigma–Aldrich. Water-soluble oat spelt xylan was prepared as described by He et al. [20]. 4-MUX2 (4-methylumbelliferyl-β-D-xylobioside) was kindly provided by Dr Wim Nerinckx (Laboratory of Glycobiology, University of Ghent, Ghent, Belgium).
Affinity matrix preparation
B. subtilis and A. niger GH11 xylanases were purified from Grindamyl H 640 and Biobake 710 enzyme preparations respectively, and N-hydroxysuccinimide-activated Sepharose 4 Fast Flow matrix was used for the preparation of affinity matrices with these two enzymes (7 and 30–ml respectively) according to Gebruers et al. [21].
Purification of xylanase inhibitors from wheat
Wheat wholemeal (1–kg) was suspended in 5–litres of aqueous 0.1% (w/v) L-ascorbic acid solution, extracted overnight at 7 °C and centrifuged at 10000–g for 30–min at 7 °C. L-Ascorbic acid reduced the oxidation of phenolic compounds during the extraction. Calcium chloride (2–g/l) was added to the supernatant, and the pH was raised to 8.5 by adding 2–M NaOH. The extract was left overnight at 7 °C, and the resulting precipitate (containing pectins) was removed by centrifugation at 10000–g for 30–min at 7 °C. The pH of the supernatant was adjusted to 4.5 by adding 2–M HCl. Proteins with xylanase-inhibition activity in the supernatant were retained by CEC (cation-exchange chromatography) on a SP Sepharose Big Beads column (180–mm×130–mm, equilibrated with 25–mM sodium acetate buffer, pH–4.5). The bound protein fraction was eluted in one step with 1–litre of 1–M NaCl solution, dialysed against deionized water for 48–h at 7 °C and freeze-dried, resulting in the CEC fraction (approx. 7.76–g of protein). Portions (6–g) of the CEC fraction were extracted with 25–mM sodium acetate buffer, pH–5.0, containing 100–ml of 0.2–M NaCl, centrifuged at 10000–g for 30–min at 7 °C and filtered through a paper filter. The filtrate was loaded on the B. subtilis xylanaseaffinity column (10–mm×70–mm, equilibrated with 25–mM sodium acetate buffer, pH–5.0, containing 0.2–M NaCl) at a flow rate of 0.33–ml/min. TAXI was eluted from the column with 5–ml of 250–mM Tris solution, pH–12.0, at a flow rate of 1–ml/min, and immediately neutralized with 1–M ethanoic (acetic) acid solution. The resulting run-through was used to isolate XIP and TLXI. Portions (30–ml) of the run-through were applied on the A. niger xylanase-affinity column (16–mm×150–mm, equilibrated with 25–mM sodium acetate buffer, pH–5.0, containing 0.2–M NaCl) at a flow rate of 1–ml/min. Bound proteins were then eluted from the column with buffers of increasing pH (50–ml of 250–mM Tris, pH–10.0–12.0), at a flow rate of 3–ml/min. XIP was removed from the column in the elution step with pH–10.0. TLXI eluted when the pH was raised to 12.0. The flow scheme is shown in Supplementary Figure S1 (http://www.BiochemJ.org/bj/403/bj4030583add.htm).
Protein sequencing
To determine the N-terminal amino acid sequence, inhibitor proteins (approx. 50–μg) were separated by SDS/PAGE in a 12% polyacrylamide gel using the Hoeffer Mighty Small unit, electroblotted on to a PVDF membrane with the Trans-Blot Semi-Dry Electrophoretic Transfer Cell (Bio-Rad) (electric potential difference of 10–V) for 1–h at room temperature (20 °C) and subjected to Edman degradation. Sequence analysis was performed on a Procise cLC 491 Sequencer (Applied Biosystems).
For internal amino acid sequence determination, TLXI (500–μg) was dissolved in 0.3–ml of 70% methanoic (formic) acid containing a small amount of CNBr (two crystals; the exact amount was not critical). Protein cleavage was performed by incubating the solution in the dark for 27–h at room temperature, after which all volatiles were evaporated under a nitrogen gas stream. The resulting peptides were dissolved in 1–ml of 25–mM Tris/HCl buffer, pH–8.0, containing 1% (v/v) 2-mercaptoethanol and kept in boiling water for 5–min. The peptides were separated using reverse-phase HPLC on a Microsorb 300–Å (1–Å=0.1–nm) pore-size C8 reverse-phase column (Varian). Solvent A was MilliQ water with 0.1% trifluoroacetic acid, and solvent B was acetonitrile with 0.1% trifluoroacetic acid. Separation was performed by applying a gradient from 2 to 100% solvent B in 66–min at a flow rate of 1–ml/min. The isolated polypeptides were subjected to Edman degradation and analysed as described above.
Gene isolation and characterization
Genomic DNA was isolated from young leaves of wheat cultivar Estica using the DNeasy Plant Mini kit (Qiagen). Total RNA was extracted from young embryos (3–weeks post-anthesis) of the same cultivar by means of the Invisorb Spin Plant-RNA Mini kit (Invitek). mRNA was purified from the total RNA with the Oligotex mRNA Mini kit (Qiagen). Primers were designed based on EST (expressed sequence tag) sequences BE399034 and BE427320 corresponding to the N-terminal sequence of native TLXI. RACE (rapid amplification of cDNA ends) reactions were performed using the GeneRacer kit (Invitrogen) according to the manufacturer's instructions with HotStarTaq polymerase as enzyme. 3′-RACE and 3′-nested RACE were performed with I3race (5′-GTGCCAGACCGGCGACTG-3′) and I3nested (5′-GTGGCAGCTCGCTGACTTG-3′) as gene-specific primers respectively, whereas, for 5′-RACE and 5′-nested RACE, I5race (5′-TTGGTGGAGCACGAGCGCCAC-3′) and I5nested (5′-CCGGCCACACCGTGAAGTG-3′) respectively were used for this purpose. For PCR on genomic DNA, Iintf (5′-CAAGCGCGGCACCGCTCACCATC-3′) was used in combination with XI2 (5′-AATACCTGACACACGTGTACGG-3′). PCR products were purified using the PCRapid kit (Invitek) and subsequently sequenced on a 377 DNA Sequencer using ABI PRISM Big Dye Terminator chemistry (Applied Biosystems). Assembly of the obtained sequences (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/403/bj4030583add.htm) resulted in a contiguous sequence.
Cloning, mutagenesis and protein expression
Construction of expression plasmids
The DNA sequence encoding mature TLXI was amplified with primer combination Ximatf (5′-CACAGATCTGCACCGCTCACCATCACGAAC-3′) and Ximatrstop (5′-CACAGATCTTCATGGGCAGAAGACGATCTG-3′) using Pfu DNA polymerase (Stratagene) and PCR product Iintf/XI2 as template. The resulting PCR product was cloned in a pCR®4-TOPO® vector (Invitrogen), verified by DNA sequencing, and subsequently subcloned as a BglII fragment in the BsmBI site of expression vector pPICZαC (Invitrogen). The ligation mixture was used to transform Escherichia coli TOP10F’ cells [F’ {proAB, lacIq, lacZΔM15, Tn10 (TetR)} mcrA, Δ(mrrhsdRMS-mcrBC), 80lacZΔM15, ΔlacX74, deoR, recA1, λ-araD139, Δ(ara-leu)7697, galU, galK, rpsL(StrR), endA1, nupG].
Expression and purification of rTLXI (recombinant TLXI)
A sequence-verified pPICZαC-tlxi construct, linearized with PmeI, was used to transform competent Pichia pastoris KM71H cells (arg4 aox1::ARG4) using the EasySelect Pichia Expression kit (Invitrogen). For large-scale expression, a single colony was grown in 10–ml of buffered minimal glycerol-complex medium (pH–6.0), supplemented with 0.35–M NaCl, for 24–h at 30 °C, with shaking at 250–rev./min. The volume was increased to 500–ml in a 2–litre flask and incubated overnight under the same conditions. A pre-induction transition phase was included in which 10% glycerol/13.4% yeast nitrogen base was added to the primary culture at a ratio of 1:10 (v/v). When the cells reached an attenuance at 600–nm (D600) of 0.20–0.24 in a 1:50 dilution, the cell culture was harvested by centrifugation at 2500–g for 10–min. To induce protein expression, the cells were suspended in 100–ml of buffered minimal methanol-complex medium (pH–6.0) and cultured in baffled flasks for another 50–h at 20 °C in the presence of 1.25% (v/v) methanol. To harvest culture supernatants, yeast cells were removed by centrifugation at 2500–g for 10–min. Before purification, the medium was dialysed overnight against 25–mM sodium acetate buffer, pH–5.0. rTLXI was purified by CEC using a SP-Sepharose Fast Flow column (10–mm×200–mm) equilibrated with the same buffer. Bound proteins were eluted using a linear salt gradient of 0–1–M NaCl in 100–min (flow rate of 1–ml/min). Inhibition activity and purity of the resulting fractions were assessed using the Xylazyme-AX method (see below) and SDS/PAGE respectively.
Protein content determination
Protein concentrations were determined by the Bradford Coomassie Brilliant Blue method with BSA as standard [22]. For pure TLXI samples, protein concentrations were determined spectrophotometrically at 280–nm using a specific absorbance value of 1.457 absorbance units for 1–mg/ml TLXI (1–cm pathlength UV cell).
Xylanase inhibition assay (Xylazyme-AX method)
Inhibition activities were determined with the colorimetric Xylazyme-AX method [23]. All xylanase solutions were prepared in 25–mM sodium acetate buffer, pH–5.0, with 0.5–mg/ml BSA, and contained 2–units of xylanase per 1–ml. Xylanase units were defined as described by Gebruers et al. [23], and, under the conditions of the assay, the xylanase concentration corresponding to 1–unit was approximately 5.1–nM for the A. niger, 8.9–nM for the T. longibrachiatum (Xyn I), 2.1–nM for the T. longibrachiatum (Xyn II), 11.6–nM for the T. viride, 5.8–nM for the B. subtilis, 0.1–nM for the Thermobacillus xylanilyticus and 2.1–nM for the Penicillium funiculosum GH11 xylanases. The corresponding concentrations for the GH10 xylanases were 53.7–nM for the A. aculeatus, 36.6–nM for the A. niger, 30.9–nM for the Ps. fluorescens, 14.2–nM for the Thermobacillus xylanilyticus and 2.9–nM for the A. oryzae enzyme. Different inhibitor concentrations up to 8000–nM and up to 3000–nM were used for TLXI and rTLXI respectively. All measurements were performed in triplicate.
Electrophoresis
SDS/PAGE under non-reducing and reducing conditions was performed on 20% polyacrylamide gels with a PhastSystem unit, as described in GE Healthcare Separation Technique File 110. 2-Mercaptoethanol (5%) was used as reducing agent. The low-molecular-mass markers were from GE Healthcare. The pI value of the inhibitor protein was determined by isoelectric focusing as described in GE Healthcare Separation Technique File 100, using the PhastSystem unit with polyacrylamide gels containing ampholytes (pH–3–9). Broad-range pI markers (3.5–9.3) were used. All gels were silver-stained as described in GE Healthcare Development Technique File 210.
Glycan detection
The proteins (0.1–mg) were separated by SDS/PAGE in a 12% polyacrylamide gel (see above). Glycan detection was performed in-gel, using the Pro-Q Emerald 300 Glycoprotein Stain kit, according to the instructions of the manufacturer. To gain insight into the extent of glycosylation, TLXI was analysed by electrospray-ionization MS on an Esquire-LC/MS system (Bruker).
Deglycosylation
Enzymic N-deglycosylation
The N-linked glycans of the purified TLXI and rTLXI were removed with PNGase F (Enzymic In-solution N-Deglycosylation kit) under reducing and denaturing conditions according to the instructions of the manufacturer (Sigma–Aldrich). The different deglycosylated and control samples were analysed by SDS/PAGE followed by silver staining.
Chemical deglycosylation
TLXI and rTLXI were treated with anhydrous TFMSA (trifluoromethanesulfonic acid), which effectively cleaves N- and O-linked glycans from glycoproteins, leaving the primary structure of the protein intact [24]. Anisole was used as a scavenger to neutralize reactive groups formed during the deglycosylation reaction. Excess TFMSA was neutralized by reaction with pyridine.
Briefly, the purified inhibitors (100–μg) were freeze-dried in glass tubes for 24–h to ensure complete dryness. A 10% anisole solution in TFMSA (150–μl) was cooled to 0 °C, and then added to the inhibitor samples. The reaction was performed at 0 °C. After 3–h, the reaction mixture was placed in a methanol/solid CO2 bath and neutralized with an equal volume of ice-cold aqueous pyridine (60%). The excess pyridine was removed by gel-filtration on a PD-10 column and, after freeze-drying, the different deglycosylated samples were analysed by SDS/PAGE followed by silver staining.
Western blot and immunoprobing
Rabbit polyclonal antibodies against native TLXI were prepared, and Western blot analysis was performed on samples of TLXI (0.50–μg), rTLXI (0.56–μg), wheat CEC fraction (18–μg) and commercial thaumatin (4–μg), as described by Beaugrand et al. [25].
Kinetic analysis
Kinetic parameters were derived from reaction progress curves of TLXI and rTLXI with T. longibrachiatum GH11 xylanase (Xyn I). 4-MUX2 was used as substrate. TLXI and 4-MUX2 solutions were mixed, and the reaction was started by adding the enzyme solution, yielding a final volume of 150–μl in McIlvaine buffer (0.2–M Na2HPO4/0.1–M citric acid), pH–5.0. The mixtures were incubated for various lengths of time up to 90–min at 40 °C. The reaction was stopped by adding 15–μl of 500–mM glycine/NaOH buffer, pH–13.0, and putting the samples on ice. The hydrolysis products formed were quantified on a fluorimeter (Horiba Jobin Yvon) based on a standard curve of 4-methylumbelliferone (0–6–μM). Excitation was at 360–nm and the emission was recorded at 446–nm for 0.1–s (five trials; S.E.M. <1%). Both monochromator slit widths were 1–nm. All measurements were performed in triplicate in 75–μl capillaries.
Substrate binding
TLXI (9–μM in 1–ml of 25–mM sodium acetate buffer, pH–5.0) was mixed with soluble or insoluble oat spelt xylan (50–mg). These mixtures were shaken for 30–min at room temperature. In the case of insoluble xylan, the supernatant was removed after centrifugation at 15000–g for 10–min, and the pellet was washed twice with 1–ml of 25–mM sodium acetate buffer, pH–5.0. The inhibition activity against the T. longibrachiatum xylanase (Xyn I) in the mixture (for soluble xylan) and in the supernatant (for insoluble xylan) (50–μl) was determined with the Xylazyme-AX method (see above) and compared with the inhibition activity of the original TLXI sample. For insoluble xylan, supernatant and pellet were also analysed by SDS/PAGE.
RESULTS AND DISCUSSION
Xylanase inhibitor purification
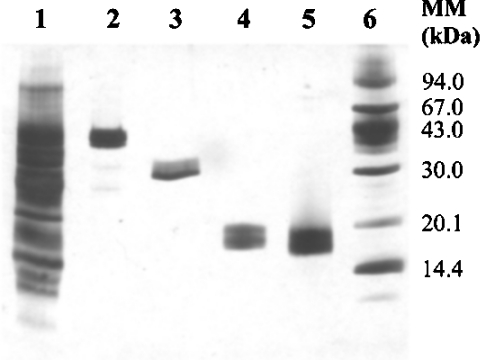
Starting from wholemeal, the two already known xylanase inhibitors, TAXI and XIP, and the novel xylanase inhibitor, TLXI, were purified using affinity chromatography with immobilized xylanases (Figure 1). The purification yields of TAXI and XIP were similar to those described earlier (14–15–mg/kg of wholemeal [9,26]). The yield of TLXI was approx. 2.5–mg/kg of wholemeal. The successful use of affinity chromatography with immobilized xylanases indicates that the complexation between TLXI and xylanase is reversible.
Figure 1. SDS/PAGE profiles of a wheat CEC fraction and the isolated xylanase inhibitors.
A CEC fraction (lane 1) was obtained by fractionation of wheat wholemeal extract by CEC. Pure TAXI (lane 2), XIP (lane 3) and TLXI (lanes 4 and 5) were obtained in this order from the CEC fraction by affinity chromatography on B. subtilis and A. niger GH11 xylanase-affinity columns. TLXI was analysed by SDS/PAGE under non-reducing (lane 4) and reducing conditions (lane 5). The size of the molecular mass (MM) markers (lane 6) is indicated on the right-hand side in kDa. The gel was silver-stained.
Molecular characterization
Primers, designed on Triticeae EST sequences, allowed us to amplify and subsequently sequence the complete tlxi gene coding sequence, including the 5′- and 3′-untranslated regions. At present, several wheat ESTs can be found in the database that show 99–100% identity with the tlxi sequence and correspond to proteins expressed in root (CK199869), grain (CD914550 and CD914871) and ovary (CD937830). In addition, EST sequences from barley (BQ760132) and durum wheat (AJ611730) are 65% and 47% identical (69% and 59% similar) respectively with the protein sequence of TLXI.
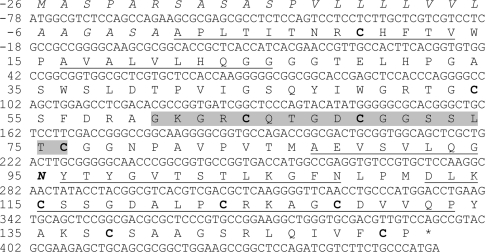
The tlxi gene contains no introns and encodes a mature protein of 151 amino acids preceded by a signal sequence of 26 amino acids (PSORT [27]). Approx. 50% of the mature protein sequence was confirmed by sequencing peptides of native TLXI obtained after CNBr cleavage. The sequence includes a potential glycosylation site at Asn95 as well as ten cysteine residues (Figure 2) that are probably involved in intramolecular disulfide bonds. The calculated molecular mass and pI of the mature protein were determined to be 15642.8 Da and 8.38 respectively.
Figure 2. Complete coding sequence of TLXI.
The first methionine residue of the signal sequence (italic) is at position −26. The mature protein starts at position 1, stops at position 151 and contains ten cysteine residues (bold) and one possible N-glycosylation site (italic bold) at position 95. Parts of the amino acid sequence were confirmed by sequencing peptides obtained after CNBr cleavage of native TLXI (underlined). The Prosite thaumatin family signature PS00316 is shaded. * indicates a stop codon.
BLASTp [28] results show that the xylanase inhibitor belongs to the thaumatin family (Pfam00314) and contains the Prosite PS00316 thaumatin family signature (Figure 2). Similarity up to 60% was found with several TLPs (thaumatin-like proteins) from various origins. TLPs are a heterogeneous family of proteins which exhibit sequence similarity with thaumatin, a sweet-tasting protein found in the arils of fruits of the African shrub Thaumatococcus daniellii [29]. Until now, thaumatin is the only protein of the family for which sweetness has been described. Members of this family have already been identified in wheat (pWIR2 [30]), barley (antifungal proteins R and S [31], TLPs 1–8 [32,33]), rye {AFP (antifreeze protein) [34]}, oats (PR-5 [35]), maize (zeamatin [36]), tobacco (osmotin [37]), many kinds of fruit (cherry [38], apple [39]) and other plants.
TLXI belongs to a group of smaller TLPs, which are found mainly in cereals. The proteins in this group all contain ten cysteine residues, which are involved in five intramolecular disulfide bridges and form the basis for high stability under extreme thermal and pH conditions [33]. Only one TLP, i.e. zeamatin, showing 43% identity with TLXI, is known to inhibit enzymes, namely α-amylases and trypsin [36]. Most of the TLPs have been shown to possess antifungal activity and therefore have been assigned to class 5 of the pathogenesis-related proteins. Their activity is thought to rely on an interaction of the acidic cleft of TLPs with 1,3-β-D-glucans of fungal membranes [40], causing membrane permeabilization. Based on its homology with TLP, it can be assumed that TLXI also plays a role in plant defence.
The tlxi gene was recombinantly expressed in P. pastoris, resulting in a 21–kDa protein with xylanase-inhibiting activity against T. longibrachiatum xylanase (XynI) (Supplementary Figure S3 at http://www.BiochemJ.org/bj/403/bj4030583add.htm). Subsequent purification yielded 4–mg of pure rTLXI per litre of culture medium.
Biochemical characterization
Native TLXI appears in SDS/PAGE under reducing as well as non-reducing conditions as a broad band at approx. 18–kDa (Figure 1). rTLXI appears in SDS/PAGE as one 21–kDa protein band. Based on isoelectric focusing, TLXI and rTLXI have a pI of 9.3 or higher (results not shown).
The reaction of the antibodies, raised against native TLXI from wheat, with the commercial thaumatin sample on a Western blot verified that TLXI is a member of the thaumatin family (Supplementary Figure S4 at http://www.BiochemJ.org/bj/403/bj4030583add.htm). On this Western blot, it also became clear that the broad band for native TLXI, seen on SDS/PAGE, consists of three to four finer protein bands, suggesting the presence of different forms with varying extents of glycosylation. The Pro-Q Emerald 300 Glycoprotein Stain kit indeed showed considerable glycosylation, not only for TLXI but also for rTLXI (results not shown). For native TLXI, this was confirmed by MS. Next to a peak at 15632.5–Da (molecular mass similar to that calculated from the amino acid sequence), peaks corresponding to TLXI proteins with different degrees of glycosylation were observed. The different molecular masses and their corresponding glycosyl moieties are listed in Table 1. The peak with the highest mass corresponded to 16638–Da, a shift in molecular mass which could be accounted for by five sugar residues: two N-acetylhexosamines (e.g. N-acetylglucosamine or N-acetylgalactosamine), one 6-deoxyhexose (e.g. fucose or rhamnose), one hexose (e.g. galactose, mannose or glucose) and one sialic acid, as determined with GlycoMod [41].
Table 1. MS data and corresponding glycosyl moieties.
The Table shows the different masses obtained with MS with their corresponding glycosyl entities as determined with GlycoMod [41]. HexNAc, N-acetylhexosamine (e.g. N-acetylglucosamine or N-acetylgalactosamine); deoxyhexose, e.g. fucose or rhamnose; hexose, e.g. galactose, mannose or glucose; NeuAc, N-acetylneuraminic acid (sialic acid).
| Molecular mass | Glycosyl moiety |
|---|---|
| 15632.5 | - |
| 15833.9 | HexNAc |
| 16040.5 | HexNAc2 |
| 16185.5 | HexNAc2–Deoxyhexose |
| 16347.0 | HexNAc2–Deoxyhexose–Hexose |
| 16638.0 | HexNAc2–Deoxyhexose–Hexose–NeuAc |
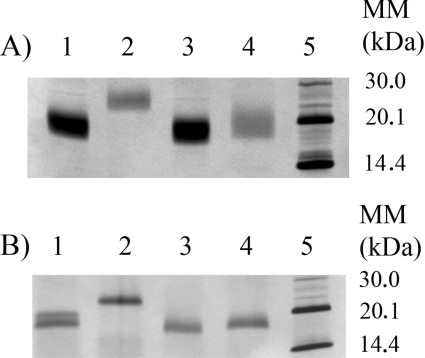
To determine the type of glycosylation, the proteins were deglycosylated, on the one hand enzymically, with PNGase F, which can only cleave N-bound sugars, and on the other hand chemically, with TFMSA, which can remove both N- and O-bound sugars (Figure 3). The enzymic deglycosylation with PNGase F had little or no effect on TLXI, unlike for rTLXI, where the SDS/PAGE molecular mass clearly decreased. TFMSA affected the molecular mass of both proteins. Indeed, TLXI no longer appeared as a broad band, and both TLXI and rTLXI clearly had a lower apparent molecular mass. From these deglycosylation experiments, we can conclude that native TLXI is O-glycosylated and few or no N-bound sugars are present. rTLXI, on the other hand, is clearly both N- and O-glycosylated. TLXI is the first member of the thaumatin family for which glycosylation is reported.
Figure 3. Deglycosylation of TLXI and rTLXI.
TLXI and rTLXI were deglycosylated enzymically with PNGase F (A) and chemically with TFMSA (B). The original TLXI and rTLXI are shown in lanes 1 and 2 respectively. Deglycosylated TLXI and rTLXI are shown in lanes 3 and 4 respectively. The sizes of the molecular-mass (MM) markers (lane 5) are indicated on the right-hand side (in kDa). The gels were silver-stained.
Inhibition specificity
To determine the xylanase specificity of TLXI, its inhibition activity towards several microbial xylanases of GH10 and GH11 was determined using the Xylazyme-AX method. TLXI was active towards most of the GH11 xylanases, but the high-pI GH11 xylanases from T. longibrachiatum (Xyn II, pI 9.0) and B. subtilis (pI 9.3), and the GH10 xylanases were unaffected by TLXI (Table 2). To be able to order the inhibited xylanases according to their sensitivity for TLXI, the [I]/[E]50 values (ratio of inhibitor concentration to enzyme concentration necessary to obtain 50% inhibition) are included in Table 2. Since TLXI binds to xylan, present in the Xylazyme-AX tablets (see below), it is clear that the results have an indicative value only, implying that one should be careful when comparing these results with those of other inhibitors. However, they give a very good indication of the xylanase specificity of TLXI and rTLXI.
Table 2. Inhibitor sensitivity of different GH10 and GH11 xylanases.
[I]/[E]50 values were determined using the Xylazyme-AX method, as described in the Experimental section, and are given in parentheses. Results are means±S.E.M. for three measurements. –, no inhibition at the highest inhibitor concentration; n.d., not determined.
| [I]/[E]50 | |||
|---|---|---|---|
| Xylanase | NCBI accession number | TLXI | rTLXI |
| GH11 | |||
| Trichoderma longibrachiatum (Xyn I) | CAA49294 | +++ (4.2±0.2) | +++ (7.8±1.2) |
| Aspergillus niger | CAA01470 | ++ (135.0±2.5) | + (1063±19) |
| Trichoderma viride | CAB60757 | ++ (170.4±1.1) | n.d. |
| Thermobacillus xylanilyticus | CAJ87325 | + (234.7±50.1) | n.d. |
| Penicillium funiculosum | CAC15487 | + (289.6±4.4) | n.d. |
| Bacillus subtilis | AAA22897 | - | n.d. |
| Trichoderma longibrachiatum (Xyn II) | CAA49293 | - | n.d |
| GH10 | |||
| Aspergillus aculeatus | AAE69552 | - | n.d. |
| Aspergillus niger | CAA03655 | - | n.d. |
| Aspergillus oryzae | BAA75475 | - | n.d. |
| Penicillium purpurogenum | AAF71268 | - | n.d. |
| Pseudomonas fluorescens | CAA33469 | - | n.d. |
| Thermobacillus xylanilyticus | CAA76420 | - | n.d. |
Its xylanase specificity supports further the assumption that TLXI plays a role in plant defence. Indeed, like TAXI, TLXI is active only against GH11 xylanases from both fungal and bacterial origin and is inactive towards GH10 xylanases [12]. Since plant xylanases are structurally similar to the microbial GH10 xylanases [5], TLXI probably does not have a regulatory role as such in planta. Its specificity clearly differs from that of XIP, which inhibits GH10 and GH11 xylanases [15].
Substrate binding
Preliminary experiments suggest interaction between TLXI and (arabino-) xylan. To confirm this assumption, a binding experiment was performed. TLXI was mixed with insoluble xylan, and, after centrifugation, the inhibition activity in the supernatant was compared with that in the original TLXI solution. Little, if any, inhibitory activity was measured in the supernatant, while the original sample clearly showed inhibition activity, and, as no SDS/PAGE inhibitor protein band was observed in the supernatant, while such a band was clearly visible when the pellet was examined, binding between TLXI and insoluble xylan was proven. For soluble xylan, the inhibition activity of the mixture of soluble xylan and TLXI was remarkably lower than that of the original TLXI sample. This demonstrates an interaction between TLXI and xylan, preventing the interaction between TLXI and xylanase. Hence, xylan could not be used to study the interaction between enzyme and inhibitor by means of classical kinetic analysis.
Kinetics and mechanism of xylanase inhibition
From the first sensitivity screen of different xylanases, the most sensitive one, i.e. T. longibrachiatum (Xyn I) xylanase, was chosen for in-depth characterization of the kinetic parameters. In view of the binding of TLXI to polymeric substrate, all analyses were performed with 4-MUX2 as substrate, to which TLXI nor rTLXI bind.
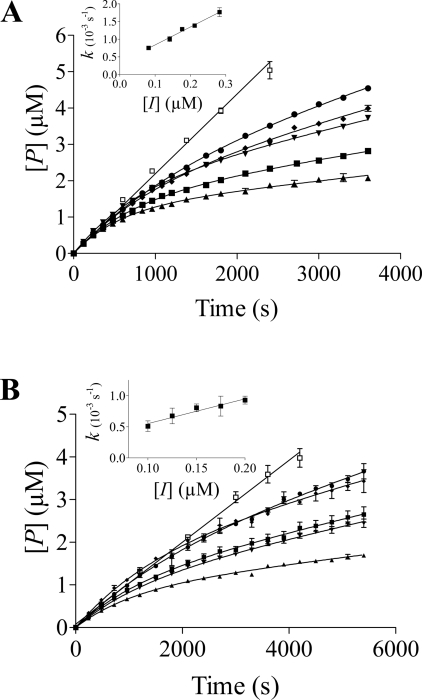
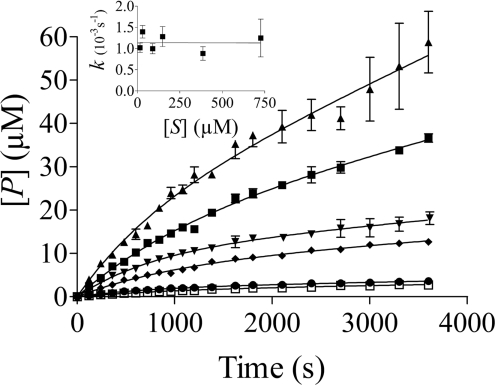
Time-dependent inhibition
As shown in Figure 4(A), xylanase inhibition by TLXI shows a clearly time-dependent approach to steady state, whereas the steady-state rate of substrate hydrolysis in the absence of inhibitor was reached instantaneously. The establishment of the equilibrium between enzyme, inhibitor and enzyme–inhibitor complex occurred over a period of approx. 30 minutes. For rTLXI, similar progress curves were obtained, but the establishment of equilibrium took even longer than for TLXI, approx. 45–min (Figure 4B). Therefore the time course for rTLXI was followed for up to 90–min, in which the reaction reached the steady-state conditions.
Figure 4. Time course of the inhibition with different inhibitor concentrations.
For different quantities of TLXI (A) and rTLXI (B), progress curves were plotted with T. longibrachiatum (Xyn I) xylanase. 4-MUX2 was used as substrate (22.7–μM). The concentrations of enzyme were 54.2 and 27.1–nM for TLXI and rTLXI respectively. The different concentrations were 283 (▲), 212 (■), 177 (▼), 141 (◆), 82 (●) and 0 (□) nM for TLXI and 200 (▲), 175 (■), 150 (▼), 125 (◆), 100 (●) and 0 (□) nM for rTLXI. The apparent first-order rate constants k for establishment of equilibrium were calculated from the progress curves and plotted against the inhibitor concentration, [I] (respective insets), and a linear correlation between [I] and k was found.
The concentrations of inhibitor which had to be used to obtain these progress curves were of similar order of magnitude as the concentration of enzyme, indicating that TLXI and rTLXI are not only slow-binding, but also tight-binding, inhibitors.
For slow tight-binding inhibitors, the velocity at any time is:
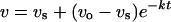 |
(1) |
where vs and vo are the steady-state and initial velocities respectively, and t is time [42].
Integration of eqn (1) gives:
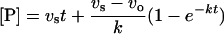 |
(2) |
where [P] is the product concentration at any time and k is the apparent first-order rate constant for the interconversion between vo and vs.
The progress curves shown in Figure 4 can be described by eqn (2). Since the increase of k with [I] is linear, as shown in the insets of Figure 4, the slow-binding inhibition arises from a simple single-step interaction between E and I in which the rate of complex formation is low [43]. The fact that the initial velocity of hydrolysis is independent of the concentration of inhibitor confirms this single-step mechanism [44].
For this mechanism, k as function of [I] is given by the following equation, provided that [S]\LtKm, under which these experiments were performed:
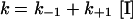 |
(3) |
The slope is the association rate constant for inhibition (k+1) and the intercept is the dissociation rate constant (k−1). From these two values, an inhibition constant (Ki=k−1/k+1) can be calculated. All of these kinetic parameters are listed in Table 3.
Table 3. Kinetic parameters for the inhibition of T. longibrachiatum xylanase (XynI).
The inhibition kinetic assays were conducted with 4-MUX2 as a substrate in McIlvaine buffer (pH–5.0) at 40 °C. The Km for this substrate for T. longibrachiatum xylanase (XynI) is 577±102–μM, the substrate concentration used was 22.7–μM and the enzyme concentration was 54.2 and 27.1–nM respectively for TLXI and rTLXI. Results are means±S.E.M. for measurements in triplicate.
| Ki (nM) | k+1 (104 M−1·s−1) | k−1 (10−4 s−1) | |
|---|---|---|---|
| TLXI | 65.1±7.3 | 0.51±0.03 | 3.31±0.56 |
| rTLXI | 38.7±16.6 | 0.40±0.06 | 1.54±0.88 |
Slow binding, or slow onset of inhibition, is a widespread phenomenon among non-protein glycosidase inhibitors [45]. For two non-cereal xylanase-inhibitor proteins, slow tight binding with xylanases has been described. In contrast with TLXI, they follow the two-step inhibition mechanism, with a rapidly formed initial collision complex, which isomerizes slowly to form the final tight complex [46,47]. The two-step inhibition mechanism is characterized by a hyperbolic increase of k with [I], instead of a linear increase as seen for TLXI.
Compared with the two known cereal xylanase-inhibitor proteins, TAXI and XIP, the association rate of TLXI is lower, while the dissociation occurs at a more similar rate. The SPR (surface plasmon resonance) data of TAXI-I, on the one hand, suggest that this is also a rather slow-binding inhibitor [48]. XIP, on the other hand, has been described as not slow binding, because an increase in pre-incubation time did not affect the inhibition activity and the SPR sensorgram did not exhibit the typical shape characteristic of the slow two-step interaction [49].
TLXI and rTLXI fulfil the requirements for tight-binding inhibition stated by Szedlacsek and Duggleby [44], i.e. that the enzyme concentration used in the study of the kinetic parameters and the Ki value are of the same order of magnitude (Table 3). However, the [I]/[E]50 values for TAXI and XIP are smaller (i.e. 0.6 for both) than the one obtained for TLXI, indicating that TAXI and XIP form even tighter complexes [12,49].
Mode of interaction
To classify the type of inhibition of a time-dependent inhibitor, it is convenient to analyse the effect of various substrate concentrations on k at a fixed inhibitor concentration (Figure 5). When [S] approaches Km, eqn (3) expands to eqn (4), (5) or (6), depending on whether the inhibition is competitive, uncompetitive or non-competitive respectively (these equations are derived with the assumption that E+S⇆ES equilibration is rapid relative to other rates) [50]:
Figure 5. Time course of the inhibition with different substrate concentrations.
For different quantities of 4-MUX2 progress curves were plotted with T. longibrachiatum (Xyn I) xylanase (54.2–nM). The concentration of TLXI was 113–nM in all progress curves. The different concentrations were 727 (▲), 386 (■), 148 (▼), 90 (◆), 30 (●) and 15 (□) μM. The apparent first-order rate constants k for establishment of equilibrium were calculated from the progress curves and plotted against the substrate concentration, [S] (inset).
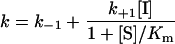 |
(4) |
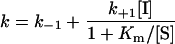 |
(5) |
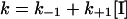 |
(6) |
The relationship between [S] and k is shown in the inset of Figure 5. Since k is independent of [S], the inhibition of TLXI is non-competitive. The corresponding mechanism is shown in the following scheme:
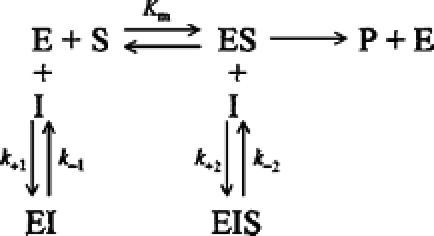 In this mechanism, Ki1 (=k−1/k+1) equals Ki2 (=k−2/k+2), and both will be replaced by Ki.
In this mechanism, Ki1 (=k−1/k+1) equals Ki2 (=k−2/k+2), and both will be replaced by Ki.
Thus the inhibition mechanism of TLXI is completely different from the mechanism of the other cereal xylanase inhibitors, as both TAXI and XIP are competitive inhibitors. The crystal structures of the complexes between xylanases and these inhibitors clearly show interactions in the active sites of the xylanases, for both inhibitors [13,16].
Inhibition constant
According to Cha [42], the steady-state velocity of a slow tight-binding inhibitor, following the non-competitive type of inhibition, is described by:
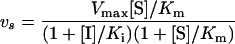 |
(7) |
where Vmax is the maximal velocity of the enzymatic reaction.
For TLXI and rTLXI respectively, Ki values of about 57.2±5.2 and 25.7±1.6–nM obtained from the progress curves with a fixed substrate concentration using eqn (7) agree well with the above-obtained Ki values (65.1 and 38.3 respectively). Again, the Ki of rTLXI is lower than the Ki of TLXI, although the [I]/[E]50 values listed in Table 2 suggest the opposite. This indicates that the overestimation of the [I]/[E]50 value made by measurements with the Xylazyme-AX method is bigger for rTLXI than for TLXI.
Another approach to determine the Ki for slow tight-binding inhibitors is the use of the Henderson equation, which accounts for the depletion of both free inhibitor and free enzyme by binding. For non-competitive inhibition, the following equation for the IC50 can be derived from the Henderson equation [42]:
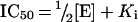 |
(8) |
where [E] is the total enzyme concentration.
The IC50 value measured with 4-MUX2 as substrate was 88.5±0.4–nM. Using this equation, an inhibition constant for TLXI of 61.4±4.0–nM was calculated, which is again in good agreement with the above-obtained results for the Ki of TLXI (65.1 and 57.2–nM).
From this IC50 value, a [I]/[E]50 value 1.6±0.1 was calculated, which is more than 2.5-fold lower than the one determined with the Xylazyme-AX method, indicating that the substrate binding of TLXI is a significant factor in the determination of the inhibition activity, when a (arabino-) xylan-containing substrate is used.
In conclusion, the present paper describes a novel typeof xylanase inhibitor from wheat. TLXI is the first TLP for which xylanase inhibition activity is described. We have demonstrated that both native TLXI and rTLXI, expressed in P. pastoris, are slow tight-binding inhibitors. In contrast with the two other cereal xylanase inhibitors, i.e. the competitive inhibitors TAXI and XIP, TLXI is a non-competitive inhibitor. Based on its xylanase specificity and its homology with TLPs, TLXI is believed to play a role in plant defence.
Online data
Acknowledgments
This study was in part carried out in framework of research project GOA/03/10 financed by the Research Fund K. U. Leuven. We gratefully acknowledge financial support from the ‘Fonds voor Wetenschappelijk Onderzoek-Vlaanderen’ (F. W. O.-Vlaanderen, Belgium) (postdoctoral fellowships of H. G. and K. G.), the ‘Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen’ (I. W. T., Brussels, Belgium) (scholarships of S. R. and E. F., Xylafun GBOU project funding) and the Research Fund K. U. Leuven (post-doctoral fellowships of J. B. and S. V. C.). Last, but not least, we thank Dr Wim Nerinckx for the synthesis of 4-MUX2.
References
- 1.Beliën T., Van Campenhout S., Robben J., Volckaert G. Microbial endoxylanases: effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol. Plant Microbe Interact. 2006;19:1072–1081. doi: 10.1094/MPMI-19-1072. [DOI] [PubMed] [Google Scholar]
- 2.Biely P. Microbial xylanolytic systems. Trends Biotechnol. 1985;3:286–290. [Google Scholar]
- 3.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brito N., Espino J. J., Gonzalez C. The endo-β-1,4-xylanase Xyn11A is required for virulence in Botrytis cinerea. Mol. Plant Microbe Interact. 2006;19:25–32. doi: 10.1094/MPMI-19-0025. [DOI] [PubMed] [Google Scholar]
- 5.Simpson D. J., Fincher G. B., Huang A. H. C., Cameron-Mills V. Structure and function of cereal and related higher plant (1→4)-β-xylan endohydrolases. J. Cereal Sci. 2002;37:111–127. [Google Scholar]
- 6.Debyser W., Delcour J. A. Inhibitors of xylanolytic and β-glucanolytic enzymes. Eur. Pat. WO 98/49278. 1997 [Google Scholar]
- 7.Debyser W., Peumans W. J., Van Damme E. J. M., Delcour J. A. Triticum aestivum xylanase inhibitor (TAXI), a new class of enzyme inhibitor affecting breadmaking performance. J. Cereal Sci. 1999;30:39–43. [Google Scholar]
- 8.Hessing M., Happe R. P. A novel class of xylanase inhibitors. Eur. Pat. EP0979830. 2000 [Google Scholar]
- 9.McLauchlan W. R., Garcia-Conesa M. T., Williamson G., Roza M., Ravestein P., Maat J. A novel class of protein from wheat which inhibits xylanases. Biochem. J. 1999;338:441–446. [PMC free article] [PubMed] [Google Scholar]
- 10.Goesaert H., Gebruers K., Brijs K., Courtin C. M., Delcour J. A. ‘TAXI’-type endoxylanase inhibitors in different cereals. J. Agric. Food Chem. 2003;51:3770–3775. doi: 10.1021/jf0262155. [DOI] [PubMed] [Google Scholar]
- 11.Goesaert H., Elliott G., Kroon P. A., Gebruers K., Courtin C. M., Robben J., Delcour J. A., Juge N. Occurrence of proteinaceous endoxylanase inhibitors in cereals. Biochim. Biophys. Acta. 2004;1696:193–202. doi: 10.1016/j.bbapap.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Gebruers K., Brijs K., Courtin C. M., Fierens K., Goesaert H., Rabijns A., Raedschelders G., Robben J., Sansen S., Sørensen J. F., et al. Properties of TAXI-type endoxylanase inhibitors. Biochim. Biophys. Acta. 2004;1696:213–221. doi: 10.1016/j.bbapap.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Sansen S., De Ranter C. J., Gebruers K., Brijs K., Courtin C. M., Delcour J. A., Rabijns A. Structural basis for inhibition of Aspergillus niger xylanase by Triticum aestivum xylanase inhibitor-I. J. Biol. Chem. 2004;279:36022–36028. doi: 10.1074/jbc.M404212200. [DOI] [PubMed] [Google Scholar]
- 14.Goesaert H., Gebruers K., Brijs K., Courtin C. M., Delcour J. A. XIP-type endoxylanase inhibitors in different cereals. J. Cereal Sci. 2003;38:317–324. doi: 10.1021/jf0262155. [DOI] [PubMed] [Google Scholar]
- 15.Juge N., Payan F., Williamson G. XIP-I, a xylanase inhibitor protein from wheat: a novel protein function. Biochim. Biophys. Acta. 2004;1696:203–211. doi: 10.1016/j.bbapap.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Payan F., Leone P., Porciero S., Furniss C., Tahir T., Williamson G., Durand A., Manzanares P., Gilbert H. J., Juge N., Roussel A. The dual nature of the wheat xylanase protein inhibitor XIP-I: structural basis for the inhibition of Family 10 and Family 11 xylanases. J. Biol. Chem. 2004;279:36029–36037. doi: 10.1074/jbc.M404225200. [DOI] [PubMed] [Google Scholar]
- 17.Beliën T., Van Campenhout S., Van Acker M., Volckaert G. Cloning and characterization of two endoxylanases from the cereal phytopathogen Fusarium graminearum and their inhibition profile against endoxylanase inhibitors from wheat. Biochem. Biophys. Res. Commun. 2005;327:407–414. doi: 10.1016/j.bbrc.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 18.Igawa T., Tokia T., Kudo T., Yamaguchi I., Kimura M. A wheat xylanase inhibitor gene, XIP-I, but not TAXI-1 is significantly induced by biotic and abiotic signals that trigger plant defense. Biosci. Biotechnol. Biochem. 2005;69:1058–1063. doi: 10.1271/bbb.69.1058. [DOI] [PubMed] [Google Scholar]
- 19.Gebruers K., Courtin C. M., Moers K., Noots I., Trogh I., Delcour J. A. The bread-making functionalities of two Aspergillus niger endoxylanases are strongly dictated by their inhibitor sensitivities. Enzyme Microb. Technol. 2005;36:417–425. [Google Scholar]
- 20.He L., Bickerstaff G. F., Paterson A., Buswell J. A. Purification and partial characterisation of two xylanases that differ in hydrolysis of soluble and insoluble xylan fractions. Enzyme Microb. Technol. 1993;15:13–18. [Google Scholar]
- 21.Gebruers K., Brijs K., Courtin C. M., Goesaert H., Proost P., Van Damme J., Delcour J. A. Affinity chromatography with immobilised endoxylanases separates TAXI- and XIP-type endoxylanase inhibitors from wheat (Triticum aestivum L.) J. Cereal Sci. 2002;36:367–375. [Google Scholar]
- 22.Bradford M. M. A rapid and sensitive method for the quantification of microgram quantities of protein utilising the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Gebruers K., Debyser W., Goesaert H., Proost P., Van Damme J., Delcour J. A. Triticum aestivum Lendoxylanase inhibitor (TAXI) consists of two inhibitors, TAXI I and TAXI II, with different. specificities. Biochem. J. 2001;353:239–244. doi: 10.1042/0264-6021:3530239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edge A. S. B. Deglycosylation of glycoproteins with trifluoromethanesulphonic acid: elucidation of molecular structure and function. Biochem. J. 2003;376:339–350. doi: 10.1042/BJ20030673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaugrand J., Gebruers K., Ververken C., Fierens E., Croes E., Goddeeris B., Courtin C. M., Delcour J. A. Antibodies against wheat xylanase inhibitors as tools for the selective identification of their homologues in other cereals. J. Cereal Sci. 2006;44:59–67. [Google Scholar]
- 26.Gebruers K., Goesaert H., Brijs K., Courtin C. M., Delcour J. A. Purification of TAXI-like endoxylanase inhibitors from wheat (Triticum aestivum L.) whole meal reveals a family of isoforms. J. Enzyme Inhib. 2002;17:61–68. doi: 10.1080/14756360290018611. [DOI] [PubMed] [Google Scholar]
- 27.Nakai K., Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 28.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selitrennikoff C. P. Antifungal proteins. Appl. Environ. Microbiol. 2001;67:2883–2894. doi: 10.1128/AEM.67.7.2883-2894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rebmann G., Mauch F., Dudler R. Sequence of a wheat cDNA encoding a pathogen-induced thaumatin-like protein. Plant Mol. Biol. 1991;17:283–285. doi: 10.1007/BF00039506. [DOI] [PubMed] [Google Scholar]
- 31.Hejgaard J., Jacobsen S., Svendsen I. Two antifungal thaumatin-like proteins from barley grain. FEBS Lett. 1991;291:127–131. doi: 10.1016/0014-5793(91)81119-s. [DOI] [PubMed] [Google Scholar]
- 32.Reiss E., Horstmann C. Drechslera teres-infected barley (Hordeum vulgare L.) leaves accumulate eight isoforms of thaumatin-like proteins. Physiol. Mol. Plant Pathol. 2001;58:183–188. [Google Scholar]
- 33.Reiss E., Schlesier B., Brandt W. cDNA sequences, MALDI–TOF analyses and molecular modelling of barley PR-5 proteins. Phytochemistry. 2006;67:1856–1864. doi: 10.1016/j.phytochem.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Yu X. M., Griffith M. Antifreeze proteins in winter rye leaves form oligomeric complexes. Plant Physiol. 1999;119:1361–1369. doi: 10.1104/pp.119.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skadsen R. W., Sathish P., Kaeppler H. F. Expression of thaumatin-like permatin PR-5 genes switches from the ovary wall to the aleurone in developing barley and oat seeds. Plant Sci. 2000;156:11–22. doi: 10.1016/s0168-9452(00)00226-0. [DOI] [PubMed] [Google Scholar]
- 36.Schimoler-O’Rourke R., Richardson M., Selitrennikoff C. P. Zeamatin inhibits trypsin and α-amylase activities. Appl. Environ. Microbiol. 2001;67:2365–2366. doi: 10.1128/AEM.67.5.2365-2366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koiwa H., Kato H., Nakatsu T., Oda J., Yamada Y., Sato F. Purification and characterization of tobacco pathogenesis-related protein PR-5d, an antifungal thaumatin-like protein. Plant Cell Physiol. 1997;38:783–791. doi: 10.1093/oxfordjournals.pcp.a029236. [DOI] [PubMed] [Google Scholar]
- 38.Fils-Lycaon B. R., Wiersma P. A., Eastwell K. C., Sautière P. A cherry protein and its gene, abundantly expressed in ripening fruit, have been identified as thaumatin-like. Plant Physiol. 1996;111:269–273. doi: 10.1104/pp.111.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krebitz M., Wagner B., Ferreira F., Peterbauer C., Campillo N., Witty M., Kolarich D., Steinkellner H., Scheiner O., Breiteneder H. Plant-based heterologous expression of Mal d 2, a thaumatin-like protein and allergen of apple (Malus domestica), and its characterization as an antifungal protein. J. Mol. Biol. 2003;329:721–730. doi: 10.1016/s0022-2836(03)00403-0. [DOI] [PubMed] [Google Scholar]
- 40.Trudel J., Grenier J., Potvin C., Asselin A. Several thaumatin-like proteins bind to β-1,3-glucans. Plant Physiol. 1998;118:1431–1438. doi: 10.1104/pp.118.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper C. A., Gasteiger E., Packer N. H. GlycoMod: a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1:340–349. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 42.Cha S. Tight-binding inhibitors-I: kinetic behavior. Biochem. Pharmacol. 1975;24:2177–2185. doi: 10.1016/0006-2952(75)90050-7. [DOI] [PubMed] [Google Scholar]
- 43.Morrison J. F., Walsh C. T. The behavior and significance of slow binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 44.Szedlacsek S. E., Duggleby R. G. Kinetics of slow and tight-binding inhibitors. Methods Enzymol. 1995;249:144–180. doi: 10.1016/0076-6879(95)49034-5. [DOI] [PubMed] [Google Scholar]
- 45.Legler G. Glycoside hydrolases: mechanistic information from studies with reversible and irreversible inhibitors. Adv. Carbohyd. Chem. Biochem. 1990;48:319–384. doi: 10.1016/s0065-2318(08)60034-7. [DOI] [PubMed] [Google Scholar]
- 46.Dash C., Vathipadiekal V., George S. P., Rao M. Slow-tight binding inhibition of xylanase by an aspartic protease inhibitor. J. Biol. Chem. 2002;277:17978–17986. doi: 10.1074/jbc.M111250200. [DOI] [PubMed] [Google Scholar]
- 47.Vathipadiekal V., Rao M. Inhibition of 1,4-β-D-xylan xylanohydrolase by the specific aspartic protease inhibitor pepstatin. J. Biol. Chem. 2004;279:47024–47033. doi: 10.1074/jbc.M407866200. [DOI] [PubMed] [Google Scholar]
- 48.Fierens K., Gils A., Sansen S., Brijs K., Courtin C. M., Declerck P. J., De Ranter C. J., Gebruers K., Rabijns A., Robben J., et al. His374 of wheat endoxylanase inhibitor TAXI-I stabilizes complex formation with glycoside hydrolase family 11 endoxylanases. FEBS J. 2005;272:5872–5882. doi: 10.1111/j.1742-4658.2005.04987.x. [DOI] [PubMed] [Google Scholar]
- 49.Flatman R., McLauchlan W. R., Juge N., Furniss C., Berrin J.-G., Hughes R. K., Manzanares P., Ladbury J. E., O'Brien R., Williamson G. Interaction defining the specificity between fungal xylanases and the xylanase-inhibiting protein XIP-I from wheat. Biochem. J. 2002;365:773–781. doi: 10.1042/BJ20020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bakker A. V., Jung S., Spencer R. W., Vinick F. J., Faraci W. S. Slow tight-binding inhibition of prolyl endopeptidase by benzoyloxycarbonyl-prolyl-prolinal. Biochem. J. 1990;271:559–562. doi: 10.1042/bj2710559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.